UBIQUITIN-SPECIFIC PROTEASE14 interacts with several cell cycle regulators to regulate endoreduplication and cell and organ growth, revealing a molecular link between endoreduplication and organ size.
Abstract
Organ growth is determined by a coordinated combination of cell proliferation and cell growth and differentiation. Endoreduplication is often coupled with cell growth and differentiation, but the genetic and molecular mechanisms that link endoreduplication with cell and organ growth are largely unknown. Here, we describe UBIQUITIN-SPECIFIC PROTEASE14 (UBP14), encoded by the DA3 gene, which functions as a negative regulator of endoreduplication. The Arabidopsis thaliana da3-1 mutant shows large cotyledons, leaves, and flowers with higher ploidy levels. UBP14 acts along with UV-B-INSENSITIVE4 (UVI4), an inhibitor of the anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase, to repress endoreduplication. Also, UBP14 functions antagonistically with CELL CYCLE SWITCH52 A1 (CCS52A1), an activator of APC/C, to regulate endoreduplication. UBP14 physically associates with UVI4 both in vitro and in vivo but does not directly interact with CCS52A1. Further results reveal that UBP14 influences the stability of cyclin A2;3 (CYCA2;3) and cyclin-dependent kinase B1;1 (CDKB1;1), two downstream components of the APC/C. Thus, our findings show how endoreduplication is linked with cell and organ growth by revealing important genetic and molecular functions for the ubiquitin-specific protease UBP14 and for the key cell cycle regulators UVI4, CCS52A1, CYCA2;3, and CDKB1;1.
INTRODUCTION
Organ growth is driven by a coordinated combination of cell proliferation, cell growth, and cell differentiation (Horiguchi et al., 2006). During organ growth, the onset of cell differentiation often correlates with a switch from the mitotic cell cycle to the endoreduplication cycle, also known as the endocycle, during which mitosis is repressed and additional rounds of DNA replication occur in the absence of cytokinesis, resulting in cells with higher ploidy levels (Breuer et al., 2014; Edgar et al., 2014). Changes in the levels of endoreduplication influence cell division and cell expansion, thereby modulating organ and body size in both animals and plants (Flemming et al., 2000; Dewitte and Murray, 2003; Dewitte et al., 2003; Sugimoto-Shirasu and Roberts, 2003; Edgar et al., 2014). For example, during leaf development, a high level of endopolyploidy is positively associated with leaf size in natural populations (Gegas et al., 2014). However, the genetic and molecular mechanisms that link endoreduplication with cell and organ growth are largely unknown in both animals and plants.
Some of the regulatory mechanisms that affect the endocycle are conserved between animals and plants. Downregulation of the activity of cyclin-dependent kinase complexes associated with mitotic cyclins is a key step in promoting endocycle (Costanzo et al., 2004; Inzé and De Veylder, 2006; Breuer et al., 2014). For example, the B-type cyclin-dependent kinase CDKB1;1 and the A-type cyclin CYCA2;3 function as a complex to inhibit the switch from the mitotic cell cycle to the endocycle in Arabidopsis thaliana (Boudolf et al., 2009). Coexpression of CDKB1;1 with CYCA2;3 causes ectopic cell division and represses endoreduplication (Imai et al., 2006; Boudolf et al., 2009). Mitotic cyclins are selectively ubiquitinated by the anaphase-promoting complex/cyclosome (APC/C), a multisubunit E3 ubiquitin ligase complex, and are degraded by the ubiquitin-proteasome proteolytic pathway (Marrocco et al., 2010). The APC/C complex contains at least 11 different subunits, including a catalytic core composed of APC2 and APC11 (Page and Hieter, 1999; Tang et al., 2001). The APC/C is activated by other essential subunits, such as the docking factor APC10/Doc1 and the CELL CYCLE DIVISION20 (CDC20)/Fizzy or CDH11/Fizzy-related (FZR) activator subunits. In Arabidopsis, three CDH11/FZR-related proteins are annotated as CELL CYCLE SWITCH52 (CCS52A1/FZR2, CCS52A2/FZR1, and CCS52B), which regulates endocycle onset and progression (Lammens et al., 2008; Larson-Rabin et al., 2009; Breuer et al., 2012). In addition to activators, APC/C function is regulated by inhibitory proteins. For example, the early mitotic inhibitor 1 (Emi1) in Homo sapiens, Mus musculus, and Xenopus laevis inhibits APC/CCDH1 activity (Di Fiore and Pines, 2007). In Arabidopsis, UV-B INSENSITIVE4 (UVI4) has been proposed as a functional homolog of Emi1 (Hase et al., 2006; Heyman et al., 2011; Iwata et al., 2011). UVI4 interacts with CCS52A1 to suppress the endocycle by inhibiting CYCA2;3 degradation (Hase et al., 2006; Heyman et al., 2011).
Ubiquitinated proteins can also be deubiquitinated by a special group of thiol proteases called deubiquitinating enzymes (DUBs) in animals and plants (Wilkinson, 1997; D’Andrea and Pellman, 1998). DUBs play important roles in generating mature ubiquitin from ubiquitin precursors and in cleaving the isopeptide bonds between covalently linked ubiquitin molecules or between ubiquitin and its attached protein (Nijman et al., 2005). Animals, plants, and fungi contain numerous DUBs, and one large class is the ubiquitin-specific proteases (UBPs). Arabidopsis has 27 UBP genes that are clustered into 14 subfamilies (Yan et al., 2000; Doelling et al., 2001; Liu et al., 2008). Several Arabidopsis UBP proteins have been shown to be active enzymes in vitro (Yan et al., 2000; Sridhar et al., 2007; Liu et al., 2008). Arabidopsis UBP14 belongs to a specific subfamily that contains one gene. T-DNA insertion mutations (ubp14 and tnn6) in Arabidopsis UBP14 result in an embryonic lethal phenotype (Doelling et al., 2001; Tzafrir et al., 2002). By contrast, the per1 mutation causes a synonymous substitution in the UBP14 gene, resulting in an inhibited root hair elongation phenotype upon phosphate deficiency (Li et al., 2010). However, this per1 mutation has not been shown to affect plant growth and development under normal growth conditions (Li et al., 2010). Therefore, the biological functions of UBP14 in postembryonic development remain largely unknown. Here, we show that UBP14, encoded by DA3, functions as a regulator of endoreduplication and cell and organ growth. Our findings define an important genetic and molecular mechanism involving the UBP14 and several cell cycle regulators that links endoreduplication with cell and organ growth.
RESULTS
DA3 Is Required for Normal Endoreduplication and Organ Growth
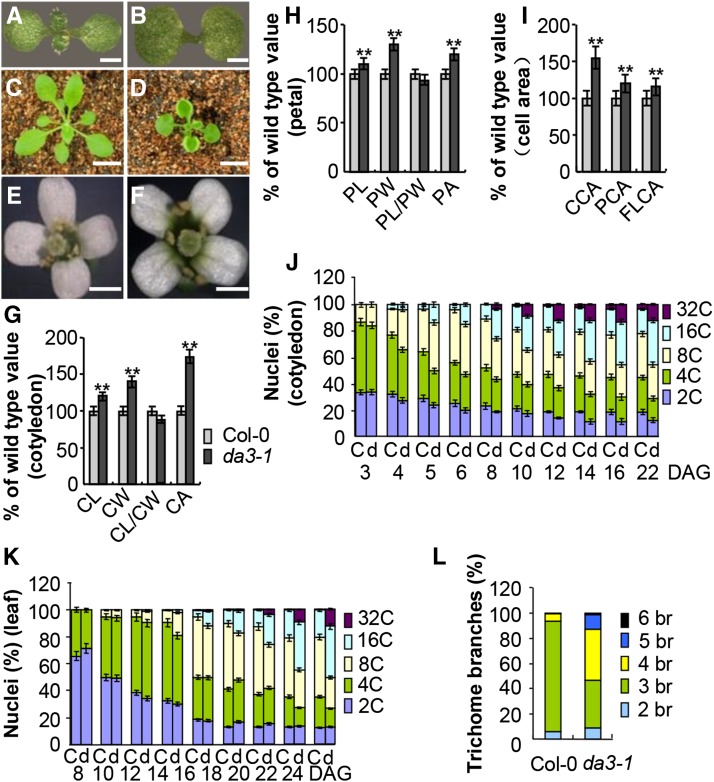
We previously identified Arabidopsis da mutants and showed that they all develop larger organs than wild-type plants (Li et al., 2008; Xia et al., 2013). Considering that endocycle defects are often associated with altered plant organ size, we examined ploidy levels of da mutants using flow cytometry. The da3-1 mutant, which has large cotyledons and flowers, showed higher ploidy levels than the wild type (Col-0) (Figures 1A, 1B, 1E to 1H, and 1J; Supplemental Figures 1 and 2). The da3-1 mutant also showed growth retardation after germination and formed curly rosette leaves (Figures 1C and 1D). Although da3-1 only produced five to six rosette leaves, the first through fourth leaves of da3-1 plants were larger than those of wild-type plants (Supplemental Figures 3B and 3C). The da3-1 mutant plants appeared smaller due to the reduced number of rosette leaves and axillary branches (Supplemental Figure 3A). In addition, da3-1 had slightly short roots with smaller root meristems (Supplemental Figure 4). These results suggest that da3-1 influences the growth in an organ-dependent manner.
Figure 1.
da3-1 Influences Endoreduplication and Cell and Organ Growth.
(A) and (B) Eight-day-old seedlings of Col-0 (A) and da3-1(B).
(C) and (D) Twenty-four-day-old seedlings of Col-0 (C) and da3-1(D).
(E) and (F) Flowers of Col-0 (E) and da3-1(F).
(G) Cotyledon length (CL), cotyledon width (CW), cotyledon length/cotyledon width, and cotyledon area (CA) of 10-d-old Col-0 and da3-1 seedlings (n = 60).
(H) Petal length (PL), petal width (PW), petal length/petal width, and petal area (PA) of Col-0 and da3-1 flowers (stage 14) (n = 50).
(I) The average area of cells in cotyledons (CCA) of 10-d-old Col-0 and da3-1 seedlings, petals (PCA) (stage 14), and the first pair of leaves (FLCA) of 21-d-old Col-0 and da3-1 seedlings . Thirty-five cotyledons (n = 35), thirty-five petals (n = 35), and thirty leaves (n = 30) were used to measure cell area.
(J) Nuclear DNA ploidy distribution of cells in Col-0 (C) and da3-1(D) cotyledons over a period of 22 DAG (n = 3). Cotyledons were harvested at the indicated times and ploidy was determined by flow cytometry.
(K) Nuclear DNA ploidy distribution of cells in the first pair of leaves of Col-0 (C) and da3-1(D) seedlings over a period of 24 DAG (n = 3). The first pair of leaves was harvested at the indicated times and ploidy was determined by flow cytometry.
(L)Trichome branch (br) distribution of Col-0 and da3-1 in the first pair of leaves of seedlings at 15 DAG (n = 200).
Values in (G) to (K) are given as means ± sd relative to the values for the respective wild-type seedlings, set at 100%. **P < 0.01 compared with the wild type (Student’s t test).
Bars =1 mm in (A) and (B), 2 cm in (C) and (D), and 1 mm in (E) and (F).
