Abstract
Techniques to detect and verify interactions between proteins in vivo have become invaluable tools in functional genomic research. While many of the initially developed interaction assays (e.g., yeast two-hybrid system and split-ubiquitin assay) usually are conducted in heterologous systems, assays relying on bimolecular fluorescence complementation (BiFC; also referred to as split-YFP assays) are applicable to the analysis of protein-protein interactions in most native systems, including plant cells. Like all protein-protein interaction assays, BiFC can produce false positive and false negative results. The purpose of this commentary is to (1) highlight shortcomings of and potential pitfalls in BiFC assays, (2) provide guidelines for avoiding artifactual interactions, and (3) suggest suitable approaches to scrutinize potential interactions and validate them by independent methods.
The identification of molecular interaction partners can provide valuable information about protein function and subcellular protein localization as well as the composition and three-dimensional architecture of protein complexes (Lalonde et al., 2008). Often, the identification of interacting proteins represents the only promising entry point into the characterization of a protein of unknown function. It is therefore unsurprising that, in the postgenomic era, methods to detect protein-protein interactions have dramatically gained importance and are increasingly being used.
Since genetic methods (e.g., two-hybrid systems, protein-fragment complementation assays; Remy and Michnick, 2015) facilitate the detection of protein interactions in vivo and do not require special reagents (such as specific antibodies for coimmunoprecipitation or affinity purification), they have quickly become popular tools to (1) screen for novel interaction partners of a given protein, (2) verify suspected interactions between proteins, and (3) characterize structural and/or sequence motifs involved in known protein interactions. However, although these methods are seemingly straightforward and potentially powerful, not all interactions detected with them are physiologically relevant. Likewise, the absence of a detectable interaction also does not necessarily mean that two proteins do not engage in intermolecular interactions under native conditions. The reasons for false positive or false negative interactions can be manifold and are often not sufficiently appreciated when researchers design and interpret protein-protein interaction experiments.
Due to the simple protocols involved and the possibility of performing them directly in plant cells, bimolecular fluorescence complementation (BiFC) assays have become increasingly popular among plant biologists to study protein-protein interactions. Here, we briefly review key aspects of BiFC, highlight potential sources of artifacts, and suggest a number of relatively simple measures to minimize the risk of identifying artifactual interactions. We hope that this commentary will increase awareness of potential pitfalls and will guide researchers in plant biology to accepted community standards for BiFC experiments and their interpretation.
PRINCIPLES OF BiFC AND POTENTIAL SOURCES OF ARTIFACTS
BiFC is based on fluorescence complementation (FC) by reconstitution of a functional fluorescent protein (FP) upon coexpression of N- and C-terminal fragments of this protein. To this end, an FP is separated into nonfluorescent N-terminal and C-terminal fragments that are translationally fused with the two proteins of interest (i.e., a pair of potentially interacting proteins). Upon interaction of the fusion proteins in living cells, the N- and C-terminal FP fragments are brought into close proximity resulting in reassembly of a functional fluorophore (Figure 1A; Hu et al., 2002; Walter et al., 2004; Bracha-Drori et al., 2004). Consequently, BiFC analyses not only allow the detection of protein-protein interactions but also provide information about the subcellular localization of the observed protein complex. It should be kept in mind that the requirement for fluorophore reconstitution is spatial proximity of the two proteins of interest, not necessarily direct interaction between them. If, for example, the two proteins of interest are part of a protein complex but do not directly interact with each other, fluorescence complementation can occur simply because the two FP fragments are sufficiently close to each other (Kerppola, 2006). Currently, reliable information is lacking on the distance between interacting proteins that is required or optimal for fluorophore reconstitution to occur. All commonly used BiFC vectors encode linker sequences (of varying lengths, but typically at least five amino acids long) between the FP fragment and the gene of interest, which may be important to provide sufficient structural flexibility of the fusion proteins to facilitate FP fragment reconstitution after interaction between the proteins of interest has occurred.
