Structures of the Os-SCL7 and Os-SCL7/DNA complex reveal the GRAS domain fold and the DNA binding and dimerization mechanisms that are key to transcriptional regulation of GRAS proteins.
Abstract
GRAS proteins belong to a plant-specific protein family with many members and play essential roles in plant growth and development, functioning primarily in transcriptional regulation. Proteins in the family are minimally defined as containing the conserved GRAS domain. Here, we determined the structure of the GRAS domain of Os-SCL7 from rice (Oryza sativa) to 1.82 Å. The structure includes cap and core subdomains and elucidates the features of the conserved GRAS LRI, VHIID, LRII, PFYRE, and SAW motifs. The structure is a dimer, with a clear groove to accommodate double-stranded DNA. Docking a DNA segment into the groove to generate an Os-SCL7/DNA complex provides insight into the DNA binding mechanism of GRAS proteins. Furthermore, the in vitro DNA binding property of Os-SCL7 and model-defined recognition residues are assessed by electrophoretic mobility shift analysis and mutagenesis assays. These studies reveal the structure and preliminary DNA interaction mechanisms of GRAS proteins and open the door to in-depth investigation and understanding of the individual pathways in which they play important roles.
INTRODUCTION
GRAS proteins form a large plant-specific protein family named for the first three identified members, GIBBERELLIC ACID INSENSITIVE (GAI), REPRESSOR of GAI, and SCARECROW (SCR). The importance of this family has been made clear through a number of functional studies (Di Laurenzio et al., 1996; Peng et al., 1997; Silverstone et al., 1998), and a remarkable number of GRAS proteins have been identified in more than 294 embryophyta species (Finn et al., 2010; Sun et al., 2012). Most of these proteins play key roles in the transcriptional regulation and signaling transduction related to plant growth and development (Bolle, 2004; Achard et al., 2006). Examples include Arabidopsis thaliana DELLA in gibberellin (GA) signaling during regulation of plant growth (Peng et al., 1997; Sun and Gubler, 2004; Murase et al., 2008), Medicago truncatula NSP1-NSP2 in DNA promoter regulation for nodulation signaling (Kaló et al., 2005; Smit et al., 2005; Hirsch and Oldroyd, 2009), Arabidopsis SCR-SHR in regulation of radial patterning for root and shoot (Pysh et al., 1999; Helariutta et al., 2000; Nakajima et al., 2001), Lilium longiflorum SCL in auxin and stress-induced signaling (Morohashi et al., 2003; Sánchez et al., 2007), At-PAT1 in phytochrome light signal transduction and plant defense (Bolle et al., 2000; Torres-Galea et al., 2006), At-LAS in axillary meristem development (Schumacher et al., 1999; Greb et al., 2003), and Petunia hybrida HAM in shoot meristem maintenance (Stuurman et al., 2002). GRAS proteins are of great interest in plant science due to their important and diverse biological roles.
By phylogenetic analysis, the GRAS family can be divided into more than 10 subfamilies, of which SCL4/7 is one important subgroup (Sun et al., 2012). In Arabidopsis, SCL4 and SCL7 have similar expression levels under normal growth conditions; however, under stress conditions, SCL7 is upregulated while SCL4 is downregulated, indicating that SCL4/7 members could function as transcription factors in response to environmental stresses (Kilian et al., 2007). Populus euphratica SCL7, an ortholog of At-SCL7, is localized in the nucleus, and overexpression of Pe-SCL7 in transgenic Arabidopsis improved its salt and drought tolerance (Ma et al., 2010). A recent study demonstrated that Helianthus annuus GRAS, a GRAS-like protein in the same major clade with SCL4/7, could alter the gibberellin content and axillary meristem outgrowth, indicating SCL4/7 members also play a role in axillary meristem development (Fambrini et al., 2015).
Based on sequence similarity and domain structure analyses, GRAS proteins contain a highly divergent N-terminal domain and a characteristic C-terminal domain commonly known as the GRAS domain. The N-terminal domains have a highly variable sequence including intrinsically disordered regions (so-called unfoldome), which may facilitate molecular recognition with different interacting partners and contribute to divergent biological functions of the GRAS family (Uversky, 2010). The major characteristic domain of GRAS proteins is the C-terminal GRAS domain, which exhibits considerable sequence similarity between GRAS proteins.
The GRAS domain is typically composed of ∼380 amino acids and contains five highly conserved motifs. The two leucine heptad repeat motifs (LRI and LRII) directly flank the VHIID motif named after the most prominent residues (Supplemental Figure 1). It is suggested that the LR motifs and VHIID motif could be important for protein or DNA interactions. In most members of the GRAS protein family, several additional amino acid motifs are invariant, including the PFYRE motif designated after the conserved amino acids and the C-terminal SAW motif characterized by three sequential units: W (X)7 G, L-W, and SAW (W (X)10 W) (Pysh et al., 1999).
Though GRAS proteins have been studied for more than a decade, with their transcription factor activity being at the heart of “green revolution” (Peng et al., 1999), the mechanism of GRAS proteins is still unclear. A structural description of GRAS proteins is needed to clarify controversy around their functional mechanism(s) and decipher their mode of action in plant-specific physiology processes. The only structure of GRAS proteins determined to date is the GAI N-terminal DELLA domain (residues 11 to 113) in complex with GA3-bound GID1A (Murase et al., 2008). Structural characterization of the GRAS domain with highly conserved motifs has been unavailable. The elucidation of the molecular mechanisms of GRAS proteins has accordingly been hampered by the lack of structural information.