Because the final size of an organ is determined by both cell number and cell size during organogenesis, we measured cell sizes in wild-type and da3-1 cotyledons, leaves, and petals. Palisade cells in da3-1 cotyledons were larger than those in wild-type cotyledons (Figure 1I; Supplemental Figure 5A). Similarly, the size of cells in da3-1 leaves and petals was significantly greater compared with wild-type leaves and petals (Figure 1I; Supplemental Figures 5B and 5C). The number of cells in cotyledons and petals was similar in both da3-1 and wild-type plants (Supplemental Figures 5D and 5E). Unexpectedly, the number of cells in the first pair of leaves in da3-1 was slightly increased compared with the wild type (Supplemental Figure 5F). These results indicate that DA3 not only restricts cell expansion but also limits cell proliferation during leaf growth.
Alterations in ploidy levels are often correlated with changes in cell size (Sugimoto-Shirasu and Roberts, 2003). Consistent with this, we observed that the increased cell size in da3-1 cotyledons, leaves, and petals was associated with high ploidy levels (Figure 1I; Supplemental Figure 2). A time-course analysis of ploidy levels in wild-type and da3-1 cotyledons showed that, at 3 d after germination (DAG), most cells in wild-type and da3-1 cotyledons had a 2C or 4C DNA content (Figure 1J). The ploidy levels of da3-1 cotyledon cells were similar to those of wild-type cotyledon cells. At 4, 5, and 6 DAG, the balance of 2C and 4C shifted toward 8C and 16C in the da3-1 mutant compared with the wild type. For example, wild-type cotyledons had ∼64% 2C and 4C cells and 36% 8C and 16C peaks at 5 DAG, whereas da3-1 cotyledons had 52% 2C and 4C cells and 48% 8C and 16C cells. At 8 DAG and subsequent time points, the 8C to 32C fractions in da3-1 were much higher than those in the wild type, and the fractions of 2C and 4C DNA were much lower in da3-1 than those in the wild type. Similarly, higher ploidy levels in da3-1 leaves and petals were observed (Figure 1K; Supplemental Figures 2C to 2F). These results strongly indicate that DA3 is required for normal endoreduplication.
High ploidy levels are often associated with an increased number of trichome branches in Arabidopsis (Heyman et al., 2011). Therefore, we examined trichomes of the wild-type and da3-1 plants. In wild-type leaves, trichomes normally had two branching points with three branches, although trichomes with two or four branches were occasionally found (Figure 1L). By contrast, 40.2, 11.8, and 1.5% of trichomes on da3-1 leaves had four, five, and six branches, respectively (Figure 1L; Supplemental Figure 6). These results further support the role of DA3 in regulation of endoreduplication.
DA3 Encodes the Ubiquitin-Specific Protease UBP14
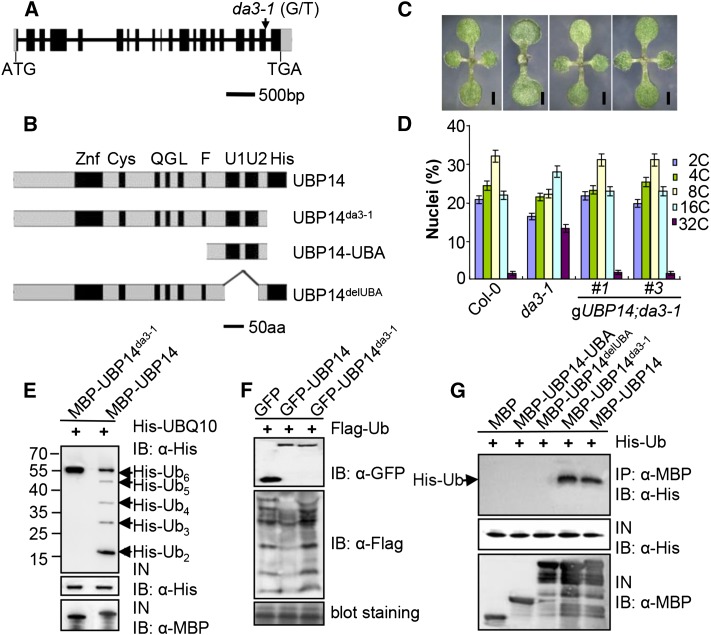
The da3-1 mutation was identified by map-based cloning in an F2 population of a cross between da3-1 and Landsberg erecta and was mapped to a 28-kb interval between markers F3H11-3 and K10D20-1 on chromosome 3 (Supplemental Figure 7A). DNA sequencing revealed that da3-1 has a G-to-T transition in the 5′-exon-intron boundary of the last intron in At3g20630 (Figure 2A; Supplemental Figure 7B). The marker At3g20630 derived cleaved amplified polymorphic sequence was developed based on this mutation in da3-1, and it cosegregated with the da3-1 phenotypes (Supplemental Figure 7C), suggesting that the At3g20630 gene corresponds to DA3, which encodes UBP14 (Figure 2B). A previous study showed that T-DNA insertion mutations (ubp14 and ttn6) in UBP14 resulted in an embryonic lethal phenotype, with the homologous embryos arresting at the globular stage (Doelling et al., 2001; Tzafrir et al., 2002). The per1 mutation resulted in a synonymous substitution in the UBP14 gene, and the per1 mutant had not been reported to influence plant growth and development under normal growth condition (Li et al., 2010). Thus, the functions of UBP14 in organ growth and development remain to be elucidated. The identity of the DA3 gene was confirmed by genetic complementation analysis using a genomic fragment (gUBP14) containing 1722 bp of 5′ flanking sequence and the At3g20630 gene. This fragment complemented the organ growth, trichome branch number, and ploidy level phenotypes of the da3-1 mutant (Figures 2C and 2D; Supplemental Figures 7E and 7F). Thus, these results indicate that At3g20630/UBP14 is the DA3 gene.
Figure 2.
DA3 Encodes the Ubiquitin-Specific Protease UBP14.
(A) DA3/UBP14 structure. The start codon (ATG) and the stop codon (TGA) for this gene are indicated. Black boxes indicate the coding sequence, gray boxes indicate the 5′ and 3′ untranslated regions, and lines between boxes indicate introns. The mutation site of da3-1 is shown by an arrow.
(B) DA3/UBP14 structure. This protein contains the Znf, Cys, His, Q, G, and L motifs and two ubiquitin-associated domains (U1 and U2). Schematic diagrams of UBP14, UBP14da3-1, and its deletion mutants are shown. aa, amino acids.
(C) Ten-day-old seedlings of Col-0, da3-1, gUBP14;da3-1#1, and gUBP14;da3-1#3 (from left to right). gUBP14;da3-1 represents da3-1 transformed with the genomic sequence of the UBP14 gene. Bars = 1 mm.
(D) Nuclear DNA ploidy distribution of cells in Col-0, da3-1, gUBP14;da3-1#1, and gUBP14;da3-1#3 cotyledons (n = 3). gUBP14;da3-1 represents da3-1 transformed with the genomic sequence of UBP14. Each bar represents the mean ± sd of three replicates.
(E) UBP14 has deubiquitination activity in vitro. MBP-UBP14 cleaved His-UBQ10 in vitro, but MBP-UBP14da3-1 did not. His-UBQ10 and cleaved His-UBQ10 proteins were detected by immunoblot analysis with an anti-His antibody.
(F) UBP14 has deubiquitination activity in vivo. N. benthamiana leaves were transformed by injection of Agrobacterium GV3101 cells harboring Pro35S:Flag-UBQ14 with Pro35S:GFP, Pro35S:GFP-UBP14, or Pro35S:GFP-UBP14da3-1 plasmids, respectively. Total ubiquitinated proteins were detected by immunoblot analysis with an anti-Flag antibody.
(G) UBA motifs are required for ubiquitin binding. His-ubiquitin was pulled down by MBP, MBP-UBP14-UBA, MBP-UBP14delUBA, MBP-UBP14da3-1, or MBP-UBP14 and then immobilized on amylose resin and analyzed by immunoblotting using an anti-His antibody.
UBP14 contains a zinc finger domain in the N-terminal region, the characteristic Cys and His boxes essential for catalysis, the conserved Q, G, L, and F domains of unknown functions, and two ubiquitin-associated domains (UBAs) that may bind ubiquitin noncovalently (Figure 2B) (Doelling et al., 2001). In da3-1, the 5′-exon-intron boundary of the last intron is changed from G to T, replacing a G that is highly conserved at plant gene splice sites. The mutation in da3-1 altered the splicing of At3g20630 mRNA and produced two main abnormal transcripts (Supplemental Figures 7B and 7D), resulting in the premature stop codon at the same position. Also, the protein encoded by the da3-1 allele lacks the His box (Figure 2B).
The Protein Encoded by the da3-1 Allele Disrupts Deubiquitination Activity
As the protein encoded by the da3-1 allele (UBP14da3-1) lacks the His box that is crucial for catalytic activity of UBP (Figure 2B), we tested whether UBP14da3-1 possessed deubiquitination activity. UBP14 and UBP14da3-1 were expressed as maltose binding protein (MBP) fusion proteins, and UBQ10 (hexamericpolyubiquitin) was expressed as a His fusion protein in Escherichia coli. As shown in Figure 2E, MBP-UBP14 was capable of cleaving the substrate His-UBQ10, but UBP14da3-1 did not cleave His-UBQ10, indicating that UBP14da3-1 disrupts the deubiquitination activity and suggesting that the His box is essential for deubiquitination. To further investigate the deubiquitination activity of UBP14 and UBP14da3-1in planta, we transiently coexpressed Pro35S:Flag-UBQ14 with Pro35S:GFP-UBP14 or Pro35S:GFP-UBP14da3-1 in Nicotiana benthamiana leaves. Transient coexpression of Pro35S:Flag-UBQ14 and Pro35S:GFP in N. benthamiana leaves was used as a negative control. Total proteins were isolated, and precipitates were detected with anti-GFP and anti-Flag antibodies, respectively. As shown in Figure 2F, the levels of total ubiquitinated proteins in leaves coexpressing Pro35S:Flag-UBQ14 and Pro35S:GFP-UBP14 were lower than those in leaves coexpressing Pro35S:Flag-UBQ14 and Pro35S:GFP, indicating that UBP14 has deubiquitination activity in planta. By contrast, the levels of total ubiquitinated proteins in leaves coexpressingPro35S:Flag-UBQ14 and Pro35S:GFP-UBP14da3-1 were not decreased, compared with those in leaves coexpressingPro35S:Flag-UBQ14 and Pro35S:GFP, further supporting the hypothesis that UBP14da3-1 disrupts the deubiquitination activity.
Ubiquitin-Associated Domains of UBP14 Are Required for Ubiquitin Binding
Because UBP14 contains two UBAs, we tested the ubiquitin binding activity of UBP14. UBP14 and UBP14da3-1 were expressed as MBP fusion proteins, and ubiquitin was expressed as a His fusion protein in E. coli. As shown in Figure 2G, both MBP-UBP14 and MBP-UBP14da3-1 bound to His-ubiquitin, indicating that UBP14 has the ubiquitin binding activity. This also suggests that the protein encoded by the da3-1 allele does not affect the ubiquitin binding activity. We then asked whether UBA domains are required for the ubiquitin binding. Both UBP14delUBA without UBA domains and UBP14-UBA with only two UBA domains were expressed as MBP fusion proteins in E. coli. As shown in Figure 2G, both UBP14delUBA and UBP14-UBA did not interact with ubiquitin. Taken together, these results indicate that the UBA domains of UBP14 are essential but not sufficient for binding ubiquitin.