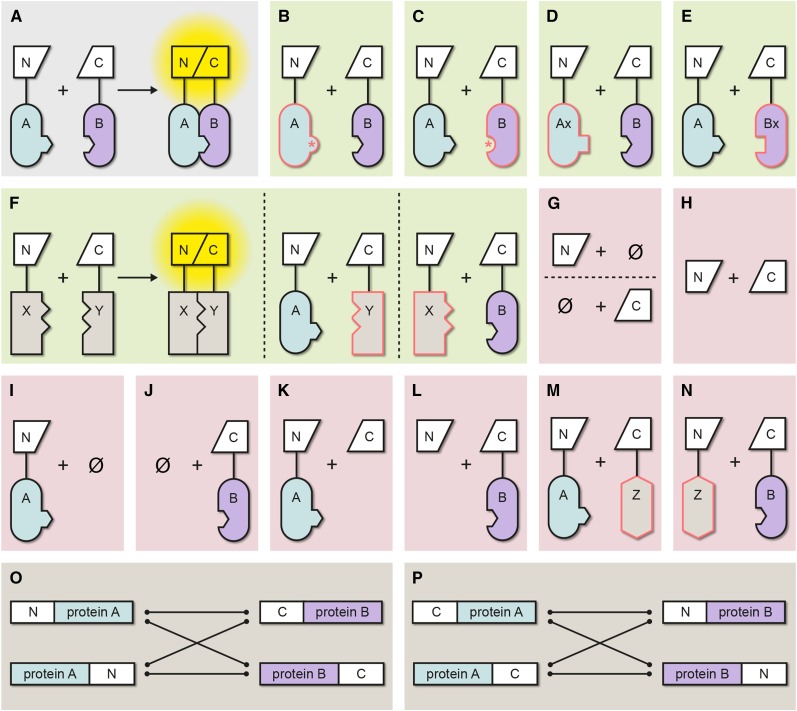
Figure 1.
Summary of Possible Negative Controls in BiFC Experiments.
(A) Interaction of proteins A and B mediates efficient fluorescence complementation and reconstitution of the FP.
(B) to (F) Appropriate negative controls (green background).
(B) Interaction domain mutated in A, thereby abolishing interaction with B.
(C) Interaction domain mutated in B, thereby abolishing interaction with A. Verification that the mutated protein is similarly stable as the wild-type form is additionally required.
(D) Ax is closely related to A (e.g., a member of the same protein family) but does no interact with B.
(E) Bx is closely related to B but does not interact with A.
(F) If none of the controls in (B) to (E) is possible, an unrelated protein, localized in the same subcellular compartment as the proteins of interest, can be used as the last resort. In this case, it is necessary also to provide evidence for this unrelated protein being part of an established interaction (X and Y) that can be reproduced by BiFC.
(G) to (N) Inappropriate negative controls (red background).
(G) Expression of either the N- or the C-terminal FP fragment alone.
(H) N- and C-terminal FP fragments are coexpressed, but without fusion to the proteins of interest.
(I) N-terminal FP fragment fused to protein A is expressed alone.
(J) C-terminal fragment fused to protein B is expressed alone.
(K) N-terminal FP fragment fused to protein A is coexpressed with the unfused C-terminal FP fragment.
(L) C-terminal FP fragment fused to protein B is coexpressed with the unfused N-terminal FP fragment.
(M) and (N) Unrelated protein Z with different subcellular localization and no positive interaction control for Z and a partner protein (see [F]) is coexpressed with A or B.
(O) and (P) Possible orientations of the protein fusions in BiFC assays. It is important to note that the orientation can have a strong impact on the propensity of spontaneous FP reconstitution (i.e., the formation of false positive interactions; Bracha-Drori et al., 2004; Horstman et al., 2014). Hence, it is essential that, for the negative controls, exactly the same orientations are used as for the positive interaction.
N, N-terminal fragment of split FP; C, C-terminal fragment of split FP; *, mutation in the interaction site; Ø, no partner protein present; red outline, no interaction with expressed partner protein possible.
Although different FPs can be employed for BiFC studies, the GFP variants eYFP (enhanced YFP) and mVenus have been most extensively used (Kerppola, 2008; Waadt et al., 2014). Several distinct sites within the eYFP (or mVenus) protein have been found to allow for efficient reconstitution after splitting into separate fragments. Commonly, eYFP is split between Ala-154 and Asp-155 located between the seventh and the eighth β-sheet (Hu et al., 2002; Walter et al., 2004), between Glu-172 and Asp-173 within the linker separating the eighth and the ninth β-sheet (Hu and Kerppola, 2003; Waadt et al., 2008), and, more recently, after residue 210 within the loop separating the tenth and the eleventh β-sheet (Ohashi et al., 2012; Gookin and Assmann, 2014). Although fragmentation at position 172 appears to result in the strongest signal intensity of reconstituted YFP fluorescence, it also enhances unwanted background fluorescence. The split after residue 154 still allows for efficient reconstitution of YFP fluorescence while giving less undesired background fluorescence (Waadt et al., 2008). Likewise, fragmentation after residue 210 has been reported to greatly diminish nonspecific complex assembly and background fluorescence signals (Ohashi et al., 2012; Gookin and Assmann, 2014).