Here, we report the crystal structure of the GRAS domain of Os-SCL7, a GRAS protein from rice (Oryza sativa). The structure demonstrates the overall structure of the GRAS domain with a Rossmann fold core subdomain and an additional cap subdomain. This structure reveals the features of the distinct conserved LRI, VHIID, LRII, PFYRE, and SAW motifs to atomic resolution. In addition, the structure reveals the dimerization mechanism, which will be significant for many GRAS proteins functioning as a dimer. Furthermore, our computationally modeled structure of the Os-SCL7/DNA complex provides preliminary insight into the mechanisms underlying how GRAS proteins interact with DNA. To validate the protein-DNA interactions suggested by the model of Os-SCL7/DNA, the DNA binding properties of Os-SCL7 mutants was examined in vitro by electrophoretic mobility shift assay (EMSA). These structural and biochemical studies present the structure and DNA interaction mechanisms of GRAS proteins and will serve as a prototype for in-depth investigation and characterization of other GRAS family members.
RESULTS
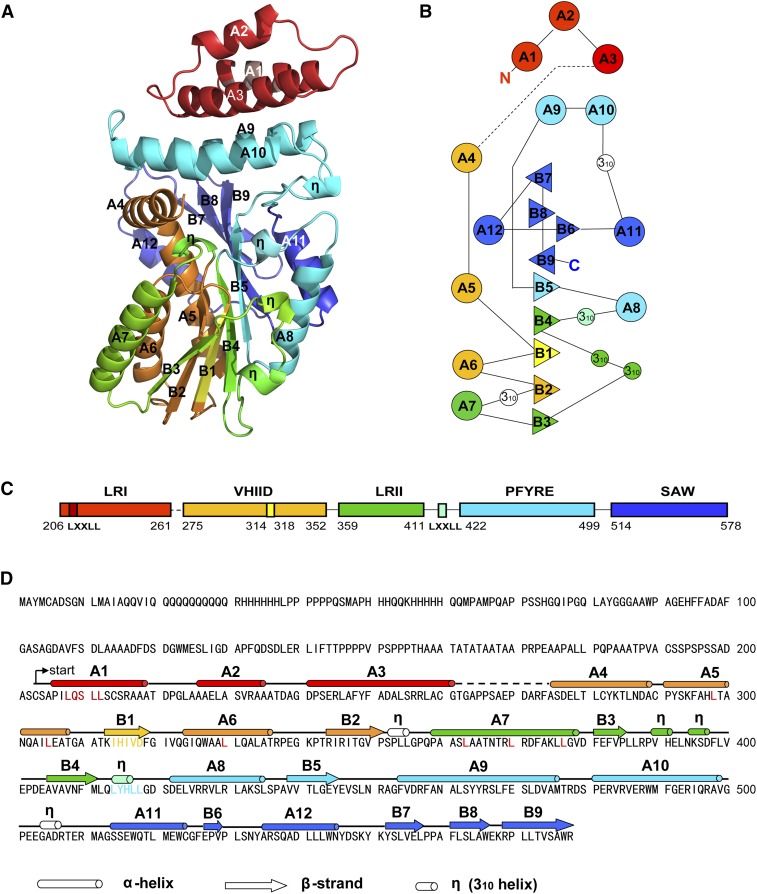
Overall Structure of the GRAS Domain Monomer of Os-SCL7
The sequence encoding the GRAS domain of Os-SCL7, residues 201 to 578, was cloned from rice, and the resulting protein was purified after heterologous expression in Escherichia coli and crystallized by vapor diffusion. The crystal structure was determined using selenomethionyl single-wavelength anomalous dispersion phasing to a resolution of 1.82 Å with Rwork of 18.09% and Rfree of 21.07% (Table 1). The monomer model contained residues 204 to 578, except loop residues 261 to 274 (Figure 1). According to previous phylogenetic and sequence analyses, the GRAS domain can be divided into five distinct conserved motifs: LRI, VHIID, LRII, PFYRE, and SAW (Supplemental Figure 1). Within the structure, the GRAS domain monomer contained an eight-stranded mixed β-sheet (B1-B9) with an accessory small β-strand B6 and 12 α-helices (A1-A12) as well as five 310 helices (η) (Figure 1D). The GRAS domain was composed of cap and core subdomains (Figure 1). The cap subdomain at the top was composed of a helical bundle formed by N-terminal α-helices A1, A2, and A3 from the LRI motif and a noncontiguous helical bundle insert A9, A10 from the PFYRE motif. The core subdomain was much larger, forming a α/β/α three-layer sandwiched Rossmann fold-like structure. The central β-sheet was formed by parallel β-strands B1-B5 and extended by antiparallel β-strands B9, B7, and B8. B6 was also a small accessory β-strand. Flanking each side of the central β-sheet were two helical layers formed by small 310 helices, α-helices A8, A11, and A5, A6, A7, and A12, respectively (Figure 1).
Table 1. Data Collection, Phasing, and Refinement Statistics for Os-SCL7 Structure.
| Data Collection | SeMet |
|---|---|
| Space group | C2221 |
| Cell parameters | |
| a, b, c (Å) | 89.19, 179.05, 59.70 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 1.822 |
| Rmerge (%) | 0.087 (0.413) |
| I/ơI | 56.97 (8.06) |
| Completeness (%) | 99.8 (100) |
| Redundancy | 10.8 (11.0) |
| Wilson B-factor (Å2) | 21.66 |
| Refinement | |
| Resolution (Å) | 1.822–33.181 |
| No. reflections | 42,785 (1,989) |
| Rwork/Rfree (%) | 18.09/21.07 |
| No. atoms | |
| Protein | 2,817 |
| Water | 362 |
| R.m.s.d. bonds (Å) | 0.019 |
| R.m.s.d. angles (°) | 1.729 |
| Ramachandran plot | |
| Favored (%) | 97.77 |
| Allowed (%) | 2.23 |
| Outliers (%) | 0.00 |
| Rotamer outliers (%) | 0.00 |
Values in parentheses refer to the highest resolution shell. R.m.s.d., root mean square deviation.