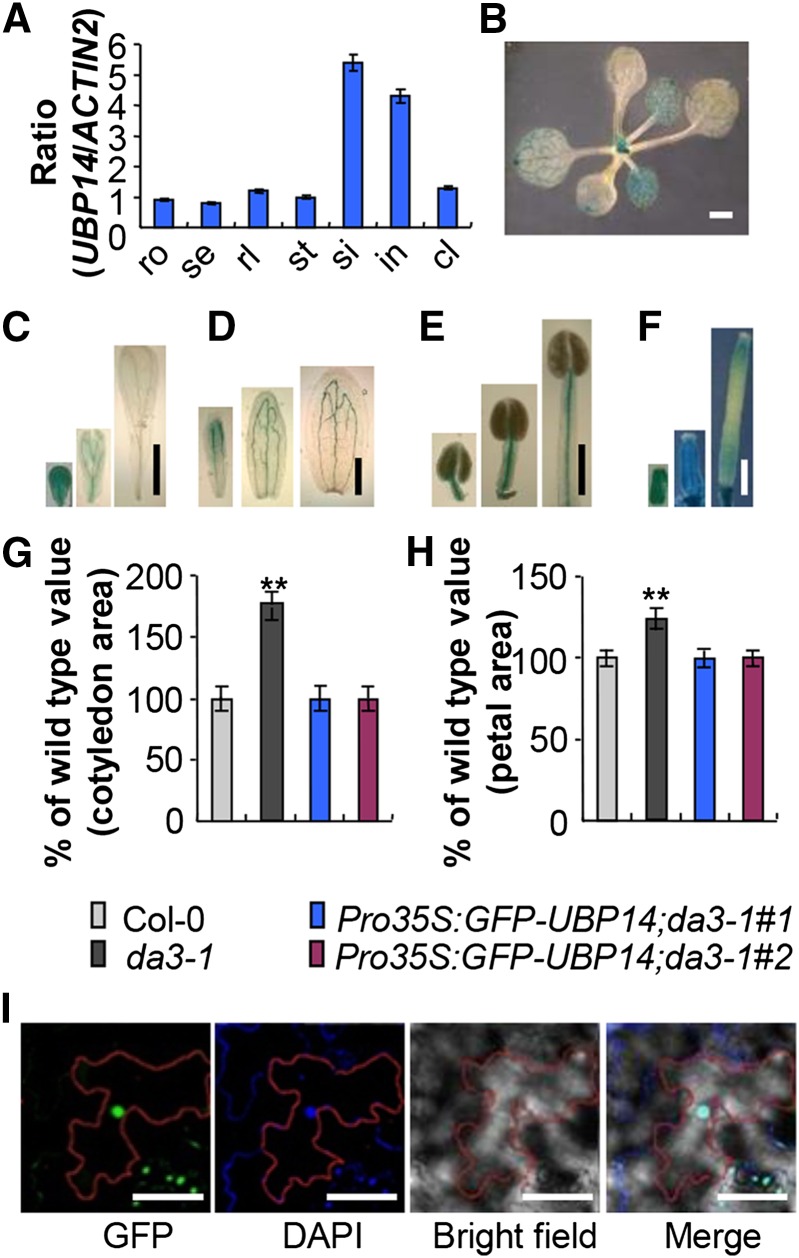
Expression and Nuclear Localization of UBP14
To investigate the expression pattern of UBP14, RNA from seedlings, roots, stems, rosette and cauline leaves, inflorescences, and siliques was analyzed by quantitative real-time RT-PCR. UBP14 transcripts were detected in all plant organs tested (Figure 3A), consistent with a previous report and the public available expression data at AtGeneExpress (data not shown) (Doelling et al., 2001). To investigate tissue-specific expression patterns of UBP14, we generated UBP14promoter:GUS fusion (ProUBP14:GUS.) transgenic plants and examined the GUS activity of 20 independent ProUBP14:GUS transgenic lines. In seedlings, GUS activity was observed in both cotyledons and leaves (Figure 3B). We then compared the GUS expression patterns of ProUBP14:GUS and ProCYCB:CDB-GUS developing leaves and found that UBP14 was expressed in both the proliferation and expansion phases during leaf development (Supplemental Figure 8). In floral organs, GUS activity was detected in developing sepals, petals, stamens, and carpels (Figures 3C to 3F).
Figure 3.
Expression Pattern and Subcellular Localization of UBP14.
(A) Quantitative real-time RT-PCR analysis of DA3 expression in the indicated Arabidopsis tissues. Total RNA was isolated from roots (ro), seedlings (se), rosette leaves (rl), stems (st), siliques (si), inflorescences (in), and cauline leaves (cl). Data shown are means ± sd of three biological replicates.
(B) to (F) UBP14 expression activity was monitored by ProUBP14:GUS transgene expression. Twenty GUS-expressing lines were observed, and all showed a similar pattern, although they differed slightly in the intensity of the staining. Histochemical analysis of GUS activity in 12-d-old seedlings (B), the developing petals (C), the developing sepals (D), the developing stamens (E), and the developing carpels (F).
(G) Cotyledon area of seedlings of Col-0, da3-1, Pro35S:GFP-UBP14;da3-1#1, and Pro35S:GFP-UBP14;da3-1#2 (n = 55). Pro35S:GFP-UBP14;da3-1 represents da3-1 transformed with Pro35S:GFP-UBP14.
(H) Petal area of Col-0, da3-1, Pro35S:GFP-UBP14;da3-1#1, and Pro35S:GFP-UBP14;da3-1#2 (n = 65). Pro35S:GFP-UBP14;da3-1 is da3-1 transformed with Pro35S:GFP-UBP14.
(I) Subcellular localization of GFP-UBP14 in epidermal cells of Pro35S:GFP-UBP14;da3-1 leaves. GFP fluorescence of GFP-UBP14, DAPI staining, and bright-field and merged images (from left to right) are shown. The red lines indicate the outline of an epidermal cell.
Values in (G) and (H) are given as means ± sd relative to the respective wild-type values, set at 100%. **P < 0.01 compared with the wild type (Student’s t test).
Bars = 1 mm in (B), 1 mm in (C) to (F), and 50 μm in (I).
To determine the subcellular localization of UBP14, we expressed a GFP-UBP14 fusion protein under the control of the 35S promoter in da3-1 plants. Transgenic lines overexpressing Pro35S:GFP-UBP14 rescued the cotyledon and flower phenotypes of da3-1 (Figures 3G and 3H; Supplemental Figure 9), indicating that the GFP-UBP14 fusion protein is functional. As shown in Figure 3I, GFP fluorescence in Pro35S:GFP-UBP14 transgenic plants was observed exclusively in nuclei. Thus, these results show that UBP14 is a nuclear-localized protein, consistent with the role of UBP14 in endoreduplication.
UBP14 Acts Genetically with UVI4 to Regulate Endoreduplication
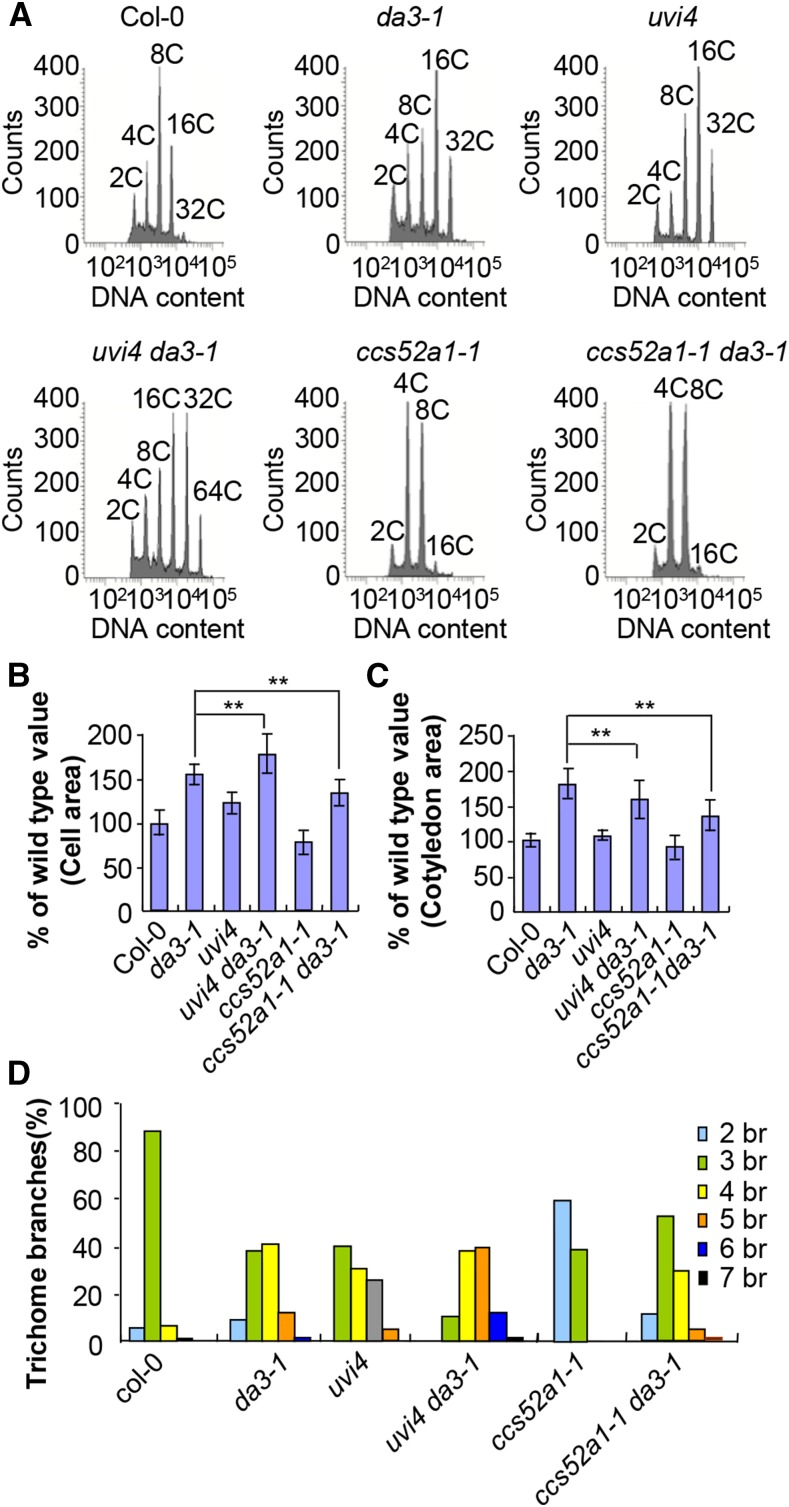
The Arabidopsis uvi4 mutant shows high ploidy levels, an increased number of trichome branches, and slightly short roots with small root meristems (Heyman et al., 2011), similar to those observed in the da3-1 mutant. To test whether UBP14 and UVI4 could function in a common genetic pathway to control endoreduplication, we crossed da3-1 with uvi4 to generate an uvi4 da3-1 double mutant. Flow cytometric analysis showed that cells in uvi4 cotyledons had higher ploidy levels than those in wild-type cotyledons (Figure 4A), consistent with previous studies (Hase et al., 2006; Heyman et al., 2011; Iwata et al., 2011). The uvi4 da3-1 cotyledons had 64C cells, and these ploidy levels were not observed in uvi4 and da3-1 single mutant cotyledons (Figure 4A). Similarly, we observed that the uvi4 da3-1 leaves had 64C cells that were not detected in uvi4 and da3-1 leaves (Supplemental Figure 10). In addition, trichomes with seven branches were observed in the leaves of the uvi4 da3-1 double mutants but not in single mutant leaves in our growth conditions (Figure 4D). These results suggest that the uvi4 mutation synergistically enhances the high ploidy level and trichome branch phenotypes of da3-1. The uvi4 mutation also increased the cell size in da3-1 (Figure 4B). However, the uvi4 da3-1 double mutants had smaller cotyledons than da3-1 (Figure 4C), indicating that uvi4 da3-1 cotyledons may contain fewer cells than da3-1 cotyledons and suggesting a possible link between endoreduplication and cell proliferation. These results suggest that UBP14 acts genetically with UVI4 to repress endoreduplication in Arabidopsis.
