Analysis of Brachypodium mutants with increased cellular steady state auxin content and root cell elongation sheds light on the importance of the Acid Growth Theory of plant cell elongation in roots.
Abstract
The long-standing Acid Growth Theory of plant cell elongation posits that auxin promotes cell elongation by stimulating cell wall acidification and thus expansin action. To date, the paucity of pertinent genetic materials has precluded thorough analysis of the importance of this concept in roots. The recent isolation of mutants of the model grass species Brachypodium distachyon with dramatically enhanced root cell elongation due to increased cellular auxin levels has allowed us to address this question. We found that the primary transcriptomic effect associated with elevated steady state auxin concentration in elongating root cells is upregulation of cell wall remodeling factors, notably expansins, while plant hormone signaling pathways maintain remarkable homeostasis. These changes are specifically accompanied by reduced cell wall arabinogalactan complexity but not by increased proton excretion. On the contrary, we observed a tendency for decreased rather than increased proton extrusion from root elongation zones with higher cellular auxin levels. Moreover, similar to Brachypodium, root cell elongation is, in general, robustly buffered against external pH fluctuation in Arabidopsis thaliana. However, forced acidification through artificial proton pump activation inhibits root cell elongation. Thus, the interplay between auxin, proton pump activation, and expansin action may be more flexible in roots than in shoots.
Coordinated cell division and expansion is crucial for plant organogenesis because cell walls restrict the movement of cells relative to each other (Cosgrove, 1999; Wolf et al., 2012). The cell wall is a complex structure of intertwined and sometimes cross-linked polymers, comprising cellulose, xyloglucans, pectins, and arabinogalactans, which resists the internal turgor pressure. Therefore, cell wall elasticity has to be regulated to permit cellular growth (Cosgrove et al., 1984; Cosgrove, 1993, 2005; Wolf et al., 2012). This is achieved through selective loosening of cell wall polymer interactions, which allows cellulose microfibrils and associated matrix polysaccharides to displace relative to each other. As cellulose microfibrils are typically arranged in a nonrandom, parallel orientation, most cells expand along one principal axis. This process is easily observed in organs with one principal growth vector, for instance, in hypocotyls or root tips. In both organs, hormones strongly influence cell elongation. Among these, auxin is most prominent because it not only orchestrates developmental programs, but also conveys environmental inputs to trigger adaptive responses such as tropisms (Sánchez-Rodríguez et al., 2010; Depuydt and Hardtke, 2011). It is generally assumed that auxin promotes cell elongation by inducing the expression of cell wall remodeling factors (Sánchez-Rodríguez et al., 2010; Wolf et al., 2012). These include expansins, which are considered facilitators of cell wall loosening by physically opening up the fiber network, thereby facilitating the access of other enzymes to their substrates (Cosgrove, 2005). Moreover, in parallel, auxin supposedly stimulates cell elongation via a nongenomic pathway, since it enhances cell elongation within minutes in classical assay systems, such as hypocotyls or coleoptiles. In these model organs, auxin treatment correlates with increased acidification of the apoplast, which presumably promotes cell elongation because central cell wall loosening factors and enzymes, e.g., expansins, polygalacturonases, endoglucanases, and pectin methylesterases, work optimally under acidic conditions. Cell wall acidification, in turn, is thought to arise from auxin-induced activation of plasma membrane-localized proton pumps (PM-H+-ATPases). This long-standing concept, named the Acid Growth Theory, was formulated in the 1970s (Rayle and Cleland, 1970, 1977, 1992; Hager et al., 1971).
Whether the Acid Growth Theory is universally applicable to plant cell expansion remains controversial. For instance, even for the classic assay systems (coleoptiles or hypocotyls), some authors have concluded that cell wall acidification indeed stimulates growth, but that this is not an auxin-dependent effect (Kutschera and Schopfer, 1985b, 1985a; Schopfer, 1989, 1993). Validation of the Acid Growth Theory is most difficult in roots, where analyses are complicated by the fact that as opposed to hypocotyls or coleoptiles, cell proliferation and cell expansion are deeply intertwined. Because both processes require variable threshold auxin activities, they are difficult to uncouple, which might account for the observation that auxin application generally inhibits or at best only slightly promotes root growth (Moloney et al., 1981; Evans et al., 1994). Likewise, Arabidopsis thaliana roots are shorter upon both a genetically imposed strong decrease as well as a strong increase in auxin production (Chen et al., 2014). Moreover, reduced mature cell length is typically accompanied by reduced meristem size and vice versa (Moubayidin et al., 2010; Scacchi et al., 2010; Rodriguez-Villalon et al., 2015), which makes it difficult to distinguish whether observed phenotypes are primarily caused by altered cell elongation or cell proliferation and/or differentiation. Thus, a paucity of clear-cut conditions and pertinent genetic material has prevented conclusive analyses of the Acid Growth Theory in Arabidopsis roots.
In this study, we took advantage of recently isolated mutants of the model grass species Brachypodium distachyon in the TAA1-RELATED2-LIKE (TAR2L) and ETHYLENE INSENSITIVE2-LIKE1 (EIN2L1) genes. TAR2L encodes an enzyme of the TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1) and TAA1-RELATED (TAR) family of proteins, which catalyze conversion of tryptophan to indole-3-pyruvic acid (IPA) in the two-step auxin biosynthesis pathway (Stepanova et al., 2011; Won et al., 2011). IPA is subsequently converted to indole-3-acetic acid (IAA), the major active auxin, by the YUCCA cytochrome P450 enzymes. Two TAR homologs exist in Brachypodium, with TAR2L dominating in the seminal root elongation zone, where cell differentiation and elongation occur (Pacheco-Villalobos et al., 2013). Bd-EIN2L1 is a homolog of Arabidopsis EIN2, an essential positive regulator of ethylene signaling (Alonso et al., 1999; Qiao et al., 2012). Both hypomorphic Brachypodium tar2lhypo and ein2l1hypo mutants display different degrees of elevated IAA levels in the seminal root elongation zone as well as dramatically enhanced cell elongation. This initially counterintuitive phenotype could be explained by the observation that the regulatory logic of the two-step auxin biosynthetic pathway is different in Brachypodium compared with Arabidopsis: Whereas ethylene positively regulates both steps in Arabidopsis, it negatively regulates the second, rate-limiting step in Brachypodium (Pacheco-Villalobos et al., 2013). The pathway intermediate IPA is metabolically linked to ethylene biosynthesis through the VAS1 enzyme, which catalyzes the formation of tryptophan from IPA using hydrophobic amino acids, mostly l-methionine, as amino group donor (Zheng et al., 2013). Because the size of the l-methionine pool limits the biosynthesis of the rate-limiting ethylene precursor 1-aminocyclopropane-1-carboxylate, not only IPA, but also ethylene biosynthesis are increased in Arabidopsis vas1 mutants. By analogy, the Brachypodium tar2lhypo mutation apparently creates a situation where reduced TAR activity leads to reduced IPA production and thereby also reduced ethylene production, while ethylene signaling is directly dampened in ein2l1hypo mutants. Thus, in both hypomorphic mutants, YUCCA genes become derepressed to different degrees and eventually more IPA is converted to IAA than in the wild type (Pacheco-Villalobos et al., 2013). Because a unique feature of both Brachypodium tar2lhypo and ein2l1hypo mutants is that their root meristem size and activity are not affected, their longer seminal roots are entirely explained by the increased mature cell length (Pacheco-Villalobos et al., 2013). Thus, cell proliferation and cell expansion are uncoupled in seminal roots of Brachypodium tar2lhypo and ein2l1hypo mutants, which both display elevated auxin levels in conjunction with greatly exaggerated cell elongation. Therefore, they offer an unprecedented opportunity to monitor the consequences of high steady state auxin levels in a monocotyledon root type.
RESULTS
Metabolic Analysis Confirms Higher Auxin Levels in tar2lhypo Root Segments Despite Reduced Tryptophan Aminotransferase Activity
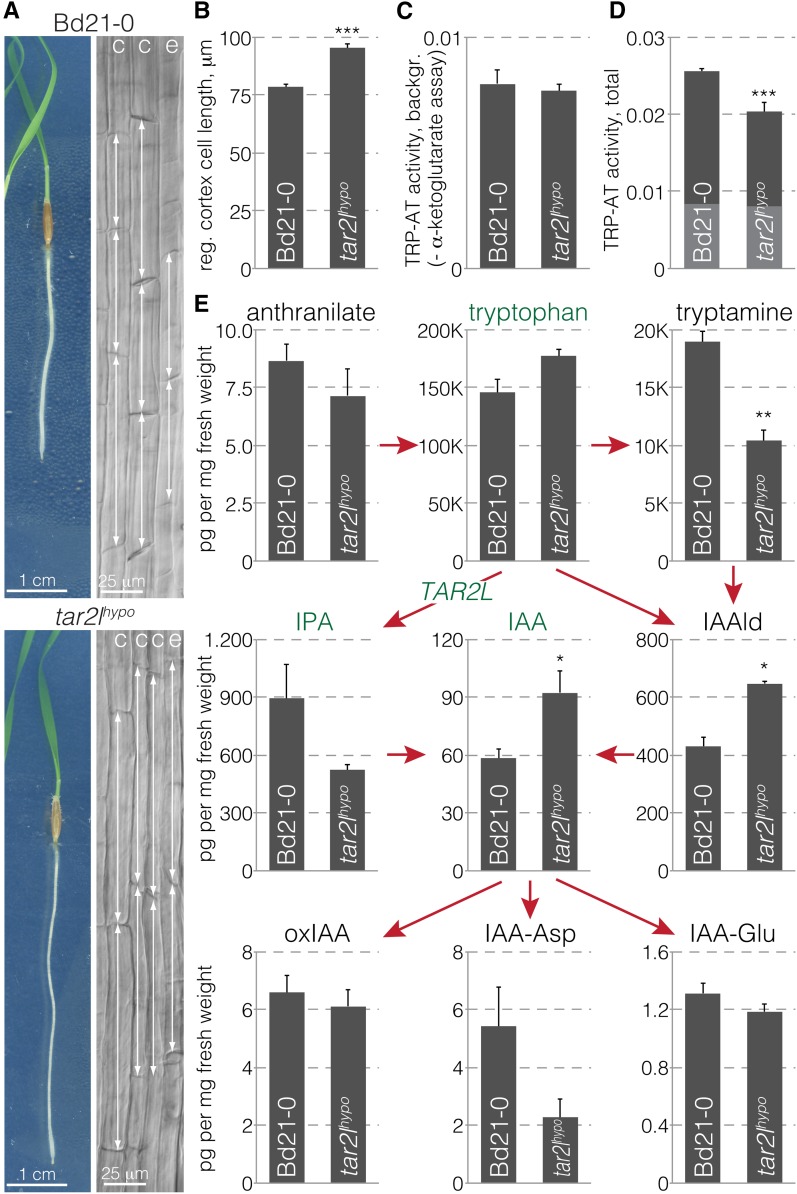
The strongly enhanced mature root cell length in Brachypodium tar2lhypo mutants (Figure 1A) (Pacheco-Villalobos et al., 2013) is apparently cell autonomous because in regenerating excised tar2lhypo root tips (Supplemental Figure 1A), newly formed cells are still longer than in its wild-type background, Bd21-0 (Figure 1B). The same is true for ein2l1hypo compared with its wild-type background Bd21-3 (Supplemental Figure 1B). In our subsequent analyses, we primarily concentrated on the tar2lhypo mutant because of the relatively strong phenotype of tar2lhypo seminal roots. For a more complete analysis of auxin metabolism in tar2lhypo, we measured the tryptophan aminotransferase background activity (Figure 1C) as well as total (Figure 1D) activity, in Bd21-0 and tar2lhypo seminal roots, which revealed an approximately 30% reduction in specific activity in the mutant (Figure 1D). Full-scale analysis of auxin biosynthesis intermediates in 1-cm segments from the root elongation zone of 4-d-old seedlings (Supplemental Figure 1C) with comparable fresh weight in Bd21-0 and tar2lhypo (Supplemental Figure 1D) produced a matching metabolic profile; that is, tryptophan levels were slightly increased while IPA levels were substantially decreased in the tar2lhypo mutant (Figure 1E). Concurrently, the abundance of a few (inactive) auxin conjugates was shifted, while as previously observed (Pacheco-Villalobos et al., 2013), the level of free auxin (IAA) was significantly increased. Also consistent with previous findings (Zheng et al., 2013), downregulation of a Brachypodium VAS1-LIKE gene (VAS1L) by RNA interference suppressed the tar2lhypo phenotype genetically (Supplemental Figures 1E and F). In summary, these observations confirm our previous finding that downregulation of TAR2L results in increased rather than decreased cellular auxin levels and causes strongly enhanced root cell elongation (Pacheco-Villalobos et al., 2013).