Figure 1.
Overall Structure, Sequence, and Secondary Structure of GRAS Domain of Os-SCL7
The five distinct conserved motifs are colored individually: LRI in red, VHIID in orange, LRII in green, PFYRE in cyan, and SAW in blue.
(A) Ribbon diagram of the GRAS domain structure; α-helices and β-strands are labeled with A and B, respectively, and 310 helices are labeled with η.
(B) Topological diagram of the secondary structure elements. Triangles represent β-strands, circles represent α-helices and smaller circles represent 310 helices. Parallel and antiparallel β-strands are represented with triangles facing different directions.
(C) Domain structure of GRAS domain with conserved motifs and sequence range labeled.
(D) The secondary structures are labeled and the different colors correspond to the five distinct motifs.
The orientation of the top cap subdomain helical bundle was approximately perpendicular to the bottom α/β/α sandwiched three-layer core. The two domains were linked by a long loop and helix A4. The long loop preceding A4 was missing in the electron density map, implying the dynamic property of the cavity between the two subdomains, whereas A4 formed a weak helix-helix interaction with A10 of the cap subdomain. The upper layer might play important roles in contacting substrate and function as a cap to shield active sites of the lower layer core subdomain from bulk solvent.
Conserved Motifs
Previous phylogenetic and sequence analysis has highlighted the importance of five distinct conserved motifs within the GRAS domain: LRI, VHIID, LRII, PFYRE, and SAW. With our high-resolution structure, the particular features of each motif could be uncovered.
(1) LRI Motif
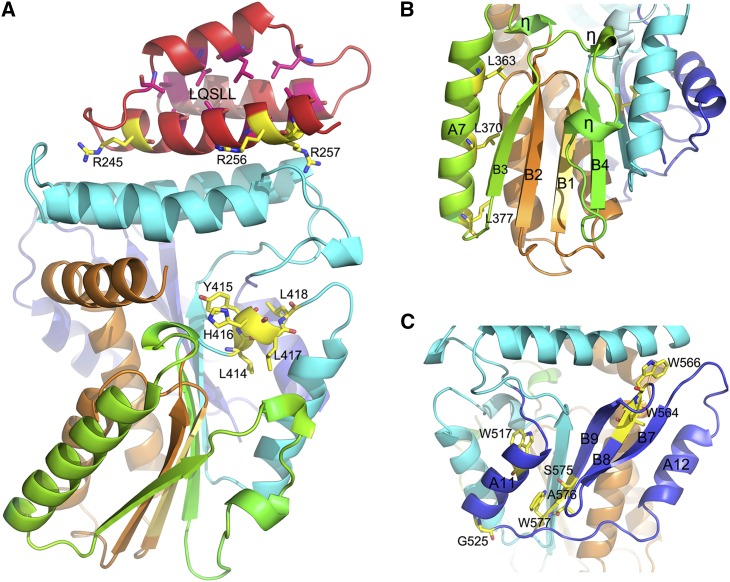
According to a body of phylogenetic analyses, the LRI domain has two units: a comparatively conserved leucine repeat unit and a putative nuclear localization signal (NLS) unit. In the Os-SCL7 structure, this motif was composed of α-helices A1, A2, and A3, forming an antiparallel three-helix bundle with a hydrophobic core in the center. Among these hydrophobic residues, L224, L229 at helix A2 and L258, L254 at helix A3 formed leucine zipper pairs, respectively. In Os-SCL7, the NLS signal with a canonical bipartite NLS sequence, featuring the basic amino acids R245 and R256/R257, was exposed on the protein surface of helix A3 (Figure 2A).
Figure 2.
Structure of Conservative Motifs of Os-SCL7.
Ribbon diagram of the GRAS domain of Os-SCL7 structure; α-helices and β-strands are labeled with A and B, respectively, and 310 helices are labeled with η. The colors correspond to the five conservative motifs. Characteristic residues are shown in ball-and-stick representation and labeled.
(A) Important residues for the NLS of the LRI motif and LXXLL motifs are labeled.
(B) Structure of LRII and VHIID motifs with major component residues labeled.
(C) Structure of SAW motif with key residues labeled.
(2) VHIID Motif
The conserved VHIID motif is composed by the sequence IHIVD in Os-SCL7. In the structure, this sequence alone formed an individual β-strand B1 as the core of the central β-sheet, flanked by B2, B3 at one side and with B4, B5 at the other side (Figure 2B). The location implies its important role in maintaining the structural integrity and stabilizing the conformation for the whole GRAS domain. Within the motif, connected with IHIVD β-strand, were α-helices A4, A5 and A6, B2 at both sides. A4 was almost perpendicular to A5 and functioned as a turn helix connecting the cap and core subdomain.
(3) LRII Motif
In the Os-SCL7 structure, the LRII domain contained helix A7, a stretch of two 310 helices and strands B3 and B4. B3 and B4 formed a β-sheet with B1 and B2 of the VHIID motif at two adjacent sides, packing the LRII motif tightly around the VHIID motif like a clip (Figure 2B). The classical leucine heptad repeat of L363, L370, and L377 positioned on helix A7 (Figure 2B) may couple the interaction of Os-SCL7 with its protein partners.