Figure 4.
Genetic Analyses of UBP14 with UVI4 and CCS52A1 in Regulation of Endoreduplication.
(A) Flow cytometric analysis of cells in Col-0, da3-1, uvi4, uvi4 da3-1, ccs52a1-1, and ccs52a1-1 da3-1 cotyledons.
(B) The average area of palisade cells in Col-0, da3-1, uvi4, uvi4 da3-1, ccs52a1-1, and ccs52a1-1 da3-1 cotyledons. Thirty cotyledons were used to measure cell area (n = 30).
(C) The average area of cotyledons of Col-0, da3-1, uvi4, uvi4 da3-1, ccs52a1-1, and ccs52a1-1 da3-1 (n = 50).
(D)Trichome branch (br) distribution in the first pair of leaves of Col-0, da3-1, uvi4, uvi4 da3-1, ccs52a1-1, and ccs52a1-1 da3-1 seedlings at 15 DAG (n = 200).
Values in (B) and (C) are given as means ± sd relative to the respective wild-type values, set at 100%. **P < 0.01 compared with the wild type (Student’s t test).
UBP14 Physically Interacts with UVI4 both in Vitro and in Vivo
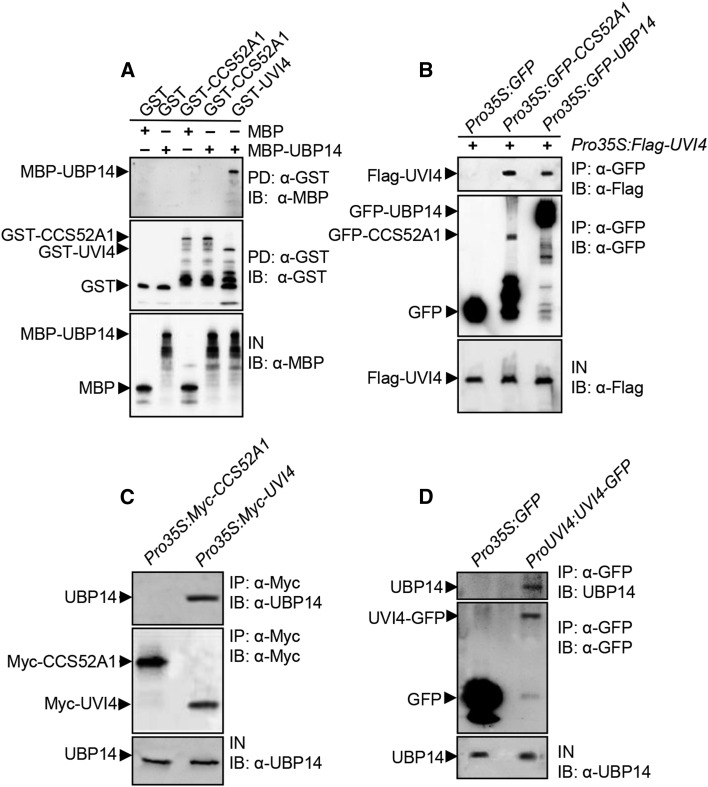
Our genetic analyses suggest that the da3-1 mutation synergistically enhanced the endoreduplication phenotype of uvi4. Several studies showed that simultaneous disruption of two or more components in a protein complex causes the enhanced phenotypes (Pérez-Pérez et al., 2009; Xia et al., 2013). We therefore investigated whether UBP14 could interact with UVI4 in an in vitro interaction/pull-down assay. UBP14 was expressed as a MBP fusion protein, while UVI4 was expressed as a GST fusion protein in E. coli. As shown in Figure 5A, MBP-UBP14 bound to GST-UVI4, indicating that UBP14 physically interacts with UVI4 in vitro.
Figure 5.
UBP14 Physically Interacts with UVI4.
(A) UBP14 physically interacts with UVI4 in vitro. MBP-UBP14 was pulled down (PD) by GST, GST-CCS52A1, or GST-UVI4 immobilized on GST beads and analyzed by immunoblotting (IB) using an anti-MBP antibody. IN represents input.
(B) UBP14 interacts with UVI4 in vivo. N. benthamiana leaves were transformed by injection of Agrobacterium GV3101 cells harboring Pro35S:Flag-UVI4 and Pro35S:GFP, Pro35S:GFP-CCS52A1, or Pro35S:GFP-UBP14 plasmids. Total proteins were immunoprecipitated (IP) with GFP-Trap-A, and the immunoblot was probed with anti-GFP and anti-Flag antibodies, respectively.
(C) UBP14 associates with UVI4 in Arabidopsis. Total proteins from Pro35S:Myc-CCS52A1 or Pro35S:Myc-UVI4 seedlings were immunoprecipitated with anti-Myc-Tag mouse mAb conjugated agarose beads, and the immunoblot was probed with anti-UBP14 and anti-Myc antibodies, respectively.
(D) UBP14 associates with UVI4 in Arabidopsis. Total proteins from Pro35S:GFP or ProUVI4:UVI4-GFP seedlings were immunoprecipitated with GFP-Trap-A, and the immunoblot was probed with anti-UBP14 and anti-GFP antibodies, respectively.
To demonstrate whether UBP14 also physically associates with UVI4 in planta, we used coimmunoprecipitation analyses to detect their interactions in vivo. We transiently coexpressed Pro35S:GFP-UBP14 with Pro35S:Flag-UVI4 in Nicotiana benthamiana leaves. Transiently expressed Pro35S:GFP and Pro35S:Flag-UVI4 were used as negative controls. Total proteins were isolated and incubated with GFP-Trap-A agarose beads to immunoprecipitate GFP-UBP14 and GFP. Immunoprecipitated proteins were detected with anti-GFP and anti-Flag antibodies, respectively. As shown in Figure 5B, Flag-UVI4 was detected in the immunoprecipitated GFP-UBP14 complex but not in the negative control (GFP), indicating that UBP14 physically associates with UVI4 in planta.
To further analyze the physical association between UBP14 and UVI4, we generated Pro35S:Myc-UVI4 transgenic Arabidopsis plants. Total proteins were isolated and incubated with agarose beads conjugated with anti-Myc-Tag mouse mAb in order to immunoprecipitate Myc-UVI4. Precipitates were detected with anti-Myc and anti-UBP14 antibodies, respectively. As shown in Figure 5C, UBP14 was detected in the immunoprecipitated Myc-UVI4 complex. We then generated ProUVI4:UVI4-GFP transgenic plants and performed an in vivo coimmunoprecipitation assay. As shown in Figure 5D, UBP14 was detected in the immunoprecipitated UVI4-GFP complex. These results confirm that UBP14 physically associates with UVI4 in Arabidopsis.
UBP14 Functions Antagonistically with CCS52A1 to Control Endoreduplication
UVI4 has been known to interact with CCS52A1, which is an activator of APC/C (Heyman et al., 2011). Because UBP14 physically associated with UVI4 in vitro and in vivo, we next tested whether UBP14 interacted directly with CCS52A1 to regulate endoreduplication. To test this, UBP14 was expressed as a MBP fusion protein, and CCS52A1 was expressed as a GST fusion protein, both in E. coli. A pull-down experiment showed that MBP-UBP14 did not directly bind to GST-CCS52A1 (Figure 5A). We then used coimmunoprecipitation analyses to detect whether UBP14 associated with CCS52A1 in Pro35S:Myc-CCS52A1 transgenic Arabidopsis plants. Total proteins were isolated and incubated with anti-Myc-Tag mouse mAb conjugated agarose beads to immunoprecipitate Myc-CCS52A1. Precipitates were detected with anti-Myc and anti-UBP14 antibodies, respectively. As shown in Figure 5C, UBP14 was not detected in the immunoprecipitated Myc-CCS52A1 complex, indicating that UBP14 might not directly interact with CCS52A1 in Arabidopsis.
UVI4 is an inhibitor of APC/C, whereas CCS52A1 is an activator of APC/C (Heyman et al., 2011). It has been proposed that the presence of the CCS52A1 activator subunit is required for UVI4 to exert its function on the APC/C activity because ccs52a1-1 is epistatic to uvi4 with respect to ploidy levels (Heyman et al., 2011). As UBP14 interacts physically and genetically with UVI4 to regulate endoreduplication in Arabidopsis, we tested whether UBP14 acted antagonistically with CCS52A1 to regulate endoreduplication. We crossed da3-1 with ccs52a1-1 and generated a ccs52a1-1 da3-1 double mutant. The ccs52a1-1 cotyledons showed decreased DNA ploidy levels, consistent with the role of CCS52A1 in activating the APC/C (Heyman et al., 2011). Ploidy levels in ccs52a1-1 da3-1 cotyledons were similar to those in ccs52a1-1 single mutant cotyledons (Figure 4A), indicating that ccs52a1-1 is epistatic to da3-1 with respect to ploidy levels. By contrast, the ccs52a1-1 mutation partially suppressed cell and cotyledon size and trichome branch number phenotypes of da3-1 (Figures 4B to 4D). Thus, these genetic analyses suggest that UBP14 may act antagonistically with CCS52A1 to control endoreduplication and cell and organ growth.
UBP14 Influences the Abundance of CYCA2;3 and CDKB1;1
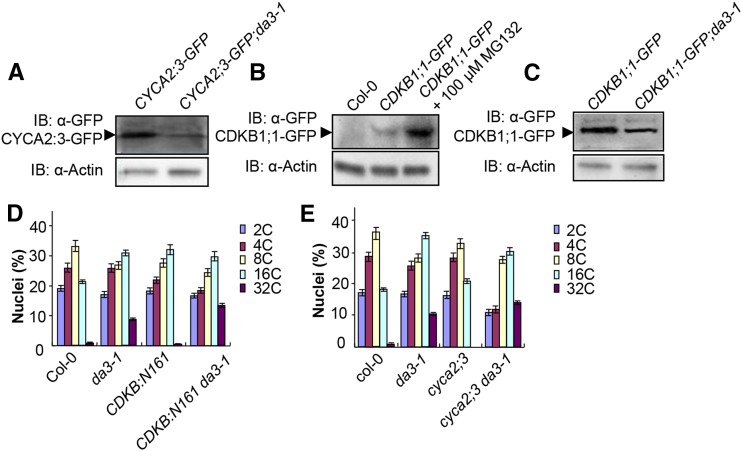
UVI4 and CCS52A1 had previously been shown to regulate the stability of CYCA2;3 (Boudolf et al., 2009; Heyman et al., 2011), a key regulator of endoreduplication. As UBP14 physically associates with UVI4 in Arabidopsis, we presumed that UBP14 would influence the abundance of CYCA2;3. We crossed da3-1 with plants harboring a β-estradiol-inducible CYCA2;3-GFP construct (Imai et al., 2006). After the selection of plants containing the CYCA2;3-GFP transgene and the da3-1 mutation, we observed the GFP signal in different genetic backgrounds. After a 16-h treatment with the inducer β-estradiol, a CYCA2;3-GFP signal was visible in the columella cells of wild-type roots but was hardly detectable in da3-1 roots (Supplemental Figure 11A). We further measured and compared the levels of CYCA2;3-GFP fusion proteins in CYCA2;3-GFP and CYCA2;3-GFP;da3-1 seedlings. After β-estradiol treatment, the levels of CYCA2;3-GFP fusion proteins in da3-1 seedlings were lower than those in wild-type seedlings (Figure 6A), but the transcript levels of CYCA2;3-GFP in da3-1 were similar to those in the wild type (Supplemental Figure 11B). These results show that UBP14 influences the stability of CYCA2;3 in Arabidopsis.
