A eukaryotic transcription factor guides DNA-dependent RNA polymerase II to recognize and transcribe RNA templates from a viroid pathogen.
Abstract
Some DNA-dependent RNA polymerases (DdRPs) possess RNA-dependent RNA polymerase activity, as was first discovered in the replication of Potato spindle tuber viroid (PSTVd) RNA genome in tomato (Solanum lycopersicum). Recent studies revealed that this activity in bacteria and mammals is important for transcriptional and posttranscriptional regulatory mechanisms. Here, we used PSTVd as a model to uncover auxiliary factors essential for RNA-templated transcription by DdRP. PSTVd replication in the nucleoplasm generates (−)-PSTVd intermediates and (+)-PSTVd copies. We found that the Nicotiana benthamiana canonical 9-zinc finger (ZF) Transcription Factor IIIA (TFIIIA-9ZF) as well as its variant TFIIIA-7ZF interacted with (+)-PSTVd, but only TFIIIA-7ZF interacted with (−)-PSTVd. Suppression of TFIIIA-7ZF reduced PSTVd replication, and overexpression of TFIIIA-7ZF enhanced PSTVd replication in planta. Consistent with the locale of PSTVd replication, TFIIIA-7ZF was found in the nucleoplasm and nucleolus, in contrast to the strictly nucleolar localization of TFIIIA-9ZF. Footprinting assays revealed that only TFIIIA-7ZF bound to a region of PSTVd critical for initiating transcription. Furthermore, TFIIIA-7ZF strongly enhanced the in vitro transcription of circular (+)-PSTVd by partially purified Pol II. Together, our results identify TFIIIA-7ZF as a dedicated cellular transcription factor that acts in DdRP-catalyzed RNA-templated transcription, highlighting both the extraordinary evolutionary adaptation of viroids and the potential of DdRPs for a broader role in cellular processes.
INTRODUCTION
Besides their canonical role in DNA-templated RNA transcription, some DNA-dependent RNA polymerases (DdRPs) have an RNA-dependent RNA polymerase (RdRP) activity. This RdRP activity of Pol II was first found to be essential for the replication of Potato spindle tuber viroid (PSTVd) (Muhlbach and Sanger, 1979) and later for transcription and replication of human hepatitis delta virus (HDV) (MacNaughton et al., 1991; Modahl et al., 2000). Recent studies showed that this RdRP activity has broader biological significance beyond pathogen infection. Under nutrient-deficient conditions, the noncoding 6S RNA of Escherichia coli binds to the DdRP and prevents it from transcribing a DNA template. In response to nutrient availability, the DdRP transcribes a short RNA from the 6S RNA template that leads to desequestration of DdRP. This step in turn enables binding to DNA promoters and synthesis of protein-coding mRNAs (Wassarman and Saecker, 2006). Mammalian Pol II can bind to a hairpin formed by the noncoding B2 RNA and use the longer strand of the hairpin sequence as a template to extend the short strand by transcription. This extension by Pol II in turn destabilizes the B2 RNA and represents a novel posttranscriptional mechanism for altering the stability of B2 RNA (Wagner et al., 2013). Thus, in addition to its function in pathogen replication, the RdRP activity of DdRPs is emerging as an essential mechanism for regulating gene expression across kingdoms.
High-resolution crystallographic structures revealed that yeast Pol II uses the same site for binding to DNA and RNA templates, leading to the hypothesis that the RdRP activity of a DdRP may represent a missing link in molecular evolution; a DdRP may have evolved first to use an RNA before switching to a DNA template (Lehmann et al., 2007). Apparently, this relic RdRP activity of a DdRP has persisted in modern life forms in catalyzing the transcription of some noncoding RNA templates to regulate gene expression and is part of the reprogramming by noncoding (viroids) and limited-coding (HDV) RNA pathogens vital for their replication. Given the slow rate and moderate processivity of the RdRP activity, it was postulated that the generation of long RNAs such as viroids likely requires cellular factors (Rackwitz et al., 1981). Identifying cellular transcription factors that enable a DdRP to switch between DNA and RNA templates is thus crucial for advancing studies on the evolution, mechanism, and function of this relic RdRP activity of DdDP. Here, we used the systemic PSTVd infection of Nicotiana benthamiana as a unique and tractable model to characterize such factors.