Recently, vectors have been developed that enable coexpression of reference FPs from the same plasmid expressing the BiFC fusion proteins (Grefen and Blatt, 2012; Gookin and Assmann, 2014). These vector sets represent potentially useful additions to the BiFC toolbox. However, thorough side-by-side comparisons of the signal intensities and signal-to-noise ratios obtained with the different vectors and differently split YFP versions (ideally using the same interacting proteins and following the “golden rules” suggested here) will be required before strong recommendations about preferred vector systems for BiFC in plant cells can be made.
BiFC approaches have not remained restricted to the analyses of single protein pairs. The development of multicolor BiFC, which is based on simultaneous reconstitution of split YFP and CFP or YFP/CFP hybrid proteins, has enabled observation of multiple (alternative) protein complexes in living cells (Hu and Kerppola, 2003; Waadt et al., 2008). Moreover, the repertoire of FC techniques has recently been extended by the dual-color trimolecular fluorescence complementation assay for the visualization of ternary protein complexes in plant cells (Offenborn et al., 2015).
An inherent feature of BiFC is the irreversibility of the FP reassembly. The reconstituted FP is stabilized by the extensive interaction interface between the two FP fragments that, in the case of YFP, comprises four new β-strand interfaces and more than 30 hydrogen bonds (Robida and Kerppola, 2009). This feature has largely prevented the use of BiFC-based techniques for the analysis of the dynamics of protein complex formation or the dynamics of protein-protein interactions. On the other hand, the extraordinary stability of the formed protein complex facilitates the visualization of even weak or transient interactions (e.g., interactions of protein kinases with their substrates) that are often difficult to detect with alternative methods. Recently, a reversible BiFC system has been reported that is based on the engineered Deinococcus radiodurans infrared fluorescent protein IFP1.4 (which is unrelated to GFP and YFP; Tchekanda et al., 2014). Unlike previous BiFC techniques, the IFP complementation assay permits, for example, the analysis of the spatiotemporal dynamics of hormone-induced signaling complexes in living yeast and mammalian cells at nanometer resolution. Current limitations of the IFP complementation assay lie in its low quantum yield, low brightness, and the requirement for exogenously added biliverdin when used in mammalian and yeast cells. Further improvement and adaptation of this system for the study of protein-protein interactions in plant cells may overcome the current shortcomings associated with irreversible BiFC complex formation.
The major limitation of current BiFC systems, which most likely is enhanced by the irreversible nature of FP reconstitution, is the tendency toward nonspecific self-assembly of the fluorophores resulting in false positive fluorescence signals. When the concentration of both eYFP fragments (or other FP fragments) at a given cellular localization exceeds a certain threshold, functional FC can occur spontaneously, in the absence of interactions between the proteins to be assayed. Indeed, early reports on BiFC applications (Hu et al., 2002; Walter et al., 2004) alerted the community to this potential source of artifacts and emphasized the requirement for appropriate controls to faithfully detect interactions that are truly biologically relevant. Rigorous controls (outlined in Figure 1) are all the more necessary, since the vast majority of BiFC assays involve (transient) overexpression of the candidate proteins. Together with the irreversibility of the FP reconstitution, overexpression of the FP fusions can lead to the fixation of transient artifactual interactions between proteins that would not normally interact at physiological concentrations. It is also important to realize that, while there is evolutionary pressure for specificity in protein-protein interactions, nonspecific interactions are only selected against if the given proteins (1) co-occur in the same subcellular compartment and (2) are expressed in the same cell type, developmental stage, and/or environmental condition. These are important aspects to verify because artifacts resulting from the interaction between proteins that would not normally encounter each other cannot be revealed by technical controls. When choosing the orientation of the protein fusion (i.e., fusion of the FP fragment to either the N terminus or the C terminus of the protein of interest; Figures 1O and 1P), it also must be ascertained that targeting signals for protein localization are not blocked (e.g., C-terminal signals for nuclear localization or localization to the endoplasmic reticulum and N-terminal transit peptides for import into plastids or mitochondria).