(4) PFYRE Motif
The PFYRE motif contains three parts sequentially: P, FY, and RE. In the Os-SCL7 structure, these residues were not well conserved. Within the structure, the PFYRE motif was composed of A8, B5, A9, and A10. A8 and B5 mainly contributed to the α/β/α sandwiched Rossmann fold of core subdomain, whereas A9 and A10 were separated from the core subdomain by a long linker and packed as long helical bundle insert above the core subdomain, forming helix-helix interactions with the A1-A3 helical bundle (Figure 1A).
(5) SAW Domain
The SAW motif is defined by the sequence signatures of WX7G, LXW, and SAW and is well conserved in Os-SCL7. In the structure, the SAW motif formed a well-folded C-terminal domain, composed of A11, A12, B7, B8, and B9 (Figure 2C). Helix A12 formed an antiparallel helix-helix interaction with A5, and thus together with A6 and A7 formed an extended α-helical layer of the α/β/α sandwiched core subdomain. Strands B7, B8, and B9 formed an antiparallel β-sheet (Figure 1A).
In total, the core subdomain was an extended Rossmann fold α/β/α sandwiched conformation composed by the tightly packed VHIID, LHII, PFYRE, and SAW motifs. Strands B1-B5 formed the central parallel β-sheet, extended by antiparallel β-sheet interaction with B9, B8, and B7. Helices A8, A11, and two consecutive 310 helices formed one outer layer of the α/β/α sandwiched core subdomain. Parallel helices A5, A6, and A7 formed the other layer, of which two short leucine zippers were positioned at the surface, formed by L298, L305, and L330 of A5 and A6 and by L331, L370, and L377 of A6 and A7 (Supplemental Figure 2). This arrangement could provide a key protein interaction module to bind protein partners and could be involved in biological function such as transcriptional activity regulation.
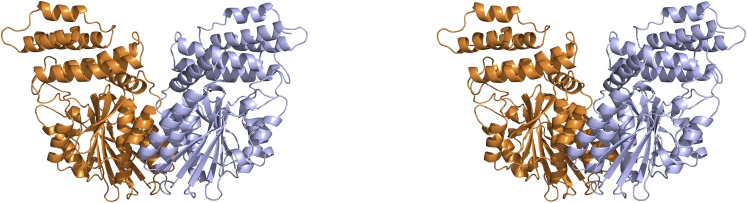
Dimer Formation and Putative DNA Binding Groove
During protein preparation, Os-SCL7 was a dimer in solution, as indicated by size-exclusion chromatography (Supplemental Figure 3). In the crystal structure, the Os-SCL7 side-by-side dimer was formed by the crystallographic two-fold symmetry axis (Figure 3). This dimer interface had a buried surface area of 1346.3 Å2, accounting for 8.47% of total monomer surface area. The dimer was primarily formed by interaction between A12 with A7′ and A6′ through helix-helix hydrophobic interaction, with residues involved including W544, L541, F373, L377, and Y534. Above the dimer interface was a large groove formed by helix A4, the loop between helix A9 and A10 at the sides, and the loop between A12 and B7 at the bottom. This groove was ∼18 Å in width and 22 Å in depth. The dimer had two highly positively charged patches, one contributed by each monomer, lining the surface of the groove (Figure 4C). This strongly implicates the groove as a site suitable for accommodating DNA.
Figure 3.
Os-SCL7 Dimer Structure Shown as Ribbon Diagram in Stereo View.
The two monomers are colored in bright orange and light blue. The helices A12, A7′, and A6′ central to dimer formation are labeled.
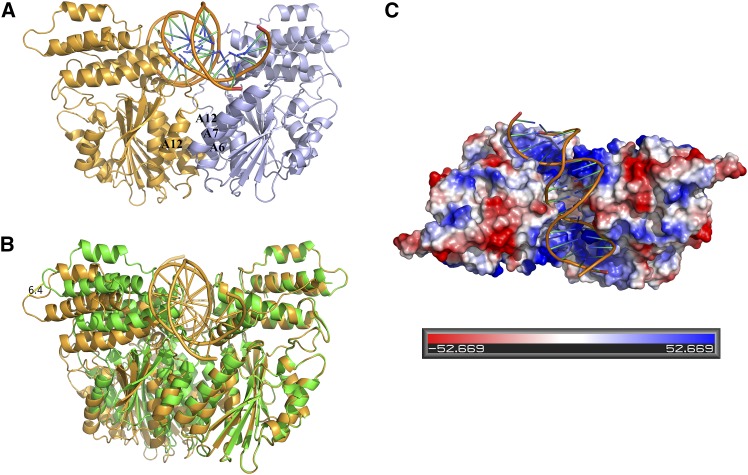
Figure 4.
Structure Model and Structural Comparison of the Os-SCL7/DNA Complex.
Ribbon diagram of the Os-SCL7/DNA structure with DNA represented in cartoon.
(A) The modeled Os-SCL7/DNA structure, with two monomers colored in bright orange and light blue. The helices A12, A7′, and A6′ related to dimer formation are labeled.
(B) Structural comparison of the modeled complex structure with the native Os-SCL7 structure. The native structure is colored in green, and the complex model is colored in orange. The distance enlarged is ∼6.4 Å as marked.
(C) Top view of surface potential drawing of the Os-SCL7/DNA complex model. Positively and negatively charged regions are colored in blue and red, respectively, with scale shown at bottom.