The 359-nucleotide covalently closed circular genome of PSTVd folds into a rod-shaped secondary structure with 27 loops interspersed among short stems (Supplemental Figure 1A). Because PSTVd does not encode any protein, all proteins critical for its life cycle must be derived from the host. Its rolling circle replication involves transcription of the (+)-strand circular genome RNA into concatemeric (−)-linear RNA, which is then used as a template to generate concatemeric (+)-linear RNAs that are then cleaved into unit length and circularized (Supplemental Figure 1B) (Flores et al., 2005; Ding, 2009). Pol II is implicated in PSTVd transcription, as shown by inhibition of PSTVd replication with α-amanitin, an inhibitor of Pol II (Muhlbach and Sanger, 1979; Schindler and Mühlbach, 1992) and by Pol II-catalyzed transcription in vitro (Rackwitz et al., 1981). However, the binding of Pol II to PSTVd in vivo, which has not been demonstrated, is required to definitively establish the role of Pol II in PSTVd replication during infection.
Our goal was to evaluate possible transcription factors that are subverted by PSTVd for reprogramming Pol II to ensure its own replication. Eiras et al. (2011) showed that TRANSCRIPTION FACTOR IIIA (TFIIIA) from Arabidopsis thaliana binds to PSTVd in vitro, but did not evaluate its functional roles. TFIIIA is conserved in eukaryotes and generally has nine zinc fingers (ZFs). It binds to the internal promoter of 5S rDNA to promote transcription of 5S rRNA and then binds to 5S rRNA to inhibit further transcription as well as to facilitate nuclear export of 5S rRNA for ribosome biogenesis (Klug, 2005). The TFIIIA gene in land plants has evolved a conserved alternative splicing mechanism to generate exon-inclusion and exon-skipping variants (Fu et al., 2009; Hammond et al., 2009; Barbazuk, 2010). The exon-skipping form encodes the full-length TFIIIA that contains nine ZFs (TFIIIA-9ZF); in contrast, the exon-inclusion form has a predicted open reading frame encoding a TFIIIA variant that has only seven ZFs and lacks the first two ZFs of the TFIIIA-9ZF (Supplemental Figure 2). Although it had been initially suggested that the exon-inclusion form of the TFIIIA mRNA may be targeted for degradation (Fu et al., 2009; Hammond et al., 2009), the recent finding that TFIIIA-7ZF protein is present in Arabidopsis (Layat et al., 2012) prompts questions about its biological function and its functional parallels with TFIIIA-9ZF, which is a well established transcription factor for 5S rRNA biogenesis and transport in animals and plants (Barbazuk, 2010).
Here, we report that the N. benthamiana Pol II interacts with the (+)- and (−)-strand PSTVd RNAs in vivo, providing the missing physical evidence necessary to fully establish the role of Pol II in PSTVd replication. We further show using various biochemical, molecular, and subcellular localization studies that the shorter variant TFIIIA-7ZF acts as a genuine transcription factor that directly guides Pol II-catalyzed transcription using PSTVd RNA genome as a template in plants. Our finding has broad implications for characterization of cellular factors that promote RNA-templated RNA transcription or replication catalyzed by DdRPs in diverse biological systems and processes.
RESULTS
The DNA-Dependent RNA Polymerase II Interacts with the (+)- and (−)-PSTVd RNA in Vivo
Although several lines of evidence support the role of Pol II in PSTVd replication (Flores et al., 2005; Ding, 2009), the most direct physical evidence showing the binding of Pol II to the (+)- and (−)-strands of PSTVd in vivo has been elusive. To address this shortcoming, we performed RNA immunoprecipitation. To pull down Pol II from N. benthamiana, we used the commercial mouse monoclonal antibody 8WG16, which was raised against the C-terminal domain of the largest subunit of Pol II that had been purified from a plant source (Thompson et al., 1989). The existence of highly conserved YSPTSPS repeats in the C-terminal domain of eukaryotic Pol II has allowed a broad use of this 8WG16 antibody for immunoblotting and immunoprecipitation experiments in various animals and plants, including Arabidopsis (Zheng et al., 2009), rice (Oryza sativa; Roy et al., 2014), and Nicotiana tabacum (Shin et al., 2014). We tested the specificity of 8WG16 for the largest subunit of Pol II in N. benthamiana. As expected, a single band corresponding to a 220-kD protein was observed (Supplemental Figure 3).