Unfortunately, a large number of reports on BiFC-based interaction analyses have been published with inappropriate and inadequate controls, including the sole expression of a singular protein fused to a YFP fragment or the coexpression of one of the YFP fragments as fusion protein with the other YFP fragment unfused (Horstman et al., 2014). Moreover, the vast majority of published BiFC studies report only qualitative results and show “representative examples” of detected interactions and thus fall short of providing quantitative interaction analyses and statistically validated data (Horstman et al., 2014). Here, we reflect on important considerations for the validation of BiFC results, discuss options for suitable controls and independent verification of detected interactions, and make recommendations for best practices in BiFC studies.
BEST PRACTICES AND RECOMMENDATIONS
Essential Controls
To date, the vast majority of plant BiFC analyses have involved transient overexpression of the fusion proteins in cells, cell cultures, or tissues. In most cases, overexpression of the proteins has been performed in a single cell type (e.g., protoplasts isolated from mesophyll cells) or in heterologous tissues (e.g., infiltrated Nicotiana benthamiana leaves). More recently, the BiFC method has been applied in intact plant tissues using native promoters to drive the expression of the fusion proteins (Smaczniak et al., 2012). Overexpression may be a reasonable approach to reach the detection limit, and heterologous expression may be necessary due to the lack of efficient transformation techniques for a given species, tissue, or cell type. However, approaches based on overexpression require stringent controls to confirm coexpression of both proteins of interest in the same tissue or cell type and under the same environmental conditions. Such data are best obtained by comparative analyses of plant lines expressing the respective promoter:reporter gene fusions or, alternatively, by high-resolution qRT-PCR analyses.
Appropriate controls, especially those that address the possibility of spontaneous FP reconstitution, are crucial to validate BiFC data and establish specificity of the observed protein-protein interaction (Figure 1). The most stringent control in a set of BiFC experiments is the combination of one of the proteins with a mutated version of its interaction partner (Figures 1B and 1C). Ideally, the interaction partner harbors a single point mutation or a small deletion in the domain that is required for the interaction to occur. Due to protein instability or missing knowledge about the amino acid residues involved in a given protein-protein interaction, it may not always be possible to use mutated versions of (one of the) interaction partners as negative control(s). In these cases, a closely related protein (e.g., a member of the same protein family) can provide an alternative negative control that is equally acceptable (Figures 1D and 1E). It is desirable that this control protein is localized in the same compartment as the protein of interest and its proper expression has been verified. Ideally, one should include an additional (positive) control that demonstrates interaction of this control protein with one of its genuine interaction partners. Such a set of controls not only provides strong evidence for the specificity of the observed interactions, it also establishes proper expression and folding of all proteins involved.
If neither a mutated protein version nor a suitable closely related protein are available as negative control, an unrelated (but, ideally, structurally similar) protein can also be used. In this case, it is particularly important that (1) the chosen protein colocalizes with the protein of interest in the same (sub)compartment of the cell, and (2) this unrelated protein is demonstrated to be capable of interacting with another protein in BiFC assays (positive control; Figure 1F). However, this should be the control of last resort: Since BiFC is a proximity-based assay, different protein structures can have a strong impact on protein reconstitution. Kodama and Hu (2012) have suggested an elegant competition-based assay that provides a more stringent alternative control that, however, has not yet been adapted for BiFC assays in plant cells. Inappropriate controls for BiFC experiments that, unfortunately, are frequently seen in the literature include, for example, combination of one of the interaction partners with an empty vector (expressing the unfused FP fragment) or the expression of only one of the fusion proteins (Horstman et al., 2014; Figures 1G to 1N). In addition, the orientation of the protein fusions (Figures 1O and 1P) is known to influence the propensity of spontaneous FP reconstitution in BiFC assays (Bracha-Drori et al., 2004; Horstman et al., 2014). Therefore, for the negative controls to be conclusive, exactly the same orientations must be used as for the positive interaction.
Since the negative controls usually exhibit much lower (or undetectably low) fluorescence emission, faithful expression of all fusion proteins must be confirmed. This can easily be done by immunoblot analysis using monoclonal antibodies against specific epitope tags that are part of the BiFC expression cassette in most of the commonly used vectors (Waadt et al., 2008). When assessing expression levels in the negative controls by immunoblotting, it must be kept in mind that mutated proteins (especially those harboring deletions) can be less stable than their wild-type counterparts and/or their subcellular localization can be altered. In most cases, protein accumulation will not be reduced so much that it would prevent fluorescence detection altogether if protein-protein interaction occurs. Overall, verification of the expression of all fusion proteins involved (including those used in the controls) should be an essential part of any BiFC study.