Modeled Os-SCL7/DNA Complex Structure
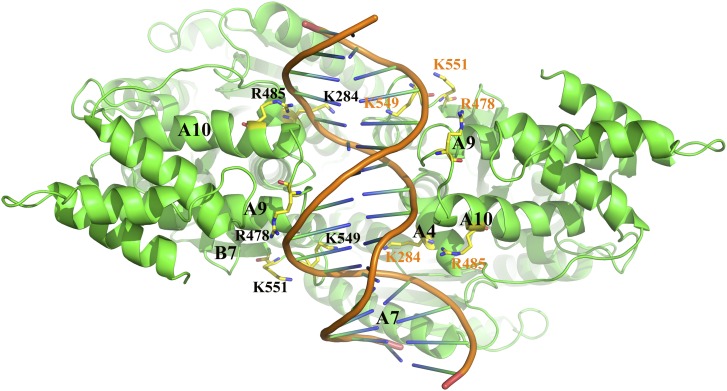
By computational simulation, a double-stranded DNA segment of 16 bp (AAAACTGAAAGGGAGA) was docked into the groove of the Os-SCL7 dimer (Figure 4A). The minor groove of DNA was deeply embedded into the groove formed by Os-SCL7 dimerization and made tight interactions with the two positively charged surface patches (Figure 4C). The DNA association was mainly contributed by helix A4, loop between A9 and A10, and loop between A12 and B7, with basic amino acids K284, R478, R485, K549, and K551 at the protein-DNA interface (Figure 5). By comparison with the dimer structure of Os-SCL7, the Os-SCL7/DNA complex structure had a conformational change upon DNA binding, with the groove expanded around 6.4 Å in width (Figure 4B).
Figure 5.
Residues Critical for Binding of Os-SCL7 to DNA.
Top view of the Os-SCL7/DNA complex, with protein and DNA represented as a ribbon and a cartoon, respectively. Residues involved in DNA binding are show in ball-and-stick representation and labeled.
In Vitro Analysis of DNA Binding Properties of Os-SCL7 and Mutants
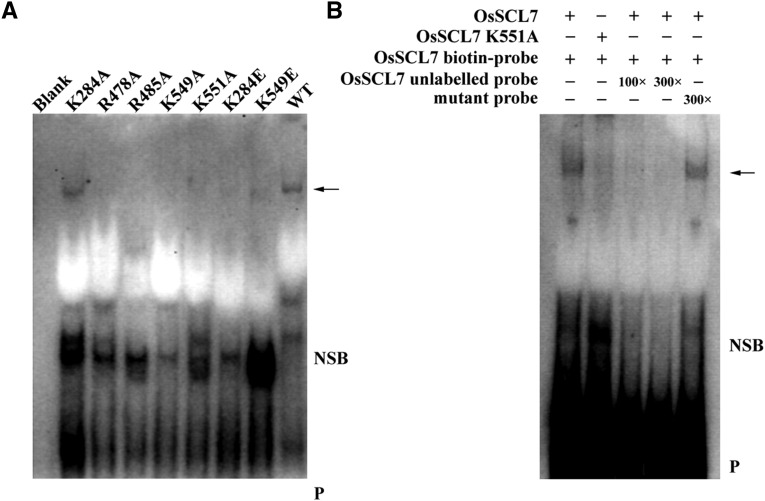
To test if the GRAS protein could directly associate in vitro with DNA, the purified GRAS domain of Os-SCL7 and biotin-labeled oligonucleotide were generated for EMSAs. The oligonucleotide sequence was the same as was used for in silico docking. A clear band shift was observed when the oligonucleotide DNA duplex was incubated with Os-SCL7, but not with the control in the EMSA competition analysis (Figure 6B), indicating a specific interaction of Os-SCL7 and our designed oligonucleotide. Therefore, our work provides direct in vitro evidence for Os-SCL7 binding to DNA and implies that Os-SCL7 may function as a transcription factor with direct DNA binding capability.
Figure 6.
EMSA Analyses of Os-SCL7 and Model-Defined Oligonucleotide.
(A) In EMSA analyses with same amounts of probe, a band shift (arrow) can be observed for Os-SCL7 and mutant K284A, but not for other mutants. Other Os-SCL7 mutants caused a dramatic reduction in binding.
(B) EMSA competition analysis. Biotin-labeled oligonucleotide probe incubated with Os-SCL7, or mutant K551A as a negative control, in the absence or presence of 100- and 300-fold molar concentration of the unlabeled probe and competitive mutant oligonucleotide probe, respectively. Arrow, DNA binding protein; NSB, nonspecific binding; P, free biotin-labeled probe.
To further assess the reliability of the DNA binding site defined by the Os-SCL7/DNA complex model, the above-mentioned binding site residues R478, R485, K551, K549, and K284 were mutated to either alanine or glutamate. EMSA analyses showed no band shift for the R478A, R485A, K551A, K549A/E, and K284E mutants, indicating diminished DNA binding capability (Figure 6A). The result is consistent with our Os-SCL7/DNA complex model.
DISCUSSION
GRAS proteins represent an essential, yet varied, set of regulatory molecules in plants whose known functional roles range from development of meristem to hormone and light signal response. However, many GRAS proteins remain functionally uncharacterized. Through intense sequence profile searches, phylogenetic analyses, and structural comparisons, it was reported that the GRAS domain and metazoan STAT shared a similar domain organization, with a SH2-like motif and tyrosine phosphorylation site in GRAS proteins suggested but not yet demonstrated (Peng et al., 1999; Richards et al., 2000). However, there is also an opposing opinion that the structure of GRAS domain shares similarity with the Rossmann fold methyltransferase superfamily involved with binding or modifying GA derivatives in GA signaling, though the plant GRAS proteins appear to lack methyltransferase activity and the residues critical for specific substrate interaction (Zhang et al., 2012). Our crystal structure of the GRAS domain of SCL7 in rice provides a high-resolution description of a GRAS domain and demonstrates that it contains a core subdomain with the Rossmann fold α/β/α sandwiched arrangement similar to that of the methyltransferase superfamily. Different from the leucine zipper-containing bZIP transcription factors, both GRAS and STAT have a discrete domain for dimerization to provide a groove for DNA binding; however, with distinct conformations, they may use different mechanisms for dimerization and DNA binding. Therefore, the structure provides a template for further structural comparison to explore its action mechanisms and untangle the controversy.