Figure 1.
Reduced Tryptophan Aminotransferase Activity in Seminal Roots of Hypomorphic tar2l Mutants Results in Higher Cellular Auxin Levels and Strongly Enhanced Cell Elongation.
(A) Four-day-old tissue culture-grown Brachypodium wild type (Bd21-0) and hypomorphic tar2l mutant (tar2lhypo) seedlings (left) and light microscopy images of their mature cortex (c) and epidermal (e) cell layers (right). Double-sided arrows point out the longitudinal dimensions of individual cells.
(B) Mature cortex cell length in roots 4 d after regeneration from isolated Bd21-0 and tar2lhypo root tips (40 cells per root, 10 roots).
(C) Tryptophan aminotransferase background activity in Bd21-0 and tar2lhypo roots.
(D) Total tryptophan aminotransferase activity in Bd21-0 and tar2lhypo roots, background portion indicated in gray.
(E) Quantification of auxin and auxin metabolites in 1-cm segments from the root elongation zone of 4-d-old Bd21-0 or tar2lhypo seedlings. Error bars indicate se of the mean (three to four biological replicates). Differences were not statistically significant (Student’s t test) unless indicated as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
High Auxin Steady State Is Associated with Remarkable Transcriptional Homeostasis of the Auxin Signaling Network
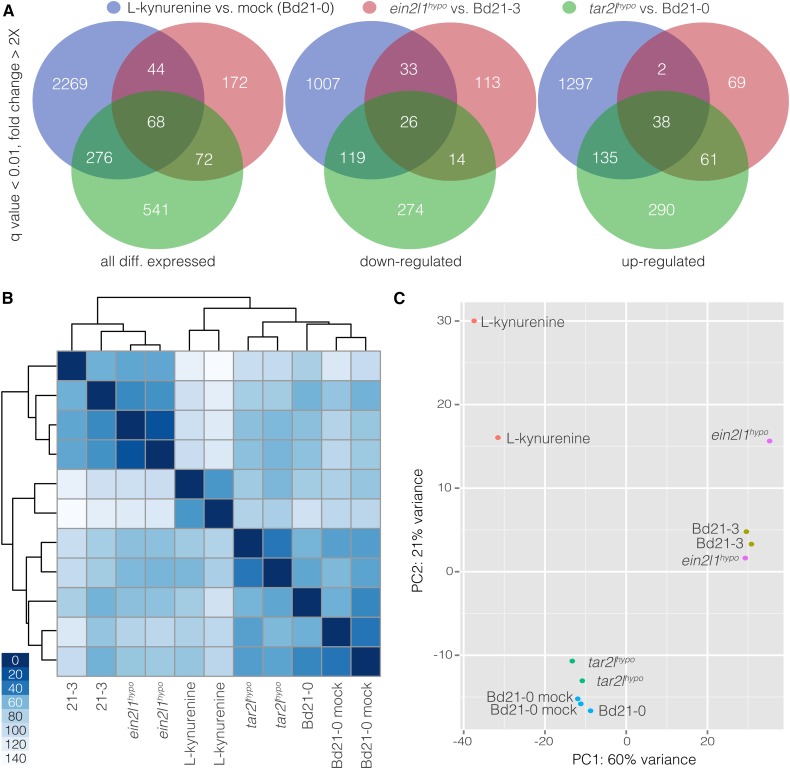
The tar2lhypo mutant offers a unique opportunity to survey a steady state high auxin concentration transcriptome that is associated with enhanced cell elongation. To this end, we performed mRNA sequencing (RNAseq) of 1-cm root elongation zone segments, grown and harvested in parallel with those used for the metabolic analysis. Complementary to this experiment, we also performed RNAseq on equivalent segments from wild-type plants that had been transferred onto medium containing l-kynurenine for 2 d. Mild concentrations of this tryptophan aminotransferase inhibitor induce higher auxin levels and enhanced cell elongation in wild-type roots, thus mimicking the tar2lhypo phenotype (Pacheco-Villalobos et al., 2013). The reads from the Bd21-0, tar2lhypo, and l-kynurenine-treated Bd21-0 samples mapped onto more than 27,000 mRNA transcripts out of the 31,679 nuclear genes annotated in the Brachypodium reference genome sequence (version 2.1) (Supplemental Data Set 1), with a pairwise overlap between samples of more than 97%. Compared with the wild type, 957 and 2657 genes were differentially expressed in tar2lhypo and l-kynurenine-treated roots, respectively (q value < 0.01, fold change > 2×) (Figure 2A) (Supplemental Data Sets 2 and 3). The higher number of differentially expressed genes in the l-kynurenine-treated samples is consistent with an organ-wide systemic effect of the treatment that includes transcriptome remodeling toward a new steady state. The overlap between the two sets was 344 genes, which represents ∼4-fold enrichment over neutral expectation (P < 0.0001, χ2 test). A similar RNAseq experiment was performed with root segments from Bd21-3 and ein2l1hypo seedlings. Again, over 27,000 transcripts were detected and 356 genes were differentially expressed (Supplemental Data Sets 1 and 4). Overlap with the tar2lhypo and l-kynurenine-treated sets was 140 and 112, respectively, which again represented high enrichment (∼12- and ∼3.5-fold, respectively) over neutral expectation (P < 0.0001, χ2 test). In summary, the RNAseq profiles indicated high overlap between the mutants and the l-kynurenine condition, with the extent of differentially expressed genes correlating with phenotype strength. Despite the similarities between their transcriptome profiles, the samples were clearly grouped apart. Both mutants were more similar to their wild-type backgrounds than to each other, and the l-kynurenine-treated samples were most distant to all others (Figure 2B). A principal component analysis confirmed that parental background was the dominant factor in the grouping of samples (Figure 2C).
Figure 2.
Differential Gene Expression in Root Segments as Determined by RNA Sequencing.
(A) Venn diagrams illustrating overlaps between the gene sets that were differentially expressed in root segments of Bd21-0 versus tar2lhypo, Bd21-3 versus ein2l1hypo, and mock-treated versus l-kynurenine-treated Bd21-0.
(B) and (C) Cluster analysis (B) and principal component (PC) (C) analysis of the different RNA sequencing samples.
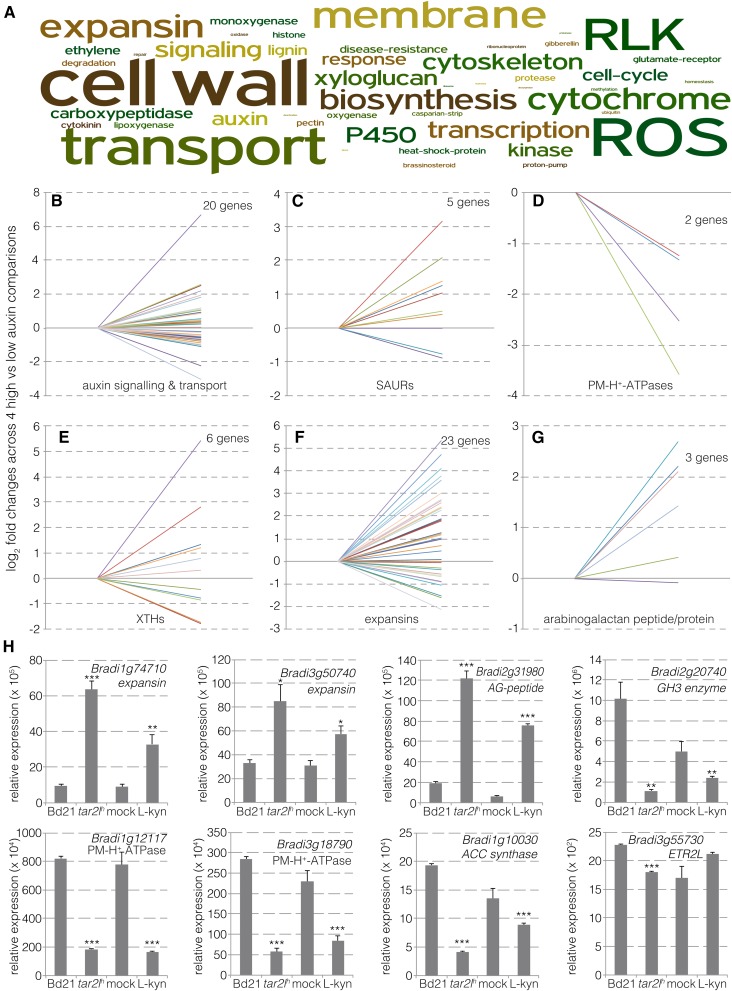
Analysis of the annotations of the differentially expressed genes revealed a rather low occurrence of genes involved in auxin or other hormone signaling pathways (Figure 3A). Significant differential expression was observed for six out of 26 annotated auxin response factors, seven out of 32 annotated AUX/IAA genes, and one out of five annotated auxin receptor genes (Figure 3B; Supplemental Data Set 5). However, the expression changes were moderate throughout. Likewise, mostly small effects were observed for the few differentially expressed genes involved in polar auxin transport, which included two auxin influx facilitators and three auxin efflux carriers. Overall, the data indicate that the transcriptional steady state of the auxin-signaling network is well buffered with respect to variation in auxin levels. Interestingly, however, some primary auxin target genes of the SMALL AUXIN UP-REGULATED (SAUR) category were differentially expressed (five out of 42 annotated genes) and mostly upregulated (four out of the five) (Figure 3C). SAUR genes are classic auxin signaling output genes, and it has been suggested that SAUR proteins antagonize posttranslational inhibition of PM-H+-ATPases (Spartz et al., 2014). Finally, with the exception of two genes that were substantially downregulated (Figure 3D), Brachypodium genes that encode PM-H+-ATPases displayed no differential expression.