Figure 6.
UBP14 Modulates the Stability of CYCA2;3 and CDKB1;1.
(A) The da3-1 mutation decreases the level of CYCA2;3-GFP. Total proteins from CYCA2;3-GFP and CYCA2;3-GFP;da3-1 seedlings induced with 50 μM β-estradiol for 16 h were subjected to immunoblot (IB) assays using anti-GFP and anti-actin antibodies, with actin used as the loading control.
(B) The proteasome inhibitor MG132 stabilizes CDKB1;1. Ten-day-old CDKB1;1-GFP seedlings were treated with or without 100 μM MG132. Col-0 seedlings were used as a negative control. Total protein extracts were subjected to immunoblot assays using anti-GFP and anti-actin antibodies.
(C) The da3-1 mutation decreases the level of CDKB1;1-GFP. Total proteins from CDKB1;1-GFP and CDKB1;1-GFP;da3-1 seedlings were subjected to immunoblot assays using anti-GFP and anti-actin antibodies.
(D) Nuclear DNA ploidy distribution of cells in Col-0, da3-1, CDKB1;1N161, and CDKB1;1N161 da3-1 cotyledons at 10 DAG as analyzed by flow cytometry (n = 3).
(E) Nuclear DNA ploidy distribution of cells in Col-0, da3-1, cyca2;3, and cyca2;3 da3-1 cotyledons at 10 DAG as analyzed by flow cytometry (n = 3).
Values in (D) and (E) are given as means ± sd of three replicates.
CYCA2;3 forms a functional complex with CDKB1;1 to repress endoreduplication in Arabidopsis (Boudolf et al., 2009). Because UBP14 and UVI4 affects the stability of CYCA2;3, we tested whether UBP14 affected the abundance of CDKB1;1. A previous study showed that the GUS activity of CDKB1;1-GUS roots was not affected by the proteasome inhibitor MG132 (Okushima et al., 2014). We first treated the CDKB-GFP seedlings with MG132 and examined CDKB1;1-GFP proteins. As shown in Figure 6B, the levels of CDKB1;1-GFP proteins were higher in MG132-treated seedlings than untreated seedlings, indicating that the ubiquitin proteasome pathway affects the stability of CDKB1;1 in seedlings. We then crossed the CDKB1;1-GFP transgenic line with da3-1 and generated CDKB1;1-GFP;da3-1 plants. The fluorescence signal of CDKB1;1-GFP in wild-type root meristems was stronger than that in da3-1 root meristems (Supplemental Figure 12A), indicating that UBP14 modulates the stability of CDKB1;1 in Arabidopsis. We further examined the levels of the CDKB1;1-GFP fusion proteins in CDKB1;1-GFP and CDKB1;1-GFP;da3-1 seedlings. The abundance of the CDKB1;1-GFP fusion proteins in da3-1 seedlings was clearly reduced in comparison with that in wild-type roots (Figure 6C), while the transcript levels of CDKB1;1-GFP in the wild type and da3-1 were similar (Supplemental Figure 12B). Taken together, these results indicate that UBP14 influences the abundance of the CDKB1;1-CYCA2;3 complex in Arabidopsis.
Because UVI4 affects the stability of CYCA2;3, and CDKB1;1 and CYCA2;3 form a complex, the CDKB1;1N161 mutation enhanced the endoreduplication phenotype of uvi4 (Boudolf et al., 2009; Heyman et al., 2011). Similarly, UBP14 influences the stability of CYCA2;3. We presumed that the CDKB1;1N161 mutation would increase the endoreduplication phenotype of da3-1. As expected, the CDKB1;1N161 enhanced the ploidy phenotype of da3-1 (Figure 6D). Similarly, the cyca2;3 mutation enhanced the endoreduplication phenotype of da3-1 (Figure 6E). It is possible that CYCA2;3, CDKB1;1, and other factors might function redundantly downstream of UBP14 to regulate endoreduplication in Arabidopsis. In the sensitized da3-1 mutant background, the mutation of downstream components such as CYCA2;3 or CDKB1;1 would result in a more severe phenotype. Similarly, simultaneous disruption of ER (ERECTA) and its downstream component MPK6 (mitogen-activated protein kinase 6) resulted in a more severe clustered flower phenotype than er-105 or mpk6 single mutants (Meng et al., 2012). Simultaneous disruption of PID (PINOID) and its downstream component ENP (ENHANCER OF PINOID) caused a stronger defect than their respective single mutants (Cheng et al., 2008). Thus, these results suggest that UBP14 may function in a common pathway with CYCA2;3 and CDKB1;1 to regulate endoreduplication.
DISCUSSION
Endoreduplication has often been associated with cell and organ growth in animals and plants (Flemming et al., 2000; Dewitte and Murray, 2003; Dewitte et al., 2003; Sugimoto-Shirasu and Roberts, 2003; Edgar et al., 2014). However, the genetic and molecular mechanisms that link endoreduplication with cell and organ growth are largely unknown. Although a number of core cell cycle regulators involved in endoreduplication have been identified, factors other than cell cycle machinery are rarely identified. In this study, we showed that DA3, which encodes the ubiquitin-specific protease UBP14, is required for normal endoreduplication and cell and organ growth in Arabidopsis. UBP14 represses endoreduplication by acting both genetically and physically with UVI4, an inhibitor of APC/C. In addition, UBP14 functions antagonistically with CCS52A1, an activator of APC/C, to regulate endoreduplication. Further results reveal that UBP14 modulates the abundance of the CDKB1;1-CYCA2;3 complex. Thus, our findings define a key genetic and molecular mechanism that links endoreduplication with cell and organ growth in Arabidopsis.
DA3 Is Required for Endoreduplication
We previously identified da mutants with large organs in Arabidopsis (Li et al., 2008; Xia et al., 2013). The da3-1 mutant produced large cotyledons, leaves (first through fourth), and flowers (Figures 1A, 1B, and 1E to 1H; Supplemental Figures 3B and 3C). The da3-1 plants also exhibited delayed growth, reduced leaf numbers, and decreased branches, resulting in a small plant phenotype (Figures 1C and 1D; Supplemental Figure 3A). The da3-1 plants had slightly shorter roots with smaller meristems than wild-type plants (Supplemental Figure 4). These results show that DA3 influences multiple growth and developmental progresses in an organ-dependent manner. Several factors have been known to affect organ growth in an organ-dependent manner. For example, loss-of-function mutations in poly(A) polymerase PAPS1 caused large flowers and small leaves (Vi et al., 2013). The simultaneous disruption of DA1, DA1-related protein 1 (DAR1), and DAR2 resulted in large seeds and flowers, small leaves, and short roots (Peng et al., 2015). Similarly, the simultaneous disruption of three redundant Arabidopsis histidine kinases (AHK2, AHK3, and AHK4) caused small leaves and large seeds in Arabidopsis (Riefler et al., 2006).
The large cotyledon, leaf, and flower phenotypes of da3-1 were primarily caused by an increase in cell size (Figure 1I). Endoreduplication is an important determinant of cell and organ size, as shown by the correlation of cell size and ploidy levels in Arabidopsis and other plant species (Dewitte et al., 2003; Breuer et al., 2014; Gegas et al., 2014). Consistent with these observations, the overall ploidy levels in da3-1 cotyledons, leaves, and flowers were increased compared with those in wild-type cotyledons, leaves, and flowers (Figures 1J and 1K; Supplemental Figure 2). A time-course analysis of ploidy levels in developing cotyledons suggested that the da3-1 mutation may cause an earlier transition to endocycle at the early stages of cotyledon development (Figure 1J). The da3-1 mutant also showed an increased proportion of cells with higher ploidy at later stages of cotyledon growth (Figure 1J). Similarly, we also observed that da3-1 leaves and petals had higher ploidy levels than wild-type leaves and petals (Figure 1K; Supplemental Figures 2C and 2D). Thus, these results indicate that DA3 is required for normal endoreduplication and suggest a linkage between cell and organ growth and the regulation of endoreduplication. The increased ploidy levels usually caused a decrease in cell proliferation. The number of cells in da3-1 cotyledons and petals was similar to that in wild-type cotyledons and petals (Supplemental Figures 5D and 5E). Unexpectedly, we found that da3-1 first pair of leaves had large and slightly more cells compared with wild-type first pair of leaves (Figures 1I; Supplemental Figure 5F). It is possible that some targets of DA3 might regulate cell expansion, whereas other targets of DA3 might influence cell proliferation during leaf growth. We also found that da3-1 had small root meristems with a reduced cell number (Supplemental Figure 4), suggesting that DA3 influences cell proliferation in an organ-dependent manner. Similar context-dependent effects were also seen for da1-ko1 dar1-1 dar2-1 and paps1-1 mutants (Vi et al., 2013; Peng et al., 2015).
DA3 Encodes the Ubiquitin-Specific Protease UBP14
DA3 encodes the ubiquitin-specific protease UBP14, one of the UBP family members in Arabidopsis (Yan et al., 2000). In Arabidopsis, 27 members of the UBP family were clustered into 14 subfamilies containing single or multiple members, and UBP14/DA3 belongs to a specific subfamily that contains one gene (Liu et al., 2008). The per1 mutation caused a synonymous substitution in the UBP14 gene, resulting in an inhibited root hair elongation phenotype upon Pi deficiency (Li et al., 2010). These results suggest a possible link between endoreduplication and root hair elongation under phosphate deficiency. T-DNA insertion mutations in Arabidopsis UBP14 caused abnormal endosperm with large polyploidy nuclei and embryonic lethal phenotypes (Doelling et al., 2001; Tzafrir et al., 2002). Endosperm in da3-1 cells had larger nucleoli than did wild-type endosperm cells (Supplemental Figure 13), consistent with the role of UBP14 in endosperm development (Doelling et al., 2001; Tzafrir et al., 2002). The da3-1 mutant also affected plant growth and development and produced viable seeds, indicating that da3-1 is not a null allele. Thus, the da3-1 mutant provides us an opportunity to understand and further investigate the roles of UBP14/DA3 in plant growth and development.
UBP14 contains a zinc finger domain, the Cys and His boxes essential for catalysis, the conserved Q, G, L, and F domains, and two UBA domains (Figure 2B). Enzymatic analysis demonstrated that DA3 had deubiquitination activity (Figure 2E). Ubiquitin binding experiments showed that DA3 bound to ubiquitin and that two UBA domains are essential but not sufficient for ubiquitin binding (Figure 2G). Thus, it is possible that UBA domains of UBP14 are required for interactions between UBP14 and ubiquitin chains of ubiquitinated proteins, which might help UBP14 to recognize and cleave the isopeptide bonds between ubiquitin and its attached target protein. The protein encoded by the da3-1 allele contains the zinc finger domain, the characteristic Cys box, the conserved Q, G, L, and F domains, and UBA domains, but lacks the His box that is essential for catalysis (Figure 2B). Enzymatic analysis showed that the protein encoded by the da3-1 allele lost deubiquitination activity (Figures 2E and 2F), indicating that the His box of DA3/UBP14 is crucial for catalysis. Interestingly, the lack of the deubiquitination activity in da3-1 did not cause embryo lethality, suggesting that the disruption of other domain activities might be required for the embryo lethal phenotype of T-DNA insertion mutants for UBP14 (Doelling et al., 2001).