The Os-SCL7 GRAS domain structure further reveals detailed features for each of the conserved motifs identified by bioinformatic studies and clarifies ambiguities on the domain assignment in previous reports. Bioinformatic studies have indicated that an additional LXXLL sequence is conserved in some GRAS proteins. It has been proposed that this LXXLL motif, with appropriate secondary structure, is a signature sequence mediating the binding to nuclear receptors for transcriptional coactivators (Heery et al., 1997). Thus, it could be a defining feature of a family of nuclear proteins, though plants may have distinct mechanisms for nuclear receptor recognition. In Os-SCL7, two LXXLL motifs, L208QSLL212 and L414YHLL418, were easily identified. L208QSLL212, a comparatively conserved LXXLL motif among the SCL7 protein family, is located on helix A1 at the N terminus and exposed on the protein surface (Figure 2A). L414YHLL418, which is highly conserved among the SCL7 protein family members from different species, forms an independent 310 helix exposed at the surface of a groove positioned between the N-terminal layer and the core subdomain. In addition, it is located at the loop between B4 and A8, linking the LRII and PFYRE motifs (Figure 2A). This location grants the L414YHLL418 motif particular flexibility and an ideal site for substrate binding. However, since mammalian nuclear receptor homologs are not known to exist in plants and the precise significance of this motif in plants awaits further investigation.
Moreover, GRAS proteins have been reported to form homodimers and heterodimers with other GRAS family members and this is likely important for their transcriptional regulation function (Itoh et al., 2002; Cui et al., 2007; Hirsch et al., 2009; Heo et al., 2011; Zhang et al., 2011; Gobbato et al., 2012; Murakami et al., 2013; Xue et al., 2015). Here, our structure of Os-SCL7 was determined in a dimer with helices A12, A7′, and A6′ at the dimer interface. This information should provide insight into the interaction mechanism for dimer formation of many GRAS proteins.
As a large family of more than 10 subfamilies with diverse functions, GRAS proteins may use different mechanisms to regulate downstream gene expression. DELLA proteins lack a DNA binding domain; however, they function as transcription activators by interacting with INDETERMINATE Domain (IDD) proteins (Yoshida et al., 2014). In addition to interacting with IDDs, At-SCL3 can directly bind with its own promoter to regulate the expression in planta (Zhang et al., 2011). Mt-NSP1 can bind to the AATTT motif on the promoters, but Mt-NSP2 may have to associate with Mt-NSP1 for transcriptional regulation (Hirsch et al., 2009). At-SHR and At-SCR have been reported to interact directly with some common promoters (Cui et al., 2007). Here, using in silico DNA docking, coupled with in vitro binding and mutagenesis assays, we further confirm the interaction of Os-SCL7 with DNA and pinpoint critical residues. Our studies provide direct evidence for Os-SCL7 functioning as a transcription factor via direct DNA binding. Furthermore, this 16-mer oligonucleotide can be found at several loci of the rice genome, suggesting downstream gene products regulated by Os-SCL7.
In summary, the GRAS protein family includes a remarkable number of important plant-specific proteins. Our studies define the three-dimensional crystal structure of the GRAS domain of Os-SCL7, providing a high-resolution atomic architecture of the GRAS domain, reveal its dimerization mechanism, and characterize its DNA binding properties. These results provide a foundation for elucidating the mechanisms and biochemical function of GRAS proteins. In turn, this will lead to a better understanding of the individual pathways in which they play important roles and how the proteins are adapted to carry out plant-specific processes.
METHODS
Protein Expression, Purification, and Crystallization
The sequence encoding the GRAS domain of GRAS protein SCL7 from rice (Oryza sativa; residues 201 to 578) was cloned into the modified pET32a vector (Novagen) (Supplemental Table 1) and overexpressed as selenomethionyl protein in the Escherichia coli strain BL21 (DE3) using methionine pathway inhibition at 289 K. Bacterial cells were lysed by ultrasonication on ice in a buffer containing 50 mM Tris-HCl, pH8.0, 500 mM NaCl, 0.2 mM EDTA, 20 mM imidazole, 2 mM β-mercaptoethanol, and 5% glycerol. Soluble N-terminally decahistidine-tagged Os-SCL7 was bound to nickel-sepharose affinity resin. The N-terminal histidine and fused thioredoxin tag were removed by cleavage with TEV protease. Protease and uncleaved protein were removed by a second binding to nickel-sepharose affinity resin. The eluted protein was further purified by size-exclusion chromatography in 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 5% glycerol. Purified Os-SCL7 was concentrated to 8 mg/mL without buffer exchange. SDS-PAGE of purified protein showed one major band at an approximate molecular mass of 41 kD, indicating pure full-length protein. Crystals were obtained by sitting drop vapor diffusion at 285K with 1 μL protein mixed with 1 μL mother liquid solution containing 10% (v/v) isopropanol and 20% (v/v) PEG4000 in 0.1 M HEPES sodium (pH 7.5) buffer. Crystals were flash-frozen in liquid nitrogen with an addition of 30% glycerol in the crystallization mother liquid as cryoprotectant.