Quantitative Analysis
For transient BiFC assays, transformation of (mesophyll) protoplasts, infiltration of N. benthamiana leaves, or particle bombardment of leaves or onion epidermis tissue can be used. Assays in transiently transformed N. benthamiana leaves, although representing a heterologous expression system, provide some advantages. Since expression of the fusion proteins usually increases over a period of up to 5 d (Waadt et al., 2008; Schlücking et al., 2013; Waadt et al., 2014), the experimenter can choose the most suitable expression level (i.e., a level that is sufficient to detect the interaction but minimizes the risk of overexpression artifacts). Also, this transient system is convenient to generate quantitative data sets of protein-protein interactions (see below).
While BiFC assays in protoplasts provide the advantage that they can be performed in cells derived from the same species as the proteins of interest (homologous system), due to the incomplete transformation of a protoplast suspension (and the variation in the transformation rate from experiment to experiment), the quantification of the interaction data is somewhat more involved. A reasonably reliable quantification would require determination of the fluorescence intensities of a larger number of (1) protoplasts that were transformed with vector combinations conferring an interaction and (2) those representing negative controls. Since protoplasts expressing negative controls may not emit a detectable (above background) fluorescence signal, this approach requires coexpression of a second reporter (FP) for identification of transformed cells. Also, protoplasts provide a shorter time frame for expression analyses (typically up to 48 h) and usually can be transformed only once. The longer expression time in infiltrated leaves (and the possibility of performing multiple infiltrations) allows avoidance of problems that might arise, for example, from different maturation times of fluorescent proteins (as known, for example, for GFP/YFP in comparison with mCherry; Khmelinskii et al., 2012).
For quantification of BiFC data from infiltrated N. benthamiana leaves, protocols that involve image acquisition from 10 randomly chosen regions of interest (encompassing at least 10 epidermal cells each; Waadt et al., 2008, 2014) that are selected blindly (i.e., without considering the actual fluorescence emission intensity of individual cells) are recommended. An important consideration for comparative BiFC studies in N. benthamiana is to infiltrate each vector combination into several leaves that come from different plants and are of the same age, size, and physiological state. For background determination, fluorescence intensity of 10 regions of interest infiltrated only with the helper plasmid (e.g., a plasmid expressing the silencing suppressor p19 of tomato bushy stunt virus; Waadt et al., 2014) should be measured and used for background subtraction from positive interactions, prior to final percentage normalization to the chosen experimental control (see below). Coexpression of a reference FP, such as CFP, can help to further minimize variation in BiFC signals due to expression differences between leaves or leaf areas. However, in general, this is not required for BiFC quantification in N. benthamiana because, if the transformation protocols are properly optimized, nearly complete cotransformation can be achieved (Waadt and Kudla, 2008). In this regard, it is important that fluorescence patterns are not confused with transformation rates: The inhomogeneous or patchy expression patterns that are occasionally obtained upon coexpression of FP fusions in N. benthamiana leaves largely result from transgene silencing rather than from inefficient cotransformation.
It is highly desirable to show results as not only “representative images” but also as quantitative data obtained from, for example, image quantitation of at least 10 randomly chosen regions of interest of infiltrated leaves (Waadt et al., 2014). Any representative image should display a complete cell, with the nucleus and the center of the cell in the focal plane. This facilitates comparison between images and identification of distinct localization patterns and helps researchers to avoid erroneous conclusions about the subcellular localization of the interaction. (In general, it is a good practice to show the observed variation in expression patterns and/or fluorescence intensities in a series of images that can be included in the supplemental materials of the article.) If the subcellular localization of the BiFC complexes is to be reliably determined, coexpression of a well characterized marker protein fused to a different, compatible FP should be performed to assess colocalization with the BiFC signal. Colocalization and fluorescence intensities can be determined by fluorescence intensity line-scan or scatterplot analyses (Held et al., 2011; Offenborn et al., 2015).