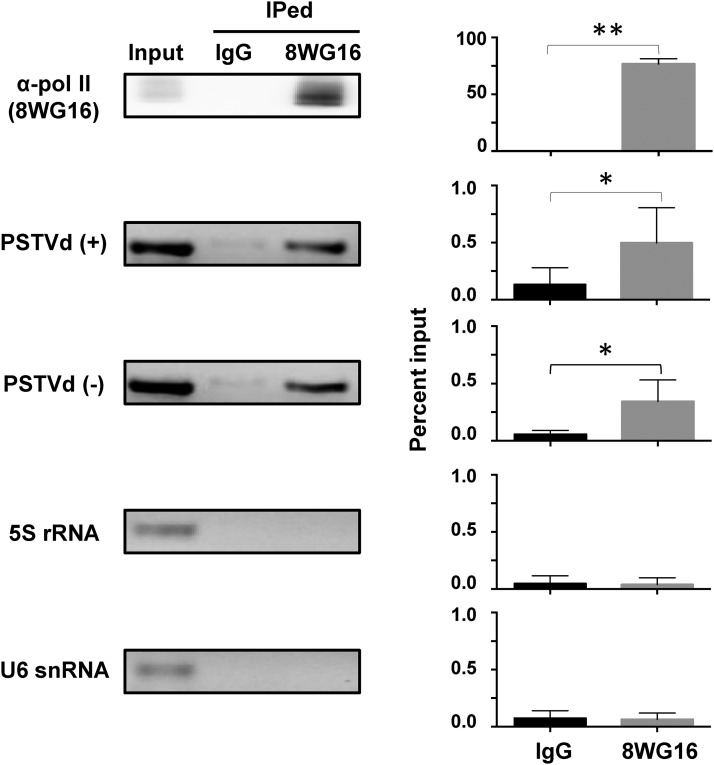
We used immunoblotting to establish the presence of Pol II after RNA immunoprecipitation and RT-PCR to demonstrate the presence of (+)- and/or (−)-strands of PSTVd RNA in the immunoprecipitated pellet. Indeed, we found reproducibly that Pol II and both polarities of PSTVd strands coexisted in the pull-down fraction, providing physical evidence for the in vivo interaction/association between Pol II and PSTVd, thus further establishing the role of Pol II in PSTVd replication (Figure 1). As negative controls, we examined both 5S rRNA and U6 snRNA, two RNAs that are known to be transcribed by Pol III in plants; these RNAs were present in the input but not in the Pol II-immunoprecipitated pellet (Figure 1).
Figure 1.
RNA Immunoprecipitation of the Pol II-PSTVd Complex.
Mouse monoclonal antibody 8WG16 (which recognizes the CTD domain of the largest subunit of Pol II purified from a plant) was used to pull down Pol II from N. benthamiana. The presence of Pol II in the immunoprecipitated (IPed) pellet from 8WG16-conjugated resin but not mouse IgG-conjugated resin was assessed by immunoblotting, while that of (+)- and (−)-strand PSTVd RNAs as well as 5S rRNA and U6 snRNA were examined by RT-PCR. The quantitation and normalization for each gel panel is shown immediately to its right. Briefly, the signals from the input lysates served as a reference representing 1% for RNA and 10% for protein for standardization. The band intensity in negative (IgG) and positive (8WG16) treatments was quantitated and normalized to the signals of inputs to obtain their percentage values. Three biological repeats were used for the two-tailed t test (**P < 0.01 and *P < 0.05).
PSTVd replication is believed to utilize an asymmetric rolling circle model (Supplemental Figure 1), where circular (+)-PSTVd serves as a template for Pol II transcription to generate concatemeric (−)-PSTVd. Therefore, we were interested in testing if Pol II binds circular (+)-PSTVd in planta. To this end, we performed RNA immunoprecipitation using 40 g of PSTVd-infected leaves. Following immunoblotting analyses to first detect the presence of the largest subunit of Pol II in the immunoprecipitated pellet (Supplemental Figure 4), we used RNA gel blot analysis to show that circular (+)-PSTVd was associated with this subunit of Pol II (Supplemental Figure 4). This result suggests that circular (+) PSTVd is the template for Pol II transcription in vivo.
N. benthamiana TFIIIA-9ZF and TFIIIA-7ZF Interact with PSTVd in Vitro and in Vivo but with Notable Differences That Have Functional Implications
Given previous reports linking TFIIIA and PSTVd binding in vitro, and the possibility of different TFIIIA isoforms in plants, we first sought to establish if indeed exon-inclusion and exon-skipping variants are expressed in N. benthamiana. We cloned the cDNAs containing the complete ORFs of the N. benthamiana TFIIIA-9ZF and TFIIIA-7ZF using RT-PCR (Supplemental Data 1). These cDNAs were used to overexpress the two TFIIIA variants in E. coli and purify them as intact proteins, without additional amino acid residues, using a chitin-based affinity chromatography/intein tag-cleavage system. We raised polyclonal antibodies against recombinant TFIIIA-7ZF, with the expectation that the serum would also recognize the TFIIIA-9ZF. Indeed, immunoblots using these antibodies demonstrated presence of the native TFIIIA-7ZF and TFIIIA-9ZF in N. benthamiana (Supplemental Figure 5). These data established that the shorter variant, TFIIIA-7ZF, is expressed in N. benthamiana mirroring the previous observation in Arabidopsis (Layat et al., 2012).