Data Collection, Structure Determination, and Refinement
The selenomethionyl single-wavelength anomalous dispersion data set for Os-SCL7 was collected at a wavelength of 0.9793 Å at 100K on the BL17U1 beamline of the Shanghai Synchrotron Radiation Facility to a diffraction limit of 1.82 Å. Diffraction images and the anomalous data set were processed and scaled with HKL2000 (Otwinowski and Minor, 1997). Single-wavelength anomalous dispersion data processing statistics are summarized in Table 1. The locations of five selenium atoms were determined and an initial model built using the AutoSolve program of the Phenix suite (Adams et al., 2002). The model was manually rebuilt with Coot (Emsley and Cowtan, 2004) and further refined in Phenix (partial electron density map shown in Supplemental Figure 4). The final model contains residues 204 to 587, except loop residues 261 to 274, with refinement statistics summarized in Table 1. The Ramachandran statistics were calculated with Procheck (Laskowski et al., 1993). Structural superimpositions were complemented by manual sequence alignment and CCP4 LSQ superposition (Winn et al., 2011). Figures were produced with Pymol (www.pymol.org). The solvent accessible surface area and buried surface area was calculated using the PISA server.
DNA Segment Docking
Autodock 4.0 with AutoDock Tool 1.5.6 was used to carry out a rigid screening for DNA binding using the crystal structure of Os-SCL7 dimer as receptor (Morris et al., 1998). A protein dimer-centered grip map of 45×64×60 grid points and 0.950 spacing was generated using the Autogrid program. An in-house DNA segment library built from the PDB was used as DNA ligands (Berman et al., 2000). In detail, all double-stranded DNA complexed protein structures from PDB were downloaded with the query parameters of base chain length and the proteins with <30% sequence identity; thus, representative DNA segments from DNA/protein structures were extracted for the in-house DNA ligand library. All DNA segments were set as rigid molecules without any rotational bond. The docking screening was performed using the Lamarchian genetic algorithm and the default parameters of the genetic algorithm were applied, except using a population size of 100 and a gene mutation of 0.2. The embedded DNA segment with the lowest energy was used as the candidate ligand. The docked protein-DNA complex was further optimized using Tinker 4.2 (http://dasher.wustl.edu/tinker) with the amber 99SB force field (Hornak et al., 2006) until the root mean square energy gradient was below 1.0 kcal mol−1 Å−1. Finally, the optimized protein-DNA complex with the minimum derivation between the x-ray protein structure and the protein structure of the protein-DNA complex was obtained as our complex model.
EMSA Analysis and Mutagenesis Assay
Point mutations (K284A/E, R478A, R485A, K549A/E, and K551A) of Os-SCL7 GRAS domain were engineered using the QuikChange site-directed mutagenesis kit (Stratagene) (Supplemental Table 1). The proteins were purified, with the quantity of purified protein estimated by BCA protein quantification kit (Viagene Biotech). The oligonucleotide probes 5′-tgaAAAACTGAAAGGGAGActggta-3′ and core complementary 5′-taccagTCTCCCTTTCAGTTTTtca-3′ were synthesized and labeled with biotin (Sangon Biotech) and then annealed for duplex self-assembly by slow cooling in a buffer of 50 mM NaCl, 10 mM Tris-HCl, pH 7.5, and 0.1 mM EDTA. EMSAs were performed using the Light Shift Chemiluminescent EMSA kit (Thermo Scientific) according to the manufacturer’s instructions. The binding reactions were performed using 6 × 10−2 nM (2.5 μg) for each protein sample incubated with 8 nM probe in binding buffer (10 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM DTT, and 5% glycerol) with protein-free sample as blank control. After 20 min of incubation at room temperature, the reactions were resolved by 6% native polyacrylamide gels with 0.5× TBE buffer. For the competition experiments, 100- or 300-fold molar excess of unlabeled probe or 300-fold molar excess of mutant probes 5′-tgaAGGCCTGTGTGGGAGActggta-3′ oligonucleotide duplex was used for the reactions and incubated for 20 min before the labeled probes were added.
Accession Numbers
Sequence date from this article can be found in the Rice Genome Annotation database under accession number LOC_Os03g51330 (Os-SCL7). The atomic coordinates and the structure factors of the Os-SCL7 structure can be found in the RCSB Protein Data Bank as entry 5HYZ.
Supplemental Data
Supplemental Figure 1. Sequence alignment of Os-SCL7 with other GRAS family members.
Supplemental Figure 2. Structure of leucine heptad repeat region of Os-SCL7.
Supplemental Figure 3. Size-exclusive chromatography for Os-SCL7 protein.
Supplemental Figure 4. The 2Fo-Fc electron-density map by Pymol of DNA binding relevant site of Os-SCL7 protein.
Supplementary Material
Acknowledgments
We thank beamline (BL17U1) staff at Shanghai Synchrotron Radiation Facility for assistance in data collection and D. Sauer for manuscript preparation. This work was financially supported by the National Nature Science Foundation of China (31270790, 31470741, and 31500218) and the National Thousand Talents Program of China.
AUTHOR CONTRIBUTIONS
S.L. and Y.Z. performed sample preparation and most of the experiments. Y.W., X.W., and L.S. perform x-ray data collection and structure determination. Z.Z. and Q.L. performed DNA docking. Y.W., S.L., and Y.Z. designed the research, analyzed data, and prepared the manuscript.