Figure 3.
Differential Expression of Auxin- and Cell Wall-Related Genes in Root Segments.
(A) Word cloud from annotations of genes differentially expressed between Bd21-0 versus tar2lhypo, or mock-treated versus l-kynurenine-treated Bd21-0.
(B) to (G) Expression changes (fold changes) for individual members of the indicated gene classes in Bd21-0 versus tar2lhypo and mock-treated versus l-kynurenine-treated Bd21-0 (Bd21-0/mock set to 1 on the left, tar2lhypo/l-kynurenine-treated Bd21-0 values on the right). Only genes that showed differential expression and a q value < 0.01 in at least one comparison are plotted. See Supplemental Data Set 5 for gene identifiers and expression values.
(H) qPCR verification of differential gene expression in independent RNA samples prepared from independent root segments (three biological replicates). Error bars indicate se of the mean. Differences were not statistically significant (Student’s t test) unless indicated as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
High Auxin Steady State Is Associated with Transcriptional Changes in Cell Wall Remodeling Factors
The majority of significantly enriched terms that stood out in a word cloud made from annotation of differentially expressed genes was related to the cell wall and associated processes (Figure 3A). Yet, only a small proportion of genes encoding cell wall remodeling proteins displayed differential expression, and these were restricted to a few groups. For example, while 6 out of 15 annotated xyloglucan endotrans-glycosylases/hydrolases were up- or downregulated at roughly equal measure (Figure 3E), no differential expression was observed among cellulose synthase genes. The most prominent differentially expressed cell wall modulators were expansins (23 out of 54 annotated expansin genes), which were significantly enriched (P = 0.0078 for the overlap between all samples, χ2 test) and are also considered classic auxin target genes. The majority (18 out of 23) displayed comparatively strong upregulation (Figure 3F). Finally, three genes encoding arabinogalactan peptides or proteins stood out because of their consistent upregulation (Figure 3G). Verification of differential expression by qPCR was performed for a selected set of genes of interest in independent RNA samples from tar2lhypo and l-kynurenine-treated root segments, confirming the RNAseq results (Figure 3H). In summary, the transcriptomic data indicate that in the presence of higher cellular auxin levels, the auxin signaling network maintains remarkable homeostasis at the transcriptional level, while the bulk of expression changes are observed in cell wall remodeling genes, notably expansins.
High Auxin Steady State Is Associated with a Specific Change in Glycosidic Cell Wall Linkages
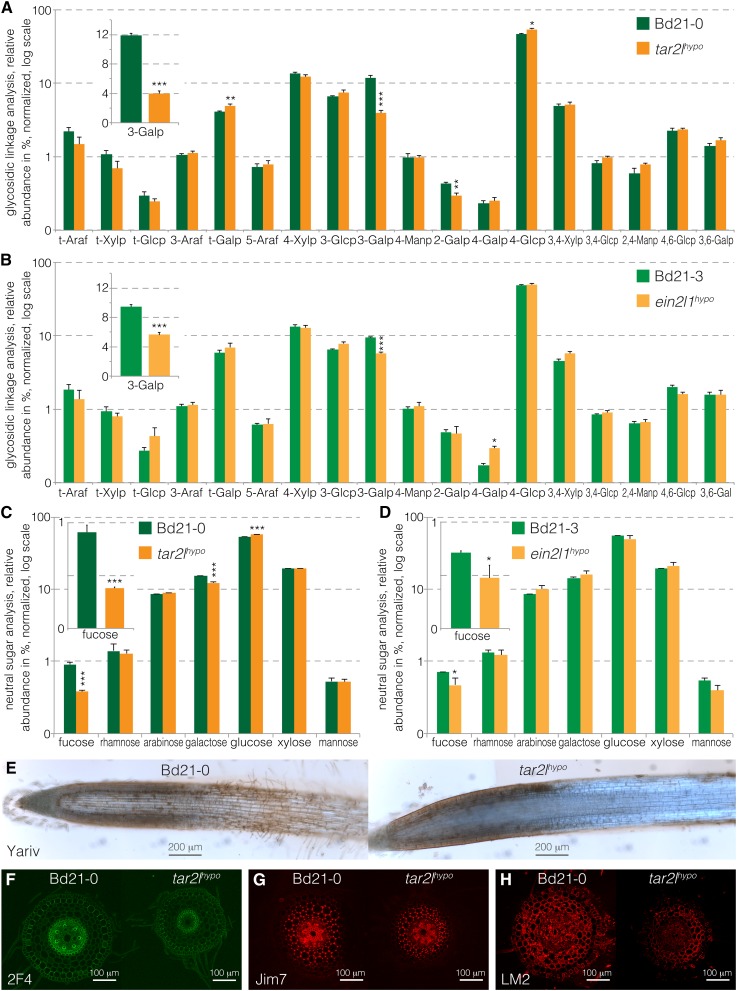
The robust changes in the arabinogalactan protein/peptide genes were of particular interest in light of our results from chemical cell wall analyses. To monitor the structural effect of altered expression in cell wall remodeling genes, we performed cell wall polysaccharide analysis of root segments from parallel samples that were grown and harvested concomitantly with the segments analyzed by RNAseq. The analysis of the glycosidic linkages occurring in the cell wall polymers detected only few significant differences between Bd21-0 and tar2lhypo, or Bd21-3 and ein2l1hypo. However, both profiles were consistent, with a specific significant decrease in 1,3-galactosyl residues in the mutants relative to their wild-type backgrounds (Figures 4A and 4B). 1,3-Linked galactose is found specifically in the glycosidic moiety of the arabinogalactan proteins (AGPs) (Seifert and Roberts, 2007; Ellis et al., 2010; Kitazawa et al., 2013; Knoch et al., 2014). 1,3-Linked galactosyl residues represented ∼10% of all linked sugar residues detected in both wild-type backgrounds and were reduced by about 2- to 3-fold in ein2l1hypo and tar2lhypo, respectively. Moreover, the analysis of the cell wall neutral sugars indicated similar relative abundance of the different monosaccharides in the mutants and wild types, with one notable exception, fucose, a minor cell wall sugar (Figures 4C and 4D). Relative fucose abundance was more than halved in tar2lhypo and reduced by about one-third in ein2l1hypo. Interestingly, just like β-1,3-linked galactose, fucose is found in AGPs (van Hengel and Roberts, 2002). Thus, the analyses point to a very specific effect of elevated cellular auxin levels on arabinogalactan complexity or abundance in Brachypodium. To confirm this observation with an alternative technique, we performed in situ Yariv staining on Bd21-0 and tar2lhypo roots. The Yariv reagent is known to specifically detect β-1,3-galactan (Yariv et al., 1967; Kitazawa et al., 2013). Staining was considerably reduced in the root elongation zone of tar2lhypo, thereby corroborating the chemical cell wall analyses (Figure 4E). Moreover, we probed transverse sections in the root elongation zone with antibodies directed against demethylesterified pectin (2F4 antibody; Figure 4F), methylesterified pectin (JIM7 antibody; Figure 4G), and arabinogalactan side chains (LM2 antibody; Figure 4H). None of these stainings showed a marked difference in epitope abundance or distribution, except that in general, the mean fluorescence signal of the LM2 antibody was reduced. This might hint to lower AGP abundance; however, it is unclear to what degree the antibody stainings are quantitative. Importantly, unlike the Yariv reagent, the LM2 antibody does not recognize the β-1,3-galactan linkages in the arabinogalactan backbone, but rather an epitope that comprises β-linked glucuronic acid, which is found at the side chain termini (Smallwood et al., 1996; Knoch et al., 2014). Therefore, collectively, the results point to reduced AGP complexity.
Figure 4.
Cell Wall Analyses of tar2lhypo and ein2l1hypo Root Segments Compared with Their Wild-Type Backgrounds.
(A) Glycosidic linkage analysis of Bd21-0 and tar2lhypo root segments.
(B) Glycosidic linkage analysis of Bd21-3 and ein2l1hypo root segments.
(C) Neutral sugar analysis of Bd21-0 and tar2lhypo root segments.
(D) Neutral sugar analysis of Bd21-3 and ein2l1hypo root segments.
(E) Yariv staining (brownish) against β-1,3-galactan linkages in AGPs on longitudinal sections of Bd21-0 and tar2lhypo root tips.
(F) 2F4 antibody staining against demethylesterified pectin (green) on transverse sections from the elongation zone of Bd21-0 and tar2lhypo root tips.
(G) JIM7 antibody staining against methylesterified pectin (red) on transverse sections from the elongation zone of Bd21-0 and tar2lhypo root tips.