A Possible Genetic and Molecular Mechanism for UBP14-Mediated Control of Endoreduplication
Genetic analyses showed that the da3-1 mutation synergistically enhanced the endoreduplication phenotype of uvi4 (Figures 4A). The simultaneous disruption of two components in a protein complex usually results in a more severe phenotype (Pérez-Pérez et al., 2009; Xia et al., 2013). Our biochemical analysis demonstrated that UBP14 physically interacted with UVI4 in vitro and in vivo (Figure 5). These results suggest that UBP14 may either function in a complex or work together with UVI4 to inhibit endoreduplication (Figure 7). It is also possible that UBP14 may function redundantly or in parallel with UVI4 to control endoreduplication. UBP14 and UVI4 might share common downstream targets (Figure 7), and UVI4 has been shown to be the inhibitor of the APC/C (Heyman et al., 2011). Thus, both UBP14 and UVI4 may inhibit the activity of APC/C, resulting in the repression of endoreduplication (Figure 7). However, UBP14 did not physically interact with a paralog of UVI4 (OSD1) and several other cell cycle regulators (SAMBA, HOBBIT, APC2, APC5-APC7, APC10, and APC11) (data not shown). It will be interesting to investigate whether UBP14 could directly interact with other components or regulators of APC/C to repress the activity of APC/C. In Arabidopsis, CCS52A1, an activator of APC/C, has been known to promote the endocycle (Lammens et al., 2008; Larson-Rabin et al., 2009; Breuer et al., 2012). Our genetic analyses showed that ccs52a1-1 is epistatic to da3-1 with respect to endoreduplication (Figure 4A), suggesting that UBP14 may act antagonistically with CCS52A1 to control endoreduplication. Biochemical analyses showed that UBP14 did not directly interact with CCS52A1 in our experimental conditions (Figures 5A and 5C). It is plausible that UBP14 and UVI4 may inhibit the activity of APC/C, while CCS52A1 promotes the activity of APC/C in Arabidopsis (Figure 7).
Figure 7.
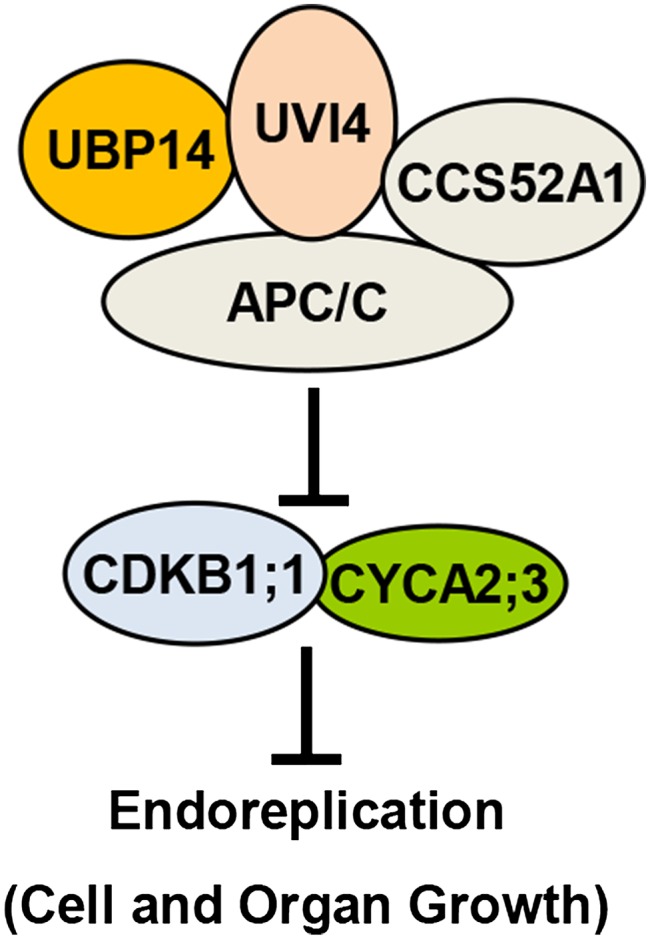
A Model for UBP14 Functions in Endoreduplication and Cell and Organ Growth.
UBP14 physically interacts with UVI4, an inhibitor of the APC/C ubiquitin ligase. UBP14 and UVI4 may work together to repress the activity of APC/C. It is also possible that UBP14 may function redundantly or in parallel with UVI4 to inhibit the activity of APC/C. UBP14 and UVI4 function antagonistically with CCS52A1 to regulate the activity of APC/C. APC/C has been shown to regulate the stability of CYCA2;3 and CDKB1;1. UBP14 influences the abundance of CYCA2;3 and CDKB1;1, which repress endoreduplication.
Because UBP14 is a functional deubiquitination enzyme and directly interacts with UVI4 (Figures 2E, 2F, and 5), it is possible that UBP14 could deubiquitinate UVI4. If so, we expected that the levels of UVI4 proteins would be reduced in the da3-1 mutant background compared with those in the wild type. We therefore generated ProUVI4:UVI4-GFP transgenic lines. We further made crosses between ProUVI4:UVI4-GFP and da3-1 and identified ProUVI4:UVI4-GFP;da3-1 plants. Surprisingly, we observed that the levels of UVI4-GFP proteins in da3-1 were increased in comparison with those in the wild type (Supplemental Figure 14), suggesting that UBP14 might not deubiquitinate UVI4. It is plausible that the da3-1 mutation might influence the levels of UVI4 proteins through a currently unknown feedback mechanism. Because UVI4 interacted with the APC/C complex (Heyman et al., 2011) and UBP14 did not physically interact with several APC proteins (data not shown), it is possible that the interaction of UBP14 with UVI4 might help UBP14 to find its substrates involved in the regulation of APC/C activity and deubiquitinate these substrates, thereby influencing the APC/C activity.
UVI4 and CCS52A1 have been shown to regulate the abundance of CYCA2;3, a target of APC/C (Boudolf et al., 2009; Heyman et al., 2011). UBP14 interacts genetically and physically with UVI4, and also functions antagonistically with CCS52A1, to regulate endoreduplication. These results suggest that UBP14 affects the stability of CYCA2;3. Consistent with this notion, we observed that the levels of CYCA2;3 proteins were lower in da3-1 seedlings than those in wild-type seedlings (Figure 6A). CYCA2;3 forms a functional complex with CDKB1;1 to repress endoreduplication in Arabidopsis (Boudolf et al., 2009). In this study, we observed that the levels of CDKB1;1 proteins were lower in da3-1 than those in the wild type (Figure 6C). Therefore, UBP14 influences the abundance of the CDKB1;1-CYCA2;3 complex, which is crucial for the regulation of endoreduplication (Boudolf et al., 2009). UBP14, UVI4, CCS52A1, CYCA2;3, and CDKB1;1 showed overlapping expression in aboveground organs in Arabidopsis (Figures 3A to 3F) (Boudolf et al., 2004; Hase et al., 2006; Imai et al., 2006; Baloban et al., 2013), supporting their genetic and biochemical relationships. However, it is noteworthy that their expression patterns in roots were not similar. For example, strong expression of UVI4 and CDKB1;1 was detected in root meristems (Hase et al., 2006; Cools et al., 2010), and expression of UBP14 was observed in the root meristem and differentiated regions (Supplemental Figure 15). By contrast, CCS52A1 was expressed around the transition zone, but not in root meristems (Vanstraelen et al., 2009). The levels of CDKB1;1-GFP in da3-1 root meristems were reduced (Supplemental Figure 12A). These results suggest that the effect of UBP14 on the stability CDKB1;1-GFP in root meristems might not require for CCS52A1. Thus, it is possible that the action of these factors in determinate organs (e.g., leaves and cotyledons) and indeterminate roots might be different. It will be interesting to investigate possible different mechanisms in the future.
The CDKB1;1N161 mutation increased the ploidy level phenotype of uvi4 (Heyman et al., 2011). Similarly, we observed that CDKB1;1N161 and cyca2;3 enhanced the ploidy level of ubp14 (Figures 6D and 6E), further suggesting that UBP14 and UVI4 have similar functions in endoreduplication. It is plausible that CYCA2;3, CDKB1;1, and other factors might function redundantly downstream of UBP14 and UVI4 to regulate endoreduplication in Arabidopsis (Figure 7). In the sensitized da3-1 mutant background, the disruption of downstream factors such as CYCA2;3 or CDKB1;1 would result in a more severe phenotype. For example, Arabidopsis ER functions upstream of MPK6, and the simultaneous disruption of ER and MPK6 caused a more severe clustered flower phenotype than er-105 or mpk6 single mutants (Meng et al., 2012). PID has been proposed to act upstream of ENP, and pid enp double mutants showed stronger defects than pid and enp single mutants (Cheng et al., 2008). Taken together, our findings reveal a key genetic and molecular mechanism for the ubiquitin-specific protease UBP14 and several key cell cycle regulators, UVI4, CCS52A1, CYCA2;3, and CDKB1;1, in linking endoreduplication with cell and organ growth (Figure 7).
METHODS
Plant Materials and Growth Conditions
All mutants used in this study were in the Columbia (Col-0) background. ccs52a1-1 (SALK_083656) and cyca2;3 (SALK_086463) were obtained from the Nottingham Arabidopsis Stock Centre and ABRC collections. The CDKB1;1-GFP (Boudolf et al., 2004), inducible CYCA2;3-GFP (Imai et al., 2006), CDKB1;1N161, and uvi4 (Heyman et al., 2011; Iwata et al., 2011) lines were obtained from Lieven De Veylder and Masaki Ito. Plants were grown under long-day conditions (16 h light/8 h dark) at 22°C. The light intensity is ∼100 μmol (m−2 s−1).
Morphological and Cellular Analysis
Cotyledons, petals (stage 14) (Smyth et al., 1990) and leaves were scanned to produce digital images. The average area of cotyledons, petals, and leaves was calculated using Image J software (http://rsb.info.nih.gov/ij/). To measure cell size, cotyledons, petals, and leaves were mounted in the clearing solution (chloral hydrate:water:glycerol = 8:3:1). Cleared samples were observed using differential interference contrast optics on a Leica microscope (Leica DM2500) and photographed with a Spot Flex Cooled CCD digital image system (SPOT Imaging).
GFP fluorescence in leaves and roots was detected using a Zeiss LSM 710 NLO confocal microscope. Plant materials were incubated with 10 µM 4′,6-diamidino-2-phenylindole (DAPI) solution to stain nuclei.
Flow Cytometry Assay
Cotyledons, petals, and leaves were chopped with a razor blade in 500 μL GS buffer (45 mM MgCl2, 30 mM sodium citrate, 20 mM MOPS, 0.1% Triton X-100) and then filtered by passing through a sieve (48 µm2 mesh). DNA was then stained by adding 10 μM DAPI. DNA content of nuclei was measured with a flow cytometer (BD FACSAria II; BD Biosciences).
Map-Based Cloning of DA3
The da3-1 mutation was mapped in the F2 population of a cross between da3-1 and Landsberg erecta. To fine-map the da3-1 mutation, new molecular markers were developed using public databases (Supplemental Table 1). We further sequenced a 28-kb interval between markers F3H11-3 and K10D20-1.