Glossary
- EMSA
electrophoretic mobility shift assay
- NLS
nuclear localization signal
- GA
gibberellin
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94. [DOI] [PubMed] [Google Scholar]
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. (2002). PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58: 1948–1954. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. (2000). The Protein Data Bank. Nucleic Acids Res. 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C. (2004). The role of GRAS proteins in plant signal transduction and development. Planta 218: 683–692. [DOI] [PubMed] [Google Scholar]
- Bolle C., Koncz C., Chua N.H. (2000). PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 14: 1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Cui H., Levesque M.P., Vernoux T., Jung J.W., Paquette A.J., Gallagher K.L., Wang J.Y., Blilou I., Scheres B., Benfey P.N. (2007). An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller J., Malamy J.E., Pysh L., Helariutta Y., Freshour G., Hahn M.G., Feldmann K.A., Benfey P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423–433. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Fambrini M., Mariotti L., Parlanti S., Salvini M., Pugliesi C. (2015). A GRAS-like gene of sunflower (Helianthus annuus L.) alters the gibberellin content and axillary meristem outgrowth in transgenic Arabidopsis plants. Plant Biol (Stuttg.) 17: 1123–1134. [DOI] [PubMed] [Google Scholar]
- Finn R.D., et al. (2010). The Pfam protein families database. Nucleic Acids Res. 38: D211–D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbato E., et al. (2012). A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 22: 2236–2241. [DOI] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schafer E., Muller D., Herrero R., Schmitz G., Theres K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17: 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery D.M., Kalkhoven E., Hoare S., Parker M.G. (1997). A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387: 733–736. [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567. [DOI] [PubMed] [Google Scholar]
- Heo J.O., Chang K.S., Kim I.A., Lee M.H., Lee S.A., Song S.K., Lee M.M., Lim J. (2011). Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 108: 2166–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Muñoz A., Heckmann A.B., Downie J.A., Oldroyd G.E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Oldroyd G.E. (2009). GRAS-domain transcription factors that regulate plant development. Plant Signal. Behav. 4: 698–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. (2006). Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65: 712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H., Ueguchi-Tanaka M., Sato Y., Ashikari M., Matsuoka M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14: 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789. [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Macarthur M.W., Moss D.S., Thornton J.M. (1993). Procheck - a Program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291. [Google Scholar]
- Ma H.S., Liang D., Shuai P., Xia X.L., Yin W.L. (2010). The salt- and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 61: 4011–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K., Minami M., Takase H., Hotta Y., Hiratsuka K. (2003). Isolation and characterization of a novel GRAS gene that regulates meiosis-associated gene expression. J. Biol. Chem. 278: 20865–20873. [DOI] [PubMed] [Google Scholar]
- Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. (1998). Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19: 1639–1662. [Google Scholar]
- Murakami Y., Imaizumi-Anraku H., Kouchi H., Kawaguchi M., Kawasaki S. (2013). The transcription activation and homodimerization of Lotus japonicus Nod factor Signaling Pathway2 protein. Plant Signal. Behav. 8: e26457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T.P., Hakoshima T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11: 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261. [DOI] [PubMed] [Google Scholar]
- Pysh L.D., Wysocka-Diller J.W., Camilleri C., Bouchez D., Benfey P.N. (1999). The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18: 111–119. [DOI] [PubMed] [Google Scholar]
- Richards D.E., Peng J., Harberd N.P. (2000). Plant GRAS and metazoan STATs: one family? BioEssays 22: 573–577. [DOI] [PubMed] [Google Scholar]
- Sánchez C., Vielba J.M., Ferro E., Covelo G., Solé A., Abarca D., de Mier B.S., Díaz-Sala C. (2007). Two SCARECROW-LIKE genes are induced in response to exogenous auxin in rooting-competent cuttings of distantly related forest species. Tree Physiol. 27: 1459–1470. [DOI] [PubMed] [Google Scholar]
- Schumacher K., Schmitt T., Rossberg M., Schmitz G., Theres K. (1999). The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 96: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Ciampaglio C.N., Sun T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791. [DOI] [PubMed] [Google Scholar]
- Stuurman J., Jäggi F., Kuhlemeier C. (2002). Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 16: 2213–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T.P., Gubler F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55: 197–223. [DOI] [PubMed] [Google Scholar]
- Sun X., Jones W.T., Rikkerink E.H. (2012). GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling. Biochem. J. 442: 1–12. [DOI] [PubMed] [Google Scholar]
- Torres-Galea P., Huang L.F., Chua N.H., Bolle C. (2006). The GRAS protein SCL13 is a positive regulator of phytochrome-dependent red light signaling, but can also modulate phytochrome A responses. Mol. Genet. Genomics 276: 13–30. [DOI] [PubMed] [Google Scholar]
- Uversky V.N. (2010). The mysterious unfoldome: structureless, underappreciated, yet vital part of any given proteome. J. Biomed. Biotechnol. 2010: 568068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Cui H., Buer B., Vijayakumar V., Delaux P.M., Junkermann S., Bucher M. (2015). Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiol. 167: 854–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., et al. (2014). DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc. Natl. Acad. Sci. USA 111: 7861–7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Iyer L.M., Aravind L. (2012). Bacterial GRAS domain proteins throw new light on gibberellic acid response mechanisms. Bioinformatics 28: 2407–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.L., Ogawa M., Fleet C.M., Zentella R., Hu J., Heo J.O., Lim J., Kamiya Y., Yamaguchi S., Sun T.P. (2011). Scarecrow-like 3 promotes gibberellin signaling by antagonizing master growth repressor DELLA in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 2160–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.