(H) LM2 antibody staining against AGP side chains (red) on transverse sections from the elongation zone of Bd21-0 and tar2lhypo root tips. Error bars indicate se of the mean (two technical replicates per each of three biological replicates). Differences were not statistically significant (Student’s t test) unless indicated as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
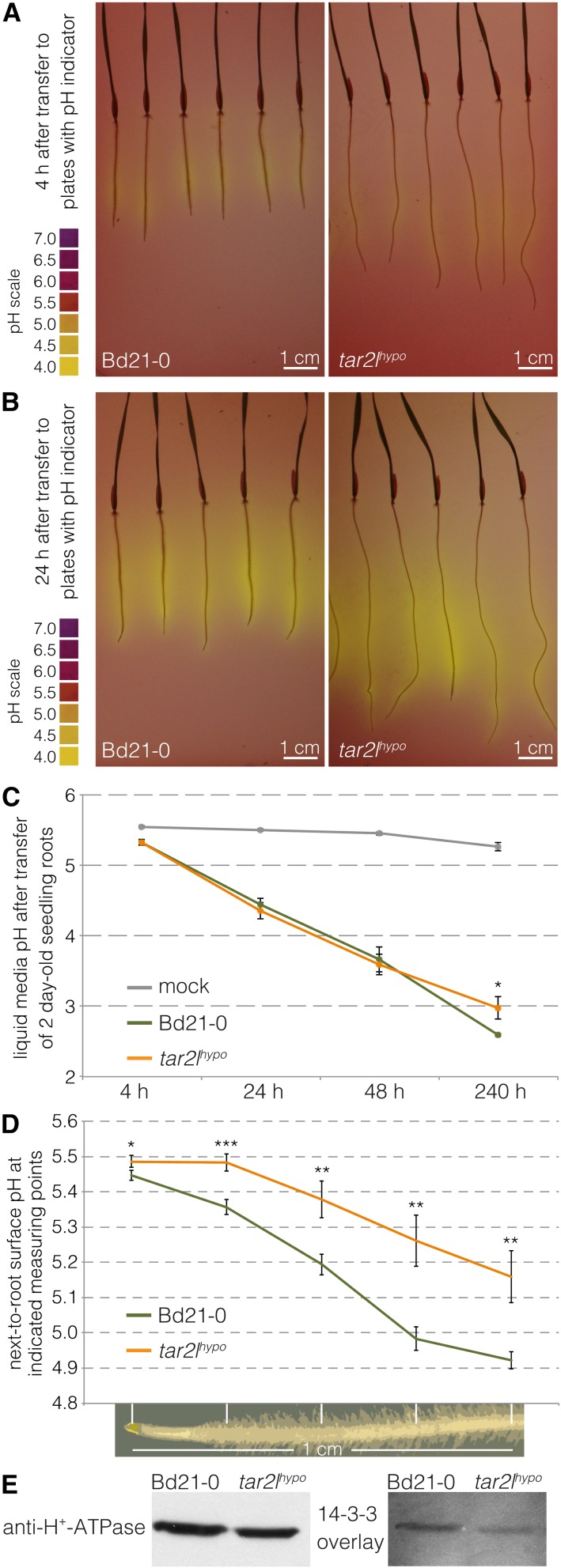
High Auxin Steady State Is Not Associated with Markedly Increased Proton Excretion
Our transcriptomic and cell wall analyses indicate that elevated cellular auxin levels in Brachypodium roots are indeed associated with differential expression of cell wall remodeling genes and matching changes in cell wall composition. To determine whether this also applies to the hallmark of the Acid Growth Theory, apoplastic acidification, we next investigated the capacity of Bd21-0 and tar2lhypo roots to acidify the medium. To this end, we first visualized rhizosphere acidification by transferring seedlings onto medium supplemented with pH indicator. Acidification was readily detected within 4 h but was not apparently stronger for tar2lhypo roots compared with Bd21-0 roots (Figures 5A and 5B). Likewise, in a quantitative assay with liquid medium, acidification could be readily followed over time; however, no difference in proton pumping activity of root tips could be detected between Bd21-0 and tar2lhypo (Figure 5C). Finally, we measured apoplast acidification more directly at the root surface using fiber optic pH microfiber sensors. To this end, five roughly equidistant measuring points from the root tip through the elongation zone were monitored along individual roots. As could be expected, these measurements revealed a gradient of increasingly acidic pH from the root tip to the differentiated cells. However, this gradient was less rather than more pronounced in tar2lhypo compared with Bd21-0 (Figure 5D). Likewise, in general, reduced rather than increased acidification was observed in ein2l1hypo roots compared with their Bd21-3 wild type background (Supplemental Figure 2A). Finally, we monitored the phosphorylation state of PM-H+-ATPases in root segments. Phosphorylation of the penultimate amino acid within the autoinhibitory C-terminal domain of PM-H+-ATPase and subsequent binding of 14-3-3 proteins is the major mechanism of enzyme activation (Palmgren et al., 1991; Portillo et al., 1991; Speth et al., 2010). We therefore monitored both the capability of 14-3-3 proteins to associate with PM-H+-ATPase in microsomal membranes of root segments (14-3-3 overlay), reflecting its phosphorylation level, and the amount of PM-H+-ATPase (Ottmann et al., 2007; Speth et al., 2010). Interestingly, phosphorylation-dependent binding of 14-3-3 proteins to the PM-H+-ATPase was reduced, rather than increased, in microsomal preparations from tar2lhypo root segments compared with Bd21-0 (Figure 5E). This is in striking contrast to the effect of auxin on PM-H+-ATPase phosphorylation in hypocotyl elongation in Arabidopsis (Takahashi et al., 2012). Collectively, these experiments suggest that higher cellular auxin levels in Brachypodium roots are not associated with proton pump activation or markedly elevated proton excretion at the mesoscopic level.
Figure 5.
Medium Acidification by Bd21-0 and tar2lhypo Root Tips.
(A) Medium acidification through proton excretion from seminal roots of 3-d-old seedlings, 4 h after transfer onto fresh medium with pH indicator.
(B) Same as (A), 24 h after transfer.
(C) Progressive acidification of liquid medium through proton excretion from seminal root tips starting with 2-d-old seedlings (six biological replicates).
(D) pH traces along the surface of root tips, measured with a fiber-optic pH microsensor at five equidistant points as indicated (10 to 12 biological replicates).
(E) Protein gel blot antibody detection of H+-ATPases in protein samples isolated from microsomes of root segments and overlay with 14-3-3 protein binding. Error bars indicate se of the mean. Differences were not statistically significant (Student’s t test) unless indicated as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
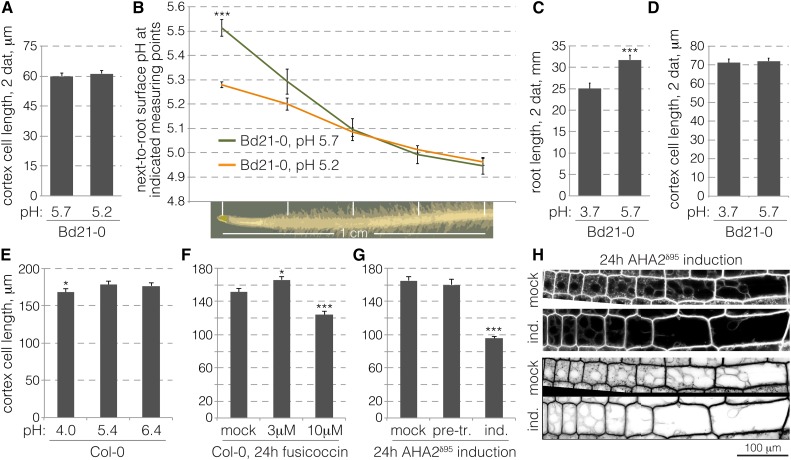
Forced Apoplastic Acidification Inhibits Root Cell Elongation
Next, to conversely determine whether acidity affects Brachypodium root cell elongation, we monitored the response of the root to externally imposed pH changes. In Arabidopsis, strong acidity eventually impairs overall root growth by inhibiting meristematic activity (Gujas et al., 2012), and the same applies to Brachypodium (see below). Reduced meristematic activity could alter mature cell length because it shifts the balance between proliferation and differentiation (Moubayidin et al., 2010; Scacchi et al., 2010). Therefore, we chose to examine mature cortex cell length after transfer of seedlings from standard medium (pH 5.7) to mildly more acidic conditions (pH 5.2), which nevertheless represent an approximately 3-fold increase in H+ concentration. Only cells formed after the transfer were scored. In these experiments, no significant length difference was observed between cells formed on either pH (Figure 6A) and overall root growth was not affected (Supplemental Figure 2B). At the same time, fiber optic pH sensor measurements along the root surface performed in parallel revealed converging pH gradients under the two conditions (Figure 6B), to approximately pH 4.9 in the root elongation zone. Medium acidification by Brachypodium root tips to pH 4.8 to 4.9 was observed repeatedly and appears to represent a lower limit in tissue culture. Therefore, we challenged roots with pH 3.7, a respective approximately 10-fold increase in acidity. Surprisingly, while overall root growth was substantially reduced at this acidic pH (Figure 6C), this was entirely attributable to reduced meristematic activity. Mature cell length was again not affected (Figure 6D). Likewise, even shortly after transfer of root tips into acidic medium, at best a small and transient significant positive effect on cell elongation could be observed (Supplemental Figure 2C). The same applies to similar experiments where root tips were transferred into medium that contained fusicoccin, a proton pump stimulant (Supplemental Figure 2D). In summary, these results suggest that cell elongation in Brachypodium roots is robustly buffered against external pH fluctuations.
Figure 6.
Root Cell Elongation in Response to External pH Variation or Forced Apoplastic Acidification.
(A) Mature cortex cell length in Bd21-0 wild-type roots 2 d after transfer of 2-d-old seedlings from standard pH (5.7) to standard pH or more acidic pH (5.2) (103 to 122 cells from six to eight roots). Only cells formed after transfer were scored.
(B) Root tip surface pH traces obtained with a fiber optic pH microsensor at five equidistant points as indicated (eight biological replicates).
(C) Root length of Bd21-0 wild-type seedlings 2 d after transfer of 2-d-old seedlings from standard pH (5.7) to standard pH or very acidic pH (3.7) (8 to 10 roots). Note that on pH 3.7, root growth is severely inhibited and meristematic activity gradually ceases.
(D) Mature cortex cell length of roots in (C) (75 to 97 cells). Only cells formed after transfer were scored.
(E) Mature cortex cell length in roots of 5-d-old Arabidopsis Col-0 wild-type seedlings grown on media with different pH (145 to 157 cells from 19 to 20 roots).
(F) Mature cortex cell length in roots of 4-d-old Arabidopsis Col-0 wild-type seedlings, 24 h after transfer from standard medium onto mock- or fusicoccin-supplemented medium. Only cells formed after transfer were scored (28 to 53 cells from six roots).
(G) Mature cortex cell length in roots of 4-d-old transgenic Arabidopsis AHA2δ95 seedlings, 24 h after transfer on medium supplemented with 1 µM dexamethasone to induce expression of the hyperactive proton pump (25 to 74 cells from five roots). Cells were scored in mock-treated roots, as well as before (pretreatment) and after induction in the same roots.