Validation by Independent Methods
In general, evidence from at least two basically independent methods is needed to draw firm conclusions about protein-protein interactions. A truly independent method does not rely on the irreversible reconstitution of a (fluorescent) protein. Ideally, the confirmatory method can also be performed in vivo, in intact plant tissue. Examples of such techniques are FRET-FLIM and split-luciferase assays (Remy and Michnick, 2006; Bayle et al., 2008; Gehl et al., 2011). If the protein-protein interaction results in a subcellular translocation of at least one of the proteins, coexpression and translocation assays can also be applied (Piljić and Schultz, 2008; Schlücking et al., 2013; Offenborn et al., 2015). Since these techniques use full-length FPs with distinct emission spectra, no FP reconstitution is required for visualization of the protein-protein interaction (hence no artificial stabilization can occur). Alternative techniques that function in vivo (albeit usually in non-plant systems) and can be used to substantiate BiFC data include the yeast two-hybrid system and the split-ubiquitin assay.
If performed with the necessary stringent controls, coimmunoprecipitation (co-IP) experiments can provide support for and further characterize protein-protein interactions detected by BiFC or other in vivo methods. Importantly, in co-IP experiments, experimental parameters that potentially modulate protein-protein interactions and/or affect their efficiency (e.g., redox milieu and buffer conditions such as the concentrations of calcium and magnesium ions) can be precisely adjusted and modified according to the subcellular origin and the physicochemical properties of the proteins of interest. It should be borne in mind that co-IP provides evidence that two proteins are in the same complex, but not necessarily that they interact directly. (However, to a lesser extent, this also holds true for BiFC [see above].)
Finally, it should not be forgotten that BiFC experiments can also produce false negative results (i.e., fail to detect an interaction that actually occurs in planta). False negative results can, for example, arise if fusion to the FP fragments results in aberrant protein folding or sterically hinders the protein-protein interaction.
SUMMARY
The most important aspects involved in the reliable detection of protein-protein interactions by BiFC assays are (1) the validation of the suitability of the expression constructs, especially those used for the negative controls; (2) the design of controls that are as similar as possible to the interaction assayed; (3) the quantitative assessment of the interaction in direct comparison to the most appropriate negative control; and (4) the verification of the BiFC data with at least one principally independent technique.
In summary, the following golden rules for BiFC experiments are suggested:
(1) Verify that the interacting proteins are coexpressed in the same subcellular compartment, tissue, cell type, and environmental conditions (by qPCR, promoter:reporter gene fusions, etc.).
(2) Perform stringent negative controls (preferably a mutated version of one of the interacting proteins carrying a defect in the interaction domain or a related protein from the same protein family).
(3) Confirm faithful expression of all fusion proteins tested, including the negative controls (e.g., by immunoblotting).
(4) Perform a quantitative analysis of the interactions (including the negative controls), especially when weak interactions are reported and/or conclusions about differences in the strength of the interactions are to be drawn (using image analysis methods).
(5) Use a truly independent method to confirm the BiFC interaction (i.e., a method not relying on the irreversible reconstitution of a split protein).
Acknowledgments
We thank Rainer Waadt (University of Heidelberg) and Jan Niklas Offenborn (University of Münster) for helpful discussion and a critical read of this manuscript as well as Jan Niklas Offenborn for help with creating the artwork. Work on protein-protein interactions in the authors’ laboratories is supported by grants from the Deutsche Forschungsgemeinschaft (FOR 2092 and BO 1482/17-1), the Human Frontiers Science Program (RGP0005/2013), and the European Research Council under the European Union’s Horizon 2020 research and innovation program (ERC-ADG-2014; Grant Agreement 669982) to R.B. and by grants from the Deutsche Forschungsgemeinschaft (KU 931/14-1 and FOR964 KU 931/8-3) to J.K.
AUTHOR CONTRIBUTIONS
R.B. and J.K. wrote the manuscript.