RNA Isolation, RT-PCR, and Quantitative Real-Time RT-PCR Analysis
Total RNA was isolated from Arabidopsis thaliana seedlings, roots, rosette leaves, stems, siliques, inflorescence, and cauline leaves using an RNAprep pure plant kit (Tiangen). Total mRNA was reverse transcribed into cDNA using SuperScript III reverse transcriptase (Invitrogen). Quantitative real-time RT-PCR analysis was performed with a Lightcycler 480 engine (Roche) using the Lightcycler 480 SYBR Green Master (Roche). ACTIN2 was used as an internal control. Relative amounts of mRNA were determined using the Cycle threshold (Ct) method. Ct values correspond to the cycle number at which the fluorescence resulting from enrichment of the PCR product reaches significant levels above the background fluorescence. The ∆Ct was calculated by subtracting the Ct values of ACTIN2 from the UBP14 Ct value. The ratios were calculated as being equal to 2−∆Ct. Three biological replicates were conducted in all experiments. The primers used for RT-PCR and quantitative real-time RT-PCR are listed in Supplemental Table 1.
Constructs and Plant Transformation
The gUBP14 construct was made using a PCR-based Gateway system. The genomic sequence of UBP14 including 1722-bp promoter and the UBP14 gene was amplified using the primers UBP14genome-F and UBP14genome-R (Supplemental Table 1). PCR products were cloned into the pCR8/GW/TOPO TA cloning vector (Invitrogen). The UBP14 genomic sequence was then subcloned into the binary vector pMDC99 by LR reaction. The plasmid gUBP14 was transformed into da3-1 mutant using Agrobacterium tumefaciens GV3101, and transformants were selected on medium containing hygromycin (30 μg/mL).
The Pro35S:UBP14 and Pro35S:GFP-UBP14 constructs were made using a PCR-based Gateway system. The specific primers used for the Pro35S:UBP14 and Pro35S:GFP-UBP14 constructs are UBP14CDS-F and UBP14CDS-R (Supplemental Table 1). PCR products were subcloned into the pCR8/GW/TOPO TA cloning vector (Invitrogen) using TOPO enzyme. UBP14 was then subcloned into the Gateway binary vector pMDC32 containing the 35S promoter or pMDC43 containing the 35S promoter and the GFP gene. The plasmid Pro35S:UBP14 was transformed into Col-0 plants, and the plasmid Pro35S:GFP-UBP14 was transformed into Col-0 and da3-1 plants using Agrobacterium GV3101. Transformants were grown on medium containing hygromycin(30 μg/mL).
The 1722-bp promoter sequence of UBP14 was amplified using the primers UBP14pro-F and UBP14pro-R (Supplemental Table 1). PCR products were cloned into pCR8/GW/TOPO TA cloning vector (Invitrogen) using TOPO enzyme. The UBP14 promoter was then subcloned into the binary vector pMDC164 with the GUS reporter gene by the LR reaction to generate the ProUBP14:GUS construct. The plasmid ProUBP14:GUS was transformed into Col-0 plants using Agrobacterium GV3101, and transformants were grown on medium containing hygromycin (30 μg/mL).
The ProUVI4:UVI4-GFP construct was made using a PCR-based Gateway system. The UVI4 genomic fragment containing 1016 bp of 5′ flanking sequence and the UVI4 genomic sequence was amplified using the primers UVI4107-F and UVI4107-R (Supplemental Table 1). PCR products were cloned into the pCR8/GW/TOPO TA cloning vector (Invitrogen). The UVI4 genomic fragment was then subcloned into the binary vector pMDC107 with the GFP gene by LR reaction. The plasmid ProUVI4:UVI4-GFP was transformed into Col-0 plants using Agrobacterium GV3101, and transformants were grown on medium containing hygromycin (30 μg/mL).
GUS Staining
Seedlings (ProUBP14:GUS) were incubated in acetone on ice for 5 min and then washed twice with sterile water. Seedlings and floral organs (ProUBP14:GUS) were stained in a GUS staining solution [1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, 100 mM Na3PO4 buffer, 3 mM each K3Fe (CN)6/K4Fe(CN)6, 10 mM EDTA, and 0.1% Nonidet P-40] and incubated at room temperature for 4 h. After GUS staining, chlorophyll was removed using 70% ethanol.
Deubiquitination Assays
The coding sequences of UBP14 and UBP14da3-1 were cloned into the BamHI and SalI sites of the pMAL-C2 vector to generate MBP-UBP14 and MBP-UBP14da3-1 constructs, respectively. Specific primers for MBP-UBP14 and MBP-UBP14da3-1 constructs are MBPUBP14-F/R and MBPUBP14-F/MR, respectively (Supplemental Table 1). The UBQ10 gene was amplified using the primers UBQ10-F and UBQ10-R (Supplemental Table 1). PCR products were cloned into the BamHI and HindIII sites of the pET28A vector to generate the His-UBQ10 construct. Bacterial lysates containing MBP-UBP14 or MBP-UBP14da3-1 were cleared by centrifugation and incubated with amylose resin (New England Biolabs) at 4°C for 30 min. Beads were washed by the column buffer (20 mM Tris, pH 7.4, 200 mM NaCl, and 1 mM EDTA) and equilibrated by the reaction buffer (50 mM Tris, pH 7.4, 20 mM DTT, 5 mM MgCl2, and 2 mM ATP). One microgram of His-UBQ10 was incubated with 2 µg of UBP14 or MBP-UBP14da3-1 in the reaction buffer for 8 h at 30°C. Cleaved ubiquitin products were detected by immunoblot analysis with anti-His (Abmart) antibody.
UBQ14 was cloned into the Gateway binary vector pW1216 containing the 35S promoter and Flag to generate the Pro35S:Flag-UBQ14 construct. The specific primers for the Pro35S:Flag-UBQ14 construct were UBQ14-F/R. Nicotiana benthamiana leaves were transformed by injection of Agrobacterium GV3101 cells harboring Pro35S:Flag-UBQ14 with Pro35S:GFP-UBP14, Pro35S:GFP-UBP14da3-1, or Pro35S:GFP plasmids. Total proteins were separated in 10% SDS-polyacrylamide gel and detected by immunoblot analysis with anti-Flag (Abmart) and anti-GFP (Abmart) antibodies, respectively.
In Vitro Protein-Protein Interaction
The coding sequence of CCS52A1 was cloned into SmaI and NotI sites of the pGEX-4T-1 vector and XbaI and HindIII sites of the pMAL-C2 vector to generate GST-CCS52A1 and MBP-CCS52A1 constructs, respectively. The coding sequence of UVI4 was subcloned into EcoRI and SalI sites of the pGEX-4T-1 vector to generate the GST-UVI4 construct. The specific primers for GST-CCS52A1, GST-UVI4, and MBP-CCS52A1 were CCS52A1GST-F/R, UVI4GST-F/R, and CCS52A1MBP-F/R, respectively (Supplemental Table 1).
To investigate protein-protein interaction, bacterial lysates containing ∼20 µg of MBP-UBP14 fusion proteins were mixed with lysates that contain ∼30 µg of GST-CCS52A1 or GST-UVI4 fusion proteins. Bacterial lysates with ∼30 µg of MBP-CCS52A1 fusion proteins was combined with lysates containing ∼30 µg of GST-UVI4 fusion proteins. Amylose resin (20 µL; New England Biolabs) was added into each combined solution with continued rocking at 4°C for 1 h. Beads were washed five times with TGH buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, pH 8.0, 1% Triton X-100, 10% glycerol, 1 mM PMSF, and 1× Complete protease inhibitor cocktail [Roche]), and the isolated proteins were separated on a 10% SDS-polyacrylamide gel and detected by immunoblot analysis with anti-GST (Abmart) and anti-MBP antibodies (Abmart), respectively.
Coimmunoprecipitation
The coding sequence of CCS52A1 was cloned into the KpnI and SacI sites of the pCAMBIA1300-221-Myc vector to generate the transformation plasmid Pro35S:Myc-CCS52A1. The coding sequence of UVI4 was cloned into the KpnI and BamHI sites of the pCAMBIA1300-221-Myc vector to generate the transformation plasmid Pro35S:Myc-UVI4. The coding sequences of CCS52A1 and UVI4 were subcloned into the pCR8/GW/TOPO TA cloning vector (Invitrogen) using TOPO enzyme. CCS52A1 was then cloned into the Gateway binary vector pMDC43 containing the 35S promoter and the GFP gene to generate the Pro35S:GFP-CCS52A1 construct. UVI4 was cloned into the Gateway binary vector pW1266 containing the 35S promoter and sequences encoding the Flag octapeptide tag to generate the Pro35S:Flag-UVI4 construct. The specific primers for Pro35S:Myc-CCS52A1, Pro35S:Myc-UVI4, Pro35S:GFP-CCS52A1, and Pro35S:Flag-UVI4 constructs were Myc-CCS52A1-F/R, Myc-UVI4-F/R, CCS52A1CDS-F/R, and UVI4CDS-F/R, respectively (Supplemental Table 1).
N. benthamiana leaves were transformed by injection of Agrobacterium GV3101 cells harboring different combinations of Pro35S:GFP-UBP14, Pro35S:GFP-CCS52A1, Pro35S:GFP, and Pro35S:Flag-UVI4 plasmids. Similarly, N. benthamiana leaves were transformed by injection of Agrobacterium GV3101 cells harboring Pro35S:Flag-UVI4 and Pro35S:GFP-UBP14 plasmids. Total proteins were extracted with extraction buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 20% glycerol, 2% Triton X-100, 1 mM EDTA, Complete protease inhibitor cocktail [Roche], and 20 mg/mL MG132) and incubated with GFP-Trap-A (Chromotek) for 1 h at 4°C. Beads were washed three times with wash buffer (50 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, and Complete protease inhibitor cocktail [Roche]). The immunoprecipitates were separated in 10% SDS-polyacrylamide gel and detected by immunoblot analysis with anti-GFP (Beyotime) and anti-Flag (Abmart) antibodies, respectively.
Anti-UBP14 antibodies were generated against UBP14 protein in Escherichia coli. UBP14 was expressed as a histidine and ubiquitin fusion protein (His-ubiquitin-UBP14) in E. coli BL21 (DE3) and purified by nickel-affinity chromatography. His-ubiquitin-UBP14 was then digested by deubiquitinating enzyme to take off ubiquitin and His. Rabbits were immunized with the purified UBP14 protein. Total proteins from Pro35S:Myc-UVI4 and Pro35S:Myc-CCS52A1 transgenic plants were extracted with extraction buffer and incubated with Anti-Myc-Tag mouse mAb (Abmart) for 1 h at 4°C. Total proteins from ProUVI4:UVI4-GFP transgenic plants were extracted with extraction buffer and incubated with GFP-Trap-A (Chromotek) for 1 h at 4°C. Beads were washed three times with wash buffer. The immunoprecipitates were separated in 10% SDS-polyacrylamide gel and detected by immunoblot analysis with anti-Myc (Abmart), anti-GFP (Beyotime), and anti-UBP14 antibodies, respectively.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for genes mentioned in this article are as follows: AT3G20630 (UBP14), AT2G42260 (UVI4), AT4G22910 (CCS52A1), AT1G15570 (CYCA2;3), AT3G54180 (CDKB1;1), and AT3G18780 (ACTIN2). Mutant identification information is as follows; ccs52a1-1 (SALK_083656) and cyca2;3 (SALK_086463). Expression data for UBP14 are available at AtGenExpress (http://jsp.weigelworld.org/expviz/expviz.jsp?experiment=developmentandnormalization=absoluteandprobesetcsv=At3g20630andaction=Run).