(H) Confocal microscopy of epidermal root cells in mock-treated or dexamethasone-induced AHA2δ95 seedlings, 4 h after induction, with FM4-64 staining. Vacuoles can be easily distinguished in the inverted gray-scale images. Error bars indicate se of the mean. Differences were not statistically significant (Student’s t test) unless indicated as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
To further explore the relation between apoplastic acidification and cell elongation, we turned to a model system that allows more direct manipulations, Arabidopsis. Similar to Brachypodium, mature cortex cell length was scarcely sensitive to pH variations in the medium (Figure 6E), meaning that again reduced overall root growth on acidic medium (Supplemental Figure 2E) can be largely attributed to decreased meristematic activity, as previously observed (Gujas et al., 2012). Thus, apparently root cell elongation is also robustly buffered against external pH changes in Arabidopsis roots. To override this buffering effect, we sought to uncouple proton pump activity from homeostatic inputs and stimulate it at will. To this end, we again applied fusicoccin, which at low concentration again resulted at most in a small significant stimulation of cell elongation, while higher concentration clearly reduced cell elongation (Figure 6F). In a more direct, genetic approach, we investigated wild-type seedlings that carried an inducible transgene for conditional expression of the Arabidopsis PM-H+-ATPase AHA2 devoid of its C terminus (AHA2δ95). This 95-amino acid deletion removes the autoinhibitory domain of the protein, which is therefore turned into a hyperactive proton pump that is uncoupled from regulatory inputs (Regenberg et al., 1995; Axelsen et al., 1999). Similar to high fusicoccin concentrations, strong induction of the AHA2δ95 construct resulted in the cessation of meristem activity, massive root cell swelling, and eventual rupture of the root tissues. By contrast, at lower induction levels, which maintained root growth, cell elongation was strongly reduced (Figure 6G). In these conditions, some cellular swelling was observed, consistent with an increased vacuole size (Figure 6H), yet this vacuolar size increase by itself was apparently not sufficient to drive significant cell elongation. Finally, in all conditions, both fusicoccin exposure and AHA2δ95 induction resulted in reduced meristematic activity and thus reduced meristem size (Supplemental Figures 2F and 2G). Yet even when strong vacuolar swelling was induced, we did not observe longer cells.
DISCUSSION
The importance of auxin in plant development cannot be overstated. Auxin impinges on a large variety of physiological and morphological processes, for which both absolute and relative auxin levels can be determinants. For instance, this is illustrated in root development, where auxin biosynthesis, polar transport, and signaling are required for proper morphogenesis, growth, and integration of environmental signals (Hardtke and Berleth, 1998; Sabatini et al., 1999; Zhao, 2014; Adamowski and Friml, 2015). A wealth of genetic and physiological data underpins the role of auxin in root development, yet root responses to systemically applied external auxin have been difficult to interpret. While picomolar levels of auxin sometimes stimulate root growth, physiological, nanomolar concentrations in general suppress root growth (Sutcliffe and Sexton, 1969; Evans et al., 1994; Overvoorde et al., 2010). Likewise, genetically increased excess cellular auxin production through ectopic overexpression of YUCCA enzymes inhibits rather than enhances root growth (Chen et al., 2014). These results indicate that in the absence of correct tissue context, increased auxin levels fail to reveal the central role of auxin in root growth, possibly because crucial auxin gradients are overridden (Benjamins and Scheres, 2008). In summary, pertinent auxin biosynthesis, transport, or signaling mutants and transgenic lines, mostly in Arabidopsis, do not display substantially enhanced root growth. Therefore, the Brachypodium tar2lhypo and ein2l1hypo mutants represent a so far very unusual situation because here locally increased auxin levels are associated with a specific and strong stimulatory effect on root cell elongation while meristematic activity and meristem size are not affected (Pacheco-Villalobos et al., 2013). This observation also contradicts the sometimes voiced argument that root cell elongation is typically maximal and therefore cannot be stimulated further by hormone action.
A Transcriptome Associated with High Auxin Steady State
The remarkable phenotypic specificity of both Brachypodium mutants with respect to root cell elongation offered us the unique opportunity to survey a transcriptome that is associated with a high auxin steady state. Auxin-regulated genes have so far been mainly identified through their response to external auxin application. This approach has been tremendously successful in identifying the principal auxin target genes and the autoregulatory feedback in the auxin signaling networks. Most prominently, they include AUX/IAA genes, which encode repressors of auxin signaling and respond rapidly and strongly to auxin application. Compared with these classic auxin-responsive transcriptomes, components of the auxin signaling network are rare among the differentially expressed genes in our RNAseq analyses. Even when significant, their expression fold changes are very moderate throughout, typically smaller than 1.5. Overall, the differentially expressed auxin signaling genes are downregulated in our samples, which could indicate a compensatory mechanism in response to higher cellular auxin levels. Thus, at the transcriptional level, the auxin signaling network maintains a remarkably buffered homeostasis. By contrast, a number of classic auxin target genes that are considered physiologically relevant immediate outputs of auxin action are differentially expressed at higher levels in the mutants and are upregulated overall. Most notably, these include genes related to cell wall remodeling, among which substantial differential expression of genes encoding expansins is most robust. In summary, these observations suggest that our mutant transcriptomes could be indicative of physiologically relevant auxin targets.
Between the five types of transcriptomes, we not only observed increasing differential expression as a function of phenotypic strength, or, in the case of roots grown on l-kynurenine, systemic action, but also in relation to parental background. However, although all samples were harvested in parallel, the ein2l1hypo and Bd21-3 root segments were processed at a different time from the other samples, and their RNAseq was performed in a separate instrument run. Thus, it is possible that the ein2l1hypo versus tar2lhypo transcriptome comparison is to some degree not only constrained by parental background, but also by batch effect. Although this limits the validity of any derived analyses, it is noteworthy that across all possible cross-comparisons, the by far most robust differential expression was observed for a gene that encodes an arabinogalactan peptide.
A Specific Effect of High Steady State Auxin Levels on Arabinogalactan Complexity
AGPs are a group of highly diverse cell surface glycoproteins (Seifert and Roberts, 2007; Ellis et al., 2010). Their protein backbone is characterized by dipeptide motifs that comprise hydroxyproline residues, which serve as attachment points for β-1,3-linked galactose oligosaccharides. These galactans can themselves serve as secondary branch points for additional side chains, which can contain a variety of other sugars, such as arabinose or fucose. The exact roles of AGPs in plant development remain somewhat unclear, in part because of their structural variety and the resultant fuzziness of analyses, but they have been implicated in various growth-related processes (Seifert and Roberts, 2007; Ellis et al., 2010). The most clear-cut evidence for a role in root development so far comes from genetic analyses of Arabidopsis plants with altered expression of enzymes that have an experimentally proven role in the biosynthesis or degradation of arabinogalactan side chains (Knoch et al., 2014). An interesting finding from this small set of studies is that while arabinogalactans appear to be generally required for cell elongation, in mutants or transgenic lines with mildly reduced arabinogalactan complexity, root elongation is substantially stimulated (van Hengel and Roberts, 2002; Eudes et al., 2008; Knoch et al., 2013). However, whether this is mainly due to enhanced cell elongation has not been reported.
The published genetic data on the role of arabinogalactans in Arabidopsis root development match our observations for Brachypodium. In our root segments, we did not observe any changes in the expression of Brachypodium homologs of proven arabinogalactan metabolism enzymes. Yet, we observed a clear reduction in β-1,3-galactan levels in biochemical and in situ analyses of our mutants. Overall, the data suggest reduced complexity and possibly also abundance of arabinogalactans. It is conceivable that these observations could be linked to changes in AGP expression in response to higher steady state auxin levels. AGP protein backbones, which are typically approximately 100 amino acids long, are secreted and attached to the plasma membrane via a glycosylphosphatidylinositol membrane anchor that is added during their processing. The same applies to the much shorter arabinogalactan peptides, which are only 10 to 12 amino acids long (Schultz et al., 2004). Of the three differentially expressed arabinogalactan backbone genes in our data set, the one encoding an 11-amino acid arabinogalactan peptide is most dynamically and robustly overexpressed (4 to 7×) in the high cellular auxin situation across all comparisons. Perhaps this shift in arabinogalactan protein backbone length distribution to a higher proportion of short backbones could lead to a looser, less complex AGP network. Future approaches, for instance, transgenic overexpression, could be used to address this notion directly.
Root Cell Elongation Is Robustly Buffered against External pH Fluctuation
The Acid Growth Theory of plant cell elongation has been formulated with respect to the elongation of shoot organs, with an experimental focus on coleoptiles and hypocotyls. To what degree it is pertinent for root cell elongation has been controversial from the beginning because of early observations that auxin application to intact roots generally inhibits growth or has no effect (Sutcliffe and Sexton, 1969). At best, growth stimulation could be observed with very low auxin concentrations or in treatments of auxin-depleted roots (Edwards and Scott, 1977; Pilet et al., 1979; Evans et al., 1994). However, in all cases, the size of the effect was small, and it was not reported whether the effect was due to altered root meristem activity or cell elongation. In accordance with these results, the impact of auxin on proton excretion also did not match observations in shoot organs and was generally variable. For example, proton efflux upon treatment of maize (Zea mays) roots was reported for nanomolar concentrations of auxin, while proton uptake was observed with micromolar concentrations, with the caveat that these roots had been pretreated with ethylene biosynthesis inhibitors (Mulkey et al., 1982). Others suggested that growing parts of barley (Hordeum vulgare) roots take up protons, while nongrowing parts secrete them (Weisenseel et al., 1979). Finally, a recent study that monitored apoplastic pH using a fluorescent molecular marker in planta found that auxin treatment has little effect on pH in the meristem tip but leads to alkalization rather than acidification in the root cell elongation zone (Gjetting et al., 2012), corroborating similar earlier claims (Evans et al., 1980; Moloney et al., 1981; Luthen and Bottger, 1993). Eventually, for maize roots, it was concluded that the pH growth curve exhibits a broad optimum ranging from pH 4.5 to 9, that any acid-induced growth is of very short duration, that the low sensitivity of root growth to external pH is independent of both pump activity and buffering capacity of the bathing solution, and that neither incubation in acidic buffer nor stimulation of the proton pump reverts auxin-induced root growth inhibition (Luthen and Bottger, 1993).