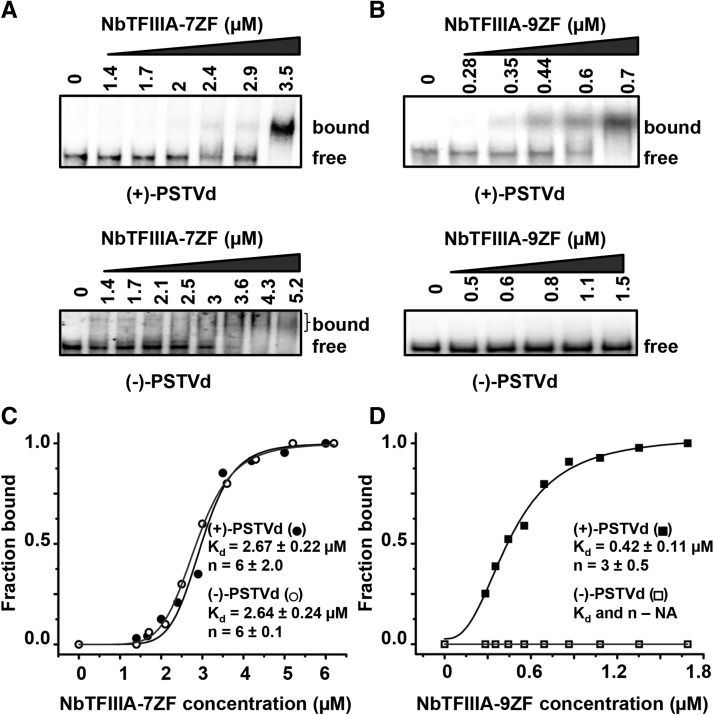
For all our studies, we employed a previously used linear PSTVd whose 5′ and 3′ ends correspond to positions 95 and 94, respectively (Eiras et al., 2011). This linear form retains full infectivity in N. benthamiana as demonstrated by successful plant infection in this study. To test whether the two TFIIIA forms bind to (+)-PSTVd in vitro, we performed electrophoretic mobility shift assays (EMSAs) using 32P-labeled PSTVd transcripts and intact, recombinant TFIIIA-7ZF and -9ZF proteins. Indeed, the interactions between PSTVd and both TFIIIA forms were detected in the EMSA experiments as evidenced by the retarded bands corresponding to the RNA-protein complexes (Figure 2; Supplemental Figure 6). TFIIIA-7ZF bound to (+)-PSTVd with a lower affinity than TFIIIA-9ZF (Kd values of 2.67 ± 0.22 µM versus 0.42 ± 0.11 µM; Figures 2C and 2D). Interestingly, only TFIIIA-7ZF was capable of binding to the (−)-PSTVd strand (Figures 2C and 2D; Supplemental Figure 6), with the affinity being nearly the same as that to the (+)-PSTVd strand. It is noteworthy that we did observe a smear but not discrete bands when we incubated 4.5 μM of TFIIIA-9ZF with (−)-PSTVd. Whether these higher order complexes have biological significance requires additional studies. These remarkable differences in binding of the two TFIIIA variants to the (+)- and (−)-PSTVd RNAs attest to their specificity of binding. The cooperative nature of the binding to both TFIIIA-7ZF and TFIIIA-9ZF is evident from the sigmoidicity of the binding curve and the computed Hill coefficients (6 and 3 with TFIIIA-7ZF and TFIIIA-9ZF, respectively; Figures 2C and 2D). The basis for this cooperativity and the stoichiometry of the protein in the RNP complex remains to be explored.
Figure 2.
Interaction of N. benthamiana TFIIIA-7ZF and -9ZF with PSTVd in EMSAs.
(A) and (B) Interaction of TFIIIA7ZF (A) and TFIIIA9ZF (B) with (+)- and (−)-PSTVd.
(C) and (D) Quantitative analyses of the binding affinities of TFIIIA-7ZF and TFIIIA-9ZF with (+)-PSTVd and with (−)-PSTVd. While one representative binding curve for each protein is shown, the reported mean and sd values were calculated from three independent experiments. NA, not applicable.