Supplemental Data
Supplemental Figure 1. da3-1 Influences Cell and Organ Growth.
Supplemental Figure 2. da3-1 Shows Increased Ploidy Levels.
Supplemental Figure 3. da3-1 Affects Plant Growth.
Supplemental Figure 4. da3-1 Influences Root Growth.
Supplemental Figure 5. da3-1 Influences Cell Size and Cell Number.
Supplemental Figure 6. da3-1 Increases the Number of Trichome Branches.
Supplemental Figure 7. Identification and Molecular Characterization of UBP14/DA3.
Supplemental Figure 8. UBP14 Is Expressed in Both the Proliferation and Expansion Phases during Leaf Development.
Supplemental Figure 9. Overexpression of GFP-DA3 Complements the Phenotypes of da3-1.
Supplemental Figure 10. UBP14 Acts Genetically with UVI4 to Regulate Endoreduplication.
Supplemental Figure 11. UBP14 Modulates the Stability of CYCA2;3.
Supplemental Figure 12. UBP14 Modulates the Stability of CDKB1;1.
Supplemental Figure 13. The Effect of da3-1 on Endosperm.
Supplemental Figure 14. The da3-1 Mutation Influences the Levels of UVI4 Proteins.
Supplemental Figure 15. UBP14 Is Expressed in Roots.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank the anonymous reviewers and the editor for their helpful comments on this article, Lieven De Veylder and John Doonan for helpful discussions, and Lieven De Veylder and Masaki Ito for kindly providing the uvi4, CYCA2;3-GFP, CDKB1;1-GFP, and CDKB1;1N161 seeds used in this study. We also thank the Arabidopsis Stock Center NASC for ccs52a1-1 and cyca2;3 seeds. This work was supported by grants from the National Natural Science Foundation of China (31425004, 91417304, 91017014, and 31221063) and the Ministry of Agriculture of China (2013ZX08009-003-003).
AUTHOR CONTRIBUTIONS
Y.L., C. Li, W.J., and Y.X. designed the research. Y.X. isolated the da3-1 mutant and conducted map-based cloning and ploidy measurements. W.J. performed genetic complementation, expression, and subcellular localization of DA3, identification of DA3 enzyme activity, pull-down and coimmunoprecipitation experiments, analysis of genetic interactions between DA3 and cell cycle regulators, and ploidy measurements. N.L. helped perform coimmunoprecipitation experiments. W.Z and C. Liu generated the anti-DA3 antibody. Y.L., W.J., C. Li, and Y.X. analyzed data. W.J. and Y.L. wrote the article. C. Li revised the article.
Glossary
- APC/C
anaphase-promoting complex/cyclosome
- DUB
deubiquitinating enzyme
- DAG
days after germination
- DAPI
4′,6-diamidino-2-phenylindole
References
- Baloban M., Vanstraelen M., Tarayre S., Reuzeau C., Cultrone A., Mergaert P., Kondorosi E. (2013). Complementary and dose-dependent action of AtCCS52A isoforms in endoreduplication and plant size control. New Phytol. 198: 1049–1059. [DOI] [PubMed] [Google Scholar]
- Boudolf V., Vlieghe K., Beemster G.T., Magyar Z., Torres Acosta J.A., Maes S., Van Der Schueren E., Inzé D., De Veylder L. (2004). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V., et al. (2009). CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol. 150: 1482–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer C., Braidwood L., Sugimoto K. (2014). Endocycling in the path of plant development. Curr. Opin. Plant Biol. 17: 78–85. [DOI] [PubMed] [Google Scholar]
- Breuer C., Morohashi K., Kawamura A., Takahashi N., Ishida T., Umeda M., Grotewold E., Sugimoto K. (2012). Transcriptional repression of the APC/C activator CCS52A1 promotes active termination of cell growth. EMBO J. 31: 4488–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Qin G., Dai X., Zhao Y. (2008). NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 21017–21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools T., Iantcheva A., Maes S., Van den Daele H., De Veylder L. (2010). A replication stress-induced synchronization method for Arabidopsis thaliana root meristems. Plant J. 64: 705–714. [DOI] [PubMed] [Google Scholar]
- Costanzo M., Nishikawa J.L., Tang X., Millman J.S., Schub O., Breitkreuz K., Dewar D., Rupes I., Andrews B., Tyers M. (2004). CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117: 899–913. [DOI] [PubMed] [Google Scholar]
- D’Andrea A., Pellman D. (1998). Deubiquitinating enzymes: a new class of biological regulators. Crit. Rev. Biochem. Mol. Biol. 33: 337–352. [DOI] [PubMed] [Google Scholar]
- Dewitte W., Murray J.A. (2003). The plant cell cycle. Annu. Rev. Plant Biol. 54: 235–264. [DOI] [PubMed] [Google Scholar]
- Dewitte W., Riou-Khamlichi C., Scofield S., Healy J.M., Jacqmard A., Kilby N.J., Murray J.A. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B., Pines J. (2007). Emi1 is needed to couple DNA replication with mitosis but does not regulate activation of the mitotic APC/C. J. Cell Biol. 177: 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Yan N., Kurepa J., Walker J., Vierstra R.D. (2001). The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J. 27: 393–405. [DOI] [PubMed] [Google Scholar]
- Edgar B.A., Zielke N., Gutierrez C. (2014). Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat. Rev. Mol. Cell Biol. 15: 197–210. [DOI] [PubMed] [Google Scholar]
- Flemming A.J., Shen Z.Z., Cunha A., Emmons S.W., Leroi A.M. (2000). Somatic polyploidization and cellular proliferation drive body size evolution in nematodes. Proc. Natl. Acad. Sci. USA 97: 5285–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegas V.C., Wargent J.J., Pesquet E., Granqvist E., Paul N.D., Doonan J.H. (2014). Endopolyploidy as a potential alternative adaptive strategy for Arabidopsis leaf size variation in response to UV-B. J. Exp. Bot. 65: 2757–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase Y., Trung K.H., Matsunaga T., Tanaka A. (2006). A mutation in the uvi4 gene promotes progression of endo-reduplication and confers increased tolerance towards ultraviolet B light. Plant J. 46: 317–326. [DOI] [PubMed] [Google Scholar]
- Heyman J., Van den Daele H., De Wit K., Boudolf V., Berckmans B., Verkest A., Alvim Kamei C.L., De Jaeger G., Koncz C., De Veylder L. (2011). Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome. Plant Cell 23: 4394–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Ferjani A., Fujikura U., Tsukaya H. (2006). Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 119: 37–42. [DOI] [PubMed] [Google Scholar]
- Imai K.K., Ohashi Y., Tsuge T., Yoshizumi T., Matsui M., Oka A., Aoyama T. (2006). The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18: 382–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé D., De Veylder L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40: 77–105. [DOI] [PubMed] [Google Scholar]
- Iwata E., Ikeda S., Matsunaga S., Kurata M., Yoshioka Y., Criqui M.C., Genschik P., Ito M. (2011). GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell 23: 4382–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens T., Boudolf V., Kheibarshekan L., Zalmas L.P., Gaamouche T., Maes S., Vanstraelen M., Kondorosi E., La Thangue N.B., Govaerts W., Inzé D., De Veylder L. (2008). Atypical E2F activity restrains APC/CCCS52A2 function obligatory for endocycle onset. Proc. Natl. Acad. Sci. USA 105: 14721–14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Rabin Z., Li Z., Masson P.H., Day C.D. (2009). FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol. 149: 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.F., Perry P.J., Prafulla N.N., Schmidt W. (2010). Ubiquitin-specific protease 14 (UBP14) is involved in root responses to phosphate deficiency in Arabidopsis. Mol. Plant. 3: 212–223. [DOI] [PubMed] [Google Scholar]
- Li Y., Zheng L., Corke F., Smith C., Bevan M.W. (2008).Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22: 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang F., Zhang H., He H., Ma L., Deng X.W. (2008).Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J. 55: 844–856. [DOI] [PubMed] [Google Scholar]
- Marrocco K., Bergdoll M., Achard P., Criqui M.C., Genschik P. (2010). Selective proteolysis sets the tempo of the cell cycle. Curr. Opin. Plant Biol. 13: 631–639. [DOI] [PubMed] [Google Scholar]
- Meng X., Wang H., He Y., Liu Y., Walker J.C., Torii K.U., Zhang S. (2012). A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24: 4948–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., Bernards R. (2005). A genomic and functional inventory of deubiquitinating enzymes. Cell 123: 773–786. [DOI] [PubMed] [Google Scholar]
- Okushima Y., Shimizu K., Ishida T., Sugimoto K., Umeda M. (2014). Differential regulation of B2-type CDK accumulation in Arabidopsis roots. Plant Cell Rep. 33: 1033–1040. [DOI] [PubMed] [Google Scholar]
- Page A.M., Hieter P. (1999). The anaphase-promoting complex: new subunits and regulators. Annu. Rev. Biochem. 68: 583–609. [DOI] [PubMed] [Google Scholar]
- Peng Y., Chen L., Lu Y., Wu Y., Dumenil J., Zhu Z., Bevan M.W., Li Y. (2015). The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell 27: 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez J.M., Candela H., Micol J.L. (2009). Understanding synergy in genetic interactions. Trends Genet. 25: 368–376. [DOI] [PubMed] [Google Scholar]
- Riefler M., Novak O., Strnad M., Schmülling T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18: 40–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis.Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar V.V., Kapoor A., Zhang K., Zhu J., Zhou T., Hasegawa P.M., Bressan R.A., Zhu J.K. (2007). Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447: 735–738. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K., Roberts K. (2003). “Big it up”: endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6: 544–553. [DOI] [PubMed] [Google Scholar]
- Tang Z., Li B., Bharadwaj R., Zhu H., Ozkan E., Hakala K., Deisenhofer J., Yu H. (2001). APC2 Cullin protein and APC11 RING protein comprise the minimal ubiquitin ligase module of the anaphase-promoting complex. Mol. Biol. Cell 12: 3839–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I., McElver J.A., Liu Cm C.M., Yang L.J., Wu J.Q., Martinez A., Patton D.A., Meinke D.W. (2002). Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol. 128: 38–51. [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M., Baloban M., Da Ines O., Cultrone A., Lammens T., Boudolf V., Brown S.C., De Veylder L., Mergaert P., Kondorosi E. (2009). APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 106: 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vi S.L., Trost G., Lange P., Czesnick H., Rao N., Lieber D., Laux T., Gray W.M., Manley J.L., Groth D., Kappel C., Lenhard M. (2013). Target specificity among canonical nuclear poly(A) polymerases in plants modulates organ growth and pathogen response. Proc. Natl. Acad. Sci. USA 110: 13994–13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson K.D. (1997). Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 11: 1245–1256. [DOI] [PubMed] [Google Scholar]
- Xia T., Li N., Dumenil J., Li J., Kamenski A., Bevan M.W., Gao F.,Li Y. (2013). The ubiquitin receptor DA1 interacts with the E3 ubiquitin ligase DA2 to regulate seed and organ size in Arabidopsis.Plant Cell 25: 3347–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N., Doelling J.H., Falbel T.G., Durski A.M., Vierstra R.D. (2000). The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol. 124: 1828–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.