Our observations largely second these conclusions for Brachypodium as well as Arabidopsis. Although it is evident from our assays that the root tips acidify the rhizosphere as could be expected, we did not detect enhanced proton excretion at the surface of mutant roots. Thus, although a few SAUR genes are upregulated under the high auxin conditions in our transcriptomes and could possibly stimulate proton pumps indirectly, similar to Arabidopsis SAUR19 (Spartz et al., 2014), this apparently does not translate into a detectable increase in proton excretion at the mesoscopic level. Likewise, the two proton pump genes that are downregulated under high auxin conditions should have little impact on overall proton pump abundance because they are only weakly expressed. The by far preponderant proton pump gene that we detected (Bradi5g24690) was expressed very robustly across all conditions tested at levels over 10 times higher than the other nine proton pump genes combined. Indeed, our protein gel blot analysis confirmed that PM-H+-ATPase abundance in tar2lhypo was comparable to the wild type or at best mildly reduced. Moreover, given that proton pump activity is mostly regulated through posttranslational modifications, one would not expect that their mild transcriptional regulation would play a major role.
Based on our data, we cannot exclude the possibility that elevated proton pumping is induced by higher cellular auxin content, but it does not propagate beyond the immediate vicinity of the cell surface because of the concomitantly increasing membrane potential. Such proton pumping could therefore not be detected by our methods. Although our observation that PM-H+-ATPase phosphorylation is decreased rather than increased in tar2lhypo argues against this scenario, it is important to note that the interpretation of our findings is constrained by the lack of single cell resolution of our observations and morphological features, such as the shorter root hairs in tar2lhypo mutants (Pacheco-Villalobos et al., 2013). Perhaps very local and transient acidification is sufficient to trigger cell elongation, which would make the observation that cell elongation in both Brachypodium and Arabidopsis roots is robustly buffered against external pH fluctuation, including imposed acidity, even more remarkable. Consistently, throughout our experiments, excess acidity eventually suppressed root growth by impairing meristematic activity rather than cell expansion. This finding suggests that compensatory mechanisms act to keep apoplastic root cell pH optimal for cell elongation. Our finding that forced apoplastic acidification, for instance, through induction of a constitutively active proton pump, strongly impairs root cell elongation underlines this notion.
In summary, our data suggest that elevated steady state auxin levels in Brachypodium seminal roots are associated with specific transcriptomic and cell wall changes. While some of those changes match expectations, e.g., enhanced expression of expansin genes, others come as a surprise, notably the specific effect on arabinogalactans. Whether all of the observed changes are direct effects of auxin or emerge indirectly, for example, from hormonal crosstalk, notably with ethylene, remains to be determined. At this point, our data provide a phenotypic reference framework for future investigations that might also clarify to what degree our observations are specific for Brachypodium. Robust buffering of root cell elongation against pH fluctuation would surely make sense in the universal biological context of root growth. Unlike the shoot system, the root system is in close contact with a solid phase environment, the soil, which imposes its pH. Roots can modify the rhizosphere pH by proton pumping to increase the solubility of essential nutrients and promote their uptake, which has an optimum in the range of pH 5.0 to 6.5. Given the variability of soil pH values and their seasonal fluctuation, it appears advantageous for the plant if root growth capacity is not dictated by the soil environment pH. From this perspective, it would make sense that the interplay between auxin, proton pump activation, and expansin action at the heart of the Auxin Growth Theory is possibly more flexible in the root than in the shoot.
METHODS
Plant Materials and Growth Conditions
The Brachypodium distachyon mutants tar2lhypo and ein2l1hypo and their respective wild-type backgrounds Bd21-0 and Bd21-3 have been described before (Pacheco-Villalobos et al., 2013). Unless indicated otherwise, analyses were performed on tissue culture-grown 4-d-old seedlings raised under previously described conditions (continuous light, 100 to 120 μE intensity, 22°C, Philips F17T8/TL741 fluorescent light bulbs) (Pacheco-Villalobos et al., 2013). Solid media were prepared using Phytagel (Sigma-Aldrich) and Murashige and Skoog (MS) salts (Sigma-Aldrich). Care was taken to place the 10-cm square Petri dishes at a slight angle from the vertical to assure seminal root growth along the agar surfaces. Roots that had grown into the plate were excluded from analysis. For metabolic profiling, chemical cell wall analysis, and RNAseq, parallel grown ∼1-cm seminal root pieces harvested from 2 to 3 mm above the root tip were sampled (Supplemental Figure 1D). To generate VAS1L RNAi knockdown lines, a DNA fragment of 422 bp containing the VAS1L 3′-UTR was amplified using the oligonucleotides attB1-BdVAS1L-F 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTGGTACAGTAACAGCCCATC-3′ and attB2-BdVAS1L-R 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGGAAGTGGCAGTTCTGTCAG-3′ and cloned into pDONR207 (Life Technologies). This 3′-UTR-specific DNA fragment was then cloned into the pANDA RNAi vector (Miki and Shimamoto, 2004). Transformation of the pANDA-BdVAS1Li plasmid into tar2lhypo embryo-derived callus was performed as described (Pacheco-Villalobos et al., 2013). Arabidopsis experiments were performed with the standard Col-0 accession under the growth conditions described above. For dexamethasone-inducible expression of AHA2 (AT4G30190) devoid of its autoinhibitory C-terminal domain (AHA2δ95) the corresponding cDNA was amplified by PCR and cloned into pTA7002 (Aoyama and Chua, 1997) via XhoI and SpeI restriction enzyme sites. Transgenic lines were obtained after transformation of Col-0 plants using standard procedures (Clough and Bent, 1998).
Tryptophan Aminotransferase Activity Assays
Seminal roots were ground in liquid nitrogen with a TissueLyser II (Qiagen). Root tissue was homogenized in a precooled mortar on ice with one volume of extraction buffer [100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer, pH 7.2, 250 mM sorbitol, 5 mM β-mercaptoethanol, 0.5% (v/v) Triton X-100, and 0.1% (w/v) phenylmethylsulfonyl fluoride). The protein extract was centrifuged at 16,000g for 30 min at 4°C. The supernatant was used for determination of tryptophan aminotransferase activity with the Salkowski reagent as described (Szkop et al., 2012). Briefly, the reactions were performed in 100 mM phosphate buffer, pH 8.0, 10 mM l-tryptophan, 10 µM pyridoxal phosphate, and 50 µg of soluble proteins. The mixture was preincubated for 3 min at 35°C. The transamination reactions were initiated by the addition of 3 mM 2-oxoglutarate and incubated for 15 min at 35°C. To estimate the basal tryptophan aminotransferase activity of the crude extracts, control samples without 2-oxoglutarate were also assayed. To terminate the reactions, 1 mL of Salkowski reagent (10 mM FeCl3 and 35% [v/v] H2SO4) was added and the samples were incubated in the dark for 10 min at room temperature. The absorbance at 530 nm of four replicates was measured.
Detection of Proton Pump Phosphorylation Status
To determine the abundance of activated PM-H+-ATPase, microsomes were prepared from root segments and analyzed with the overlay assay as described (Ottmann et al., 2007), except that RGS-(His)6-tagged 14-3-3 was applied. Bound 14-3-3 was visualized by means of an antibody raised against the RGS-His6 epitope (20 μg/mL; Qiagen; catalog no./ID 34610).
Auxin Metabolite Profiling
For full-scale profile of auxin metabolites, four independent replicate samples of pooled 1-cm root segments were analyzed. Upon harvest, samples were immediately frozen in liquid nitrogen and stored at −80°C until they were analyzed as described (Novák et al., 2012).
RT-PCR
To monitor the expression of VAS1L full-length transcript by RT-PCR, the full-length transcript was amplified using oligonucleotides 5′-ATGAGCAGCTTTGCCAAGCT-3′ and 5′-GGAAGTGGCAGTTCTGTCAG-3′. UBIQUITIN CONJUGATING ENZYME18 (see below) was used as a control.
RNA Sequencing and Data Analysis
For RNAseq, total RNA was extracted from 8 to 12 pooled root segments using RNA extraction kits (Qiagen). cDNA libraries for sequencing were then prepared with the TruSeq Stranded mRNA Library Prep Kit (Illumina) using 1 μg of RNA starting material. Sequencing was performed on HiSeq 2500 instruments (Illumina) to yield 100-bp reads. The Bd21-0, tar2lhypo, mock-treated Bd21-0, and l-kynurenine-treated Bd21-0 samples were prepared and run in parallel, multiplexed in the same sequencing lane. The Bd21-3 and ein2l1hypo samples, although grown and harvested in parallel, were processed in a separate run in the same manner. The 100-bp single reads were then mapped onto the Brachypodium primary transcripts (version 2.1, http://phytozome.jgi.doe.gov/pz/portal.html) using kallisto software (version 0.42.1, http://pachterlab.github.io/kallisto/) (Bray et al., 2016) with default settings (100 bootstrap samples). Subsequent differential expression analysis was performed using sleuth software (version 0.27.3, http://pachterlab.github.io/sleuth/), again with default settings. The word cloud was produced using the wordle online tool (www.wordle.net).
qPCR
qPCR was performed on three biological replicates with a Stratagene MxPro 3005P real-time PCR system (Stratagene), using SYBR Green to monitor DNA synthesis. Relative gene expression levels were calculated as described in Pacheco-Villalobos et al. (2013). The following oligonucleotides were used: reference gene UBIQUITIN CONJUGATING ENZYME18 (Bradi4g00660), 5′-GGAGGCACCTCAGGTCATTT-3′ and 5′-ATAGCGGTCATTGTCTTGCG-3′; EXPANSIN (Bradi1g74710), 5′-GTCCTCTACCAACAGGTGAAG-3′and 5′-AGTTCCTGGACATCTGGATC-3′; EXPANSIN (Bradi3g50740), 5′-CGCGTGCTATCAGGTTAAATGC-3′ and 5′-TCTTGTACTGGATTCTGAGGAC-3′; AG-peptide (Bradi2g31980), 5′-AGTACCCCCTTCGGTTTCGT-3′ and 5′-TGGTCGATGGACGATGCGTC-3′; GH3 enzyme (Bradi2g20740), 5′-ACCACTTACTCCGGGCTGTA-3′ and 5′-CGTGTACTCCACTAAAGACG-3′; PM-H+-ATPase (Bradi1g12117), 5′-AGATGGGAGGAAAGAGAGTC-3′ and 5′-AATGGCTAGCTGATCACCTG-3′; PM-H+-ATPase (Bradi3g18790), 5′-CCAGAGGATGAAGAACTACACG-3′ and 5′-GATCGTCATGATTGTGCCATCG-3′; ACC synthase (Bradi1g10030), 5′-CCACTGGCATCATCCAGATG-3′ and 5′-TGAACCTCGCCAATGCATTC-3′; and ETRL2 (Bradi3g55730), 5′-GCAGAAAGCTTGTGCAGATGATG-3′ and 5′-GCATGACGGCGATGTATATTGC-3′.