References
- Bayle V., Nussaume L., Bhat R.A. (2008). Combination of novel green fluorescent protein mutant TSapphire and DsRed variant mOrange to set up a versatile in planta FRET-FLIM assay. Plant Physiol. 148: 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori K., Shichrur K., Katz A., Oliva M., Angelovici R., Yalovsky S., Ohad N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40: 419–427. [DOI] [PubMed] [Google Scholar]
- Gehl C., Kaufholdt D., Hamisch D., Bikker R., Kudla J., Mendel R.R., Hänsch R. (2011). Quantitative analysis of dynamic protein-protein interactions in planta by a floated-leaf luciferase complementation imaging (FLuCI) assay using binary Gateway vectors. Plant J. 67: 542–553. [DOI] [PubMed] [Google Scholar]
- Gookin T.E., Assmann S.M. (2014). Significant reduction of BiFC non-specific assembly facilitates in planta assessment of heterotrimeric G-protein interactors. Plant J. 80: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C., Blatt M.R. (2012). A 2in1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC). Biotechniques 53: 311–314. [DOI] [PubMed] [Google Scholar]
- Held K., Pascaud F., Eckert C., Gajdanowicz P., Hashimoto K., Corratgé-Faillie C., Offenborn J.N., Lacombe B., Dreyer I., Thibaud J.-B., Kudla J. (2011). Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res. 21: 1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horstman A., Tonaco I.A.N., Boutilier K., Immink R.G.H. (2014). A cautionary note on the use of split-YFP/BiFC in plant protein-protein interaction studies. Int. J. Mol. Sci. 15: 9628–9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.-D., Kerppola T.K. (2003). Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21: 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.-D., Chinenov Y., Kerppola T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798. [DOI] [PubMed] [Google Scholar]
- Kerppola T.K. (2006). Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 7: 449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola T.K. (2008). Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37: 465–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., et al. (2012). Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat. Biotechnol. 30: 708–714. [DOI] [PubMed] [Google Scholar]
- Kodama Y., Hu C.-D. (2012). Bimolecular fluorescence complementation (BiFC): a 5-year update and future perspectives. Biotechniques 53: 285–298. [DOI] [PubMed] [Google Scholar]
- Lalonde S., Ehrhardt D.W., Loqué D., Chen J., Rhee S.Y., Frommer W.B. (2008). Molecular and cellular approaches for the detection of protein-protein interactions: latest techniques and current limitations. Plant J. 53: 610–635. [DOI] [PubMed] [Google Scholar]
- Offenborn J.N., Waadt R., Kudla J. (2015). Visualization and translocation of ternary Calcineurin-A/Calcineurin-B/Calmodulin-2 protein complexes by dual-color trimolecular fluorescence complementation. New Phytol. 208: 269–279. [DOI] [PubMed] [Google Scholar]
- Ohashi K., Kiuchi T., Shoji K., Sampei K., Mizuno K. (2012). Visualization of cofilin-actin and Ras-Raf interactions by bimolecular fluorescence complementation assays using a new pair of split Venus fragments. Biotechniques 52: 45–50. [DOI] [PubMed] [Google Scholar]
- Piljić A., Schultz C. (2008). Analysis of protein complex hierarchy in living cells. ACS Chem. Biol. 3: 749–755. [DOI] [PubMed] [Google Scholar]
- Remy I., Michnick S.W. (2006). A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods 3: 977–979. [DOI] [PubMed] [Google Scholar]
- Remy I., Michnick S.W. (2015). Mapping biochemical networks with protein fragment complementation assays. Methods Mol. Biol. 1278: 467–481. [DOI] [PubMed] [Google Scholar]
- Robida A.M., Kerppola T.K. (2009). Bimolecular fluorescence complementation analysis of inducible protein interactions: effects of factors affecting protein folding on fluorescent protein fragment association. J. Mol. Biol. 394: 391–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlücking K., Edel K.H., Köster P., Drerup M.M., Eckert C., Steinhorst L., Waadt R., Batistic O., Kudla J. (2013). A new β-estradiol-inducible vector set that facilitates easy construction and efficient expression of transgenes reveals CBL3-dependent cytoplasm to tonoplast translocation of CIPK5. Mol. Plant 6: 1814–1829. [DOI] [PubMed] [Google Scholar]
- Smaczniak C., et al. (2012). Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 109: 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchekanda E., Sivanesan D., Michnick S.W. (2014). An infrared reporter to detect spatiotemporal dynamics of protein-protein interactions. Nat. Methods 11: 641–644. [DOI] [PubMed] [Google Scholar]
- Waadt R., Kudla J. (2008). In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc. 2008: t4995. [DOI] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Waadt R., Schlücking K., Schroeder J.I., Kudla J. (2014). Protein fragment bimolecular fluorescence complementation analyses for the in vivo study of protein-protein interactions and cellular protein complex localizations. In Arabidopsis Protocols, J.J. Sanchez-Serrano and J. Salinas, eds (New York: Springer Science+Business Media), pp. 629–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]