Yariv Staining
For arabinogalactan staining with Yariv reagent, roots isolated from Bd21-0 and tar2lhypo seedlings were embedded in 6% agarose and longitudinally sectioned with a Leica-VT 1000S vibratome. Root sections were then incubated in a Yariv reagent (Biosupplies) solution (freshly prepared according to the manufacturer’s instructions) for 5 min and directly examined under a Leica DM5500B compound microscope.
Rhizosphere Acidification Assays
Media acidification assays were performed as described (Gujas et al., 2012). To visualize rhizosphere acidification, 3-d-old Brachypodium seedlings were transferred to half-strength MS-agar plates supplemented with 0.15 mM bromocresol purple (Sigma-Aldrich) (sensitivity range pH 5.2 to 6.8). The plates were incubated in the same culture chamber and scanned after 4 and 24 h. For liquid medium pH assays, 2-d-old Brachypodium seedlings were transferred on a sterile mesh attached to a tube containing 10 mL of nonbuffered half-strength liquid MS medium and 0.15 mM bromocresol green (Sigma-Aldrich) (sensitivity range pH 3.8 to 5.4). Measurement of pH was performed in a time series at 4, 24, 48, and 240 h. Three replicates consisting of eight plants per tube were measured. Negative controls (mock) without plants were measured in parallel.
Root Tip Regeneration Assays
For root tip regeneration experiments, ∼1-cm root segments from above the root tip were excised with a razor blade. The isolated root tips were then incubated on the same plates under the same conditions. De novo root tissue formation from the tips was monitored by scanning the plates after 1, 2, 3, and 4 d. Length quantification of newly formed cortex cell was performed by microscopy at 4 d after excision.
Antibody Staining of Brachypodium Root Sections
Bd21-0 and tar2lhypo plants were grown on half strength MS plates containing 1% sucrose and 0.9% agarose under long-day conditions (16 h light/8 h dark) at 22°C. Four-day-old roots were then sectioned with a vibratome (Leica VT1000 S). For immunolabeling of demethylesterified pectin, freshly cut 100-µm cross sections were first rinsed with 2F4 buffer (20 mM Tris-HCl, pH 8.2, 0.5 mM CaCl2, and 150 mM NaCl) for 10 min. The samples were then incubated with 2F4 monoclonal antibody (Plantprobes) diluted 1:250 in 2F4 buffer with 5% skim milk powder (w/v) under gentle stirring for 1 h. After washing three times with 2F4 buffer, the samples were incubated with secondary antibody (goat anti-mouse IgG (H+L), Alexa Fluor 488 conjugate, ThermoFischer A-11001) diluted 1:1000 in 2F4 buffer with 5% skim milk powder (w/v) for 3 h in the dark and then washed three times in 2F4 buffer. For immunolabeling of methylesterified homogalacturonan and arabinogalactan, the sections were incubated in 1× PBS buffer with 1% BSA and 0.05% Tween, respectively, for 1 h at room temperature, washed with 1× PBS and then incubated with JIM7 or LM2 (PLANTPROBES) diluted 1:25 in 1× PBS buffer with 1% BSA and 0.05% Tween for 1 h at room temperature. The samples were washed three times with 1× PBS and incubated with secondary antibody (donkey anti-rat Cy3; Jackson ImmunoResearch) diluted 1:500 in 1× PBS for 1 to 3 h in the dark. The samples were washed three times with 1× PBS buffer after incubation. Z-stacks were acquired using a Zeiss LSM 510 Meta confocal microscope.
Analysis of Cell Wall Polysaccharides
Biological replicates consisted of two pools of root segments harvested from 300 to 400 seedlings per genotype. The biological material was freeze-dried overnight and ground to a fine powder using a Mixer Mill MM 400 (Retsch). Cell wall preparation was performed by incubating the samples three consecutive times in 95% ethanol at 65°C for 30 min, followed by a treatment in chloroform:methanol (2:1, v/v) at room temperature for 1 h under gentle agitation. The insoluble material was then successively washed in 70% (two times 1.5 h), 80% (1 h), and 95% (2 h) ethanol and dried under vacuum (SpeedVac Plus; Savant) after a final wash in acetone. The resulting alcohol-insoluble residue was resuspended in 500 μL of an α-amylase solution (5 units/mL; Sigma-Aldrich) in 0.01 M phosphate buffer, pH 7.0, and incubated for 24 h at 37°C under continuous stirring (Mélida et al., 2009). The resulting destarched cell wall residue was washed three times with 70% ethanol, followed by three times with acetone, and stored at room temperature for further analysis. Neutral sugar composition was determined after sulfuric acid hydrolysis (Saeman et al., 1954). For this purpose, the cell wall samples were resuspended in 72% sulfuric acid and kept in this solution for 3 h at room temperature before being heated at 100°C for 3 h. Myo-inositol was used as an internal standard. The samples were then passed through 0.2-µm nylon filters and diluted 5× with deionized water. The hydrolysates were then subjected to high-performance anion-exchange chromatography using a Dionex Carbopac PA1 column and a Dionex HPLC fitted with a pulsed amperometric detector (Dionex ICS 3000 system). The samples were eluted over 20 min with deionized water and the neutral monosaccharides were detected following postcolumn addition of 300 mM sodium hydroxide at a flow rate of 0.5 mL/min. Glycosidic linkage analyses were performed using 0.8 mg of cell wall samples. The latter were swollen in 400 μL dry dimethyl sulfoxide for 3 h with stirring at room temperature prior to sugar derivatization to permethylated alditol acetates and gas chromatography-mass spectrometry analysis, as described earlier (Mélida et al., 2013). Neutral sugar and linkage analyses were performed in triplicate.
Fusicoccin Treatment and AHA2δ95 Induction
Col-0 or AHA2δ95 seeds were stratified for 4 d in water at 4°C and then germinated and grown for 4 d in half-strength MS medium adjusted to pH 5.7. For fusicoccin treatment, Col-0 seedlings were transferred to solid medium supplemented with DMSO (mock), or either 3 or 10 µM fusicoccin (Sigma-Aldrich) and grown for an additional 24 h. For analysis of forced apoplastic acidification, AHA2δ95 seedlings were transferred to solid medium supplemented with DMSO (mock) or 1 µM dexamethasone (Sigma-Aldrich) and grown for an additional 24 h. Propidium iodide-stained roots were then analyzed under a Zeiss LSM780 confocal microscope.
pH Microelectrode Measurements
For root surface pH measurements, we used a pHOptica micro fiber optic pH system (World Precision Instruments) with pH1-micro-AOT-06-059 fiber optic microsensors (PreSens) (pH range 4.0 to 9.0). Five equidistant points along the root tip surface were measured in 4-d-old seedlings grown in tissue culture. Media pH was verified in a distant location from the seedlings.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under accession numbers Bradi2g04290 (TAR2L), Bradi4g08380 (EIN2L1), Bradi2g04860 (VAS1L), and AT4G30190 (AHA2). The RNAseq raw data are available at the National Center for Biotechnology Information Sequence Read Archive under accession number SRP072551.
Supplemental Data
Supplemental Figure 1. Supplemental illustrations of Brachypodium phenotypes.
Supplemental Figure 2. Supplemental illustrations of pH experiments.
Supplemental Data Set 1. RNAseq quantification and read counts for all samples.
Supplemental Data Set 2. Gene expression level comparison of tar2lhypo versus Bd21-0.
Supplemental Data Set 3. Gene expression level comparison of l-kynurenine-treated versus mock-treated Bd21-0.
Supplemental Data Set 4. Gene expression level comparison of ein2l1hypo versus Bd21-3.
Supplemental Data Set 5. Gene expression level data for selected genes of interest plotted in Figures 3B to 3G.
Supplementary Material
Acknowledgments
This work was funded by Swiss National Science Foundation Grant CR32I3_156724 awarded to C.S.H. K.L. acknowledges the Swedish Governmental Agency for Innovation Systems (VINNOVA) and the Swedish Research Council (VR). O.N. acknowledges the Czech Science Foundation (GA14-34792S) and the Ministry of Education, Youth, and Sports of the Czech Republic-NPU I program with project LO1204.
AUTHOR CONTRIBUTIONS
D.P.-V., S.W., C.O., K.L., V.B., and C.S.H. designed the research. D.P.-V., S.M.D.-M., A.v.d.S., T.T., Y.H.K., B.G., O.N., N.J., and Z.L. performed the research. All authors analyzed data. D.P.-V. and C.S.H. wrote the paper with input from the other authors.
Glossary
- AGP
arabinogalactan protein
- MS
Murashige and Skoog
- IPA
indole-3-pyruvic acid
- IAA
indole-3-acetic acid
Footnotes
Articles can be viewed without a subscription.
References
- Adamowski M., Friml J. (2015). PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152. [DOI] [PubMed] [Google Scholar]
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612. [DOI] [PubMed] [Google Scholar]
- Axelsen K.B., Venema K., Jahn T., Baunsgaard L., Palmgren M.G. (1999). Molecular dissection of the C-terminal regulatory domain of the plant plasma membrane H+-ATPase AHA2: mapping of residues that when altered give rise to an activated enzyme. Biochemistry 38: 7227–7234. [DOI] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465. [DOI] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34: 525–527. [DOI] [PubMed] [Google Scholar]
- Chen Q., Dai X., De-Paoli H., Cheng Y., Takebayashi Y., Kasahara H., Kamiya Y., Zhao Y. (2014). Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55: 1072–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (1993). Wall extensibility: its nature, measurement and relationship to plant cell growth. New Phytol. 124: 1–23. [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (1999). Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 391–417. [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6: 850–861. [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J., Van Volkenburgh E., Cleland R.E. (1984). Stress relaxation of cell walls and the yield threshold for growth: demonstration and measurement by micro-pressure probe and psychrometer techniques. Planta 162: 46–54. [DOI] [PubMed] [Google Scholar]
- Depuydt S., Hardtke C.S. (2011). Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21: R365–R373. [DOI] [PubMed] [Google Scholar]
- Edwards K.L., Scott T.K. (1977). Rapid-growth responses of corn root segments: Effect of auxin on elongation. Planta 135: 1–5. [DOI] [PubMed] [Google Scholar]
- Ellis M., Egelund J., Schultz C.J., Bacic A. (2010). Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol. 153: 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes A., Mouille G., Thévenin J., Goyallon A., Minic Z., Jouanin L. (2008). Purification, cloning and functional characterization of an endogenous beta-glucuronidase in Arabidopsis thaliana. Plant Cell Physiol. 49: 1331–1341. [DOI] [PubMed] [Google Scholar]
- Evans M.L., Mulkey T.J., Vesper M.J. (1980). Auxin action on proton influx in corn roots and its correlation with growth. Planta 148: 510–512. [DOI] [PubMed] [Google Scholar]
- Evans M.L., Ishikawa H., Estelle M.A. (1994). Responses of Arabidopsis roots to auxin studied with high temporal resolution - Comparison of wild-type and auxin-response mutants. Planta 194: 215–222. [Google Scholar]
- Gjetting K.S., Ytting C.K., Schulz A., Fuglsang A.T. (2012). Live imaging of intra- and extracellular pH in plants using pHusion, a novel genetically encoded biosensor. J. Exp. Bot. 63: 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujas B., Alonso-Blanco C., Hardtke C.S. (2012). Natural Arabidopsis brx loss-of-function alleles confer root adaptation to acidic soil. Curr. Biol. 22: 1962–1968. [DOI] [PubMed] [Google Scholar]
- Hager A., Menzel H., Krauss A. (1971). [Experiments and hypothesis concerning the primary action of auxin in elongation growth]. Planta 100: 47–75. [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Berleth T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17: 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa K., Tryfona T., Yoshimi Y., Hayashi Y., Kawauchi S., Antonov L., Tanaka H., Takahashi T., Kaneko S., Dupree P., Tsumuraya Y., Kotake T. (2013). β-Galactosyl Yariv reagent binds to the β-1,3-galactan of arabinogalactan proteins. Plant Physiol. 161: 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoch E., Dilokpimol A., Geshi N. (2014). Arabinogalactan proteins: focus on carbohydrate active enzymes. Front. Plant Sci. 5: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoch E., et al. (2013). A β-glucuronosyltransferase from Arabidopsis thaliana involved in biosynthesis of type II arabinogalactan has a role in cell elongation during seedling growth. Plant J. 76: 1016–1029. [DOI] [PubMed] [Google Scholar]
- Kutschera U., Schopfer P. (1985a). Evidence against the acid-growth theory of auxin action. Planta 163: 483–493. [DOI] [PubMed] [Google Scholar]
- Kutschera U., Schopfer P. (1985b). Evidence for the acid-growth theory of fusicoccin action. Planta 163: 494–499. [DOI] [PubMed] [Google Scholar]
- Luthen H., Bottger M. (1993). The role of protons in the auxin-induced root-growth inhibition: a critical reexamination. Bot. Acta 106: 58–63. [Google Scholar]
- Mélida H., Sandoval-Sierra J.V., Diéguez-Uribeondo J., Bulone V. (2013). Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryot. Cell 12: 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mélida H., García-Angulo P., Alonso-Simón A., Encina A., Alvarez J., Acebes J.L. (2009). Novel type II cell wall architecture in dichlobenil-habituated maize calluses. Planta 229: 617–631. [DOI] [PubMed] [Google Scholar]
- Miki D., Shimamoto K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45: 490–495. [DOI] [PubMed] [Google Scholar]
- Moloney M.M., Elliott M.C., Cleland R.E. (1981). Acid growth effects in maize roots: Evidence for a link between auxin-economy and proton extrusion in the control of root growth. Planta 152: 285–291. [DOI] [PubMed] [Google Scholar]
- Moubayidin L., Perilli S., Dello Ioio R., Di Mambro R., Costantino P., Sabatini S. (2010). The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr. Biol. 20: 1138–1143. [DOI] [PubMed] [Google Scholar]
- Mulkey T.J., Kuzmanoff K.M., Evans M.L. (1982). Promotion of growth and hydrogen ion efflux by auxin in roots of maize pretreated with ethylene biosynthesis inhibitors. Plant Physiol. 70: 186–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O., Hényková E., Sairanen I., Kowalczyk M., Pospíšil T., Ljung K. (2012). Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 72: 523–536. [DOI] [PubMed] [Google Scholar]
- Ottmann C., Marco S., Jaspert N., Marcon C., Schauer N., Weyand M., Vandermeeren C., Duby G., Boutry M., Wittinghofer A., Rigaud J.L., Oecking C. (2007). Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol. Cell 25: 427–440. [DOI] [PubMed] [Google Scholar]
- Overvoorde P., Fukaki H., Beeckman T. (2010). Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2: a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco-Villalobos D., Sankar M., Ljung K., Hardtke C.S. (2013). Disturbed local auxin homeostasis enhances cellular anisotropy and reveals alternative wiring of auxin-ethylene crosstalk in Brachypodium distachyon seminal roots. PLoS Genet. 9: e1003564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren M.G., Sommarin M., Serrano R., Larsson C. (1991). Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H(+)-ATPase. J. Biol. Chem. 266: 20470–20475. [PubMed] [Google Scholar]
- Pilet P.E., Elliott M.C., Moloney M.M. (1979). Endogenous and exogenous auxin in the control of root growth. Planta 146: 405–408. [DOI] [PubMed] [Google Scholar]
- Portillo F., Eraso P., Serrano R. (1991). Analysis of the regulatory domain of yeast plasma membrane H+-ATPase by directed mutagenesis and intragenic suppression. FEBS Lett. 287: 71–74. [DOI] [PubMed] [Google Scholar]
- Qiao H., Shen Z., Huang S.S., Schmitz R.J., Urich M.A., Briggs S.P., Ecker J.R. (2012). Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle D.L., Cleland R. (1970). Enhancement of wall loosening and elongation by acid solutions. Plant Physiol. 46: 250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayle D.L., Cleland R. (1977). Control of plant cell enlargement by hydrogen ions. Curr. Top. Dev. Biol. 11: 187–214. [DOI] [PubMed] [Google Scholar]
- Rayle D.L., Cleland R.E. (1992). The Acid Growth Theory of auxin-induced cell elongation is alive and well. Plant Physiol. 99: 1271–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenberg B., Villalba J.M., Lanfermeijer F.C., Palmgren M.G. (1995). C-terminal deletion analysis of plant plasma membrane H(+)-ATPase: yeast as a model system for solute transport across the plant plasma membrane. Plant Cell 7: 1655–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Villalon A., Gujas B., van Wijk R., Munnik T., Hardtke C.S. (2015). Primary root protophloem differentiation requires balanced phosphatidylinositol-4,5-biphosphate levels and systemically affects root branching. Development 142: 1437–1446. [DOI] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. [DOI] [PubMed] [Google Scholar]
- Saeman J.F., Moore W.E., Mitchell R.L., Millett M.A. (1954). Techniques for the determination of pulp constituents by quantitative paper chromatography. TAPPI Journal 37: 336–343. [Google Scholar]
- Sánchez-Rodríguez C., Rubio-Somoza I., Sibout R., Persson S. (2010). Phytohormones and the cell wall in Arabidopsis during seedling growth. Trends Plant Sci. 15: 291–301. [DOI] [PubMed] [Google Scholar]
- Scacchi E., Salinas P., Gujas B., Santuari L., Krogan N., Ragni L., Berleth T., Hardtke C.S. (2010). Spatio-temporal sequence of cross-regulatory events in root meristem growth. Proc. Natl. Acad. Sci. USA 107: 22734–22739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P. (1989). pH-dependence of extension growth in Avena coleoptiles and its implications for the mechanism of auxin action. Plant Physiol. 90: 202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P. (1993). Determination of auxin-dependent pH changes in coleoptile cell walls by a null-point method. Plant Physiol. 103: 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz C.J., Ferguson K.L., Lahnstein J., Bacic A. (2004). Post-translational modifications of arabinogalactan-peptides of Arabidopsis thaliana. Endoplasmic reticulum and glycosylphosphatidylinositol-anchor signal cleavage sites and hydroxylation of proline. J. Biol. Chem. 279: 45503–45511. [DOI] [PubMed] [Google Scholar]
- Seifert G.J., Roberts K. (2007). The biology of arabinogalactan proteins. Annu. Rev. Plant Biol. 58: 137–161. [DOI] [PubMed] [Google Scholar]
- Smallwood M., Yates E.A., Willats W.G.T., Martin H., Knox J.P. (1996). Immunochemical comparison of membrane-associated and secreted arabinogalactan-proteins in rice and carrot. Planta 198: 452–459. [Google Scholar]
- Spartz A.K., Ren H., Park M.Y., Grandt K.N., Lee S.H., Murphy A.S., Sussman M.R., Overvoorde P.J., Gray W.M. (2014). SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth C., Jaspert N., Marcon C., Oecking C. (2010). Regulation of the plant plasma membrane H+-ATPase by its C-terminal domain: what do we know for sure? Eur. J. Cell Biol. 89: 145–151. [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Robles L.M., Novak O., He W., Guo H., Ljung K., Alonso J.M. (2011). The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J.F., Sexton R. (1969). Cell differentiation in the root in relation to physiological function. In Root Growth: Proceedings of the 15th Easter School in Agricultural Sciences, University of Nottingham, W.J. Whittington, ed (London: Butterworths), pp. 80–102. [Google Scholar]
- Szkop M., Sikora P., Orzechowski S. (2012). A novel, simple, and sensitive colorimetric method to determine aromatic amino acid aminotransferase activity using the Salkowski reagent. Folia Microbiol. (Praha) 57: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Hayashi K., Kinoshita T. (2012). Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 159: 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel A.J., Roberts K. (2002). Fucosylated arabinogalactan-proteins are required for full root cell elongation in arabidopsis. Plant J. 32: 105–113. [DOI] [PubMed] [Google Scholar]
- Weisenseel M.H., Dorn A., Jaffe L.F. (1979). Natural H currents traverse growing roots and root hairs of barley (Hordeum vulgare L.). Plant Physiol. 64: 512–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Hématy K., Höfte H. (2012). Growth control and cell wall signaling in plants. Annu. Rev. Plant Biol. 63: 381–407. [DOI] [PubMed] [Google Scholar]
- Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., Kasahara H., Kamiya Y., Chory J., Zhao Y. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 18518–18523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yariv J., Lis H., Katchalski E. (1967). Precipitation of arabic acid and some seed polysaccharides by glycosylphenylazo dyes. Biochem. J. 105: 1C–2C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2014). Auxin biosynthesis. Arabidopsis Book 12: e0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Guo Y., Novák O., Dai X., Zhao Y., Ljung K., Noel J.P., Chory J. (2013). Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat. Chem. Biol. 9: 244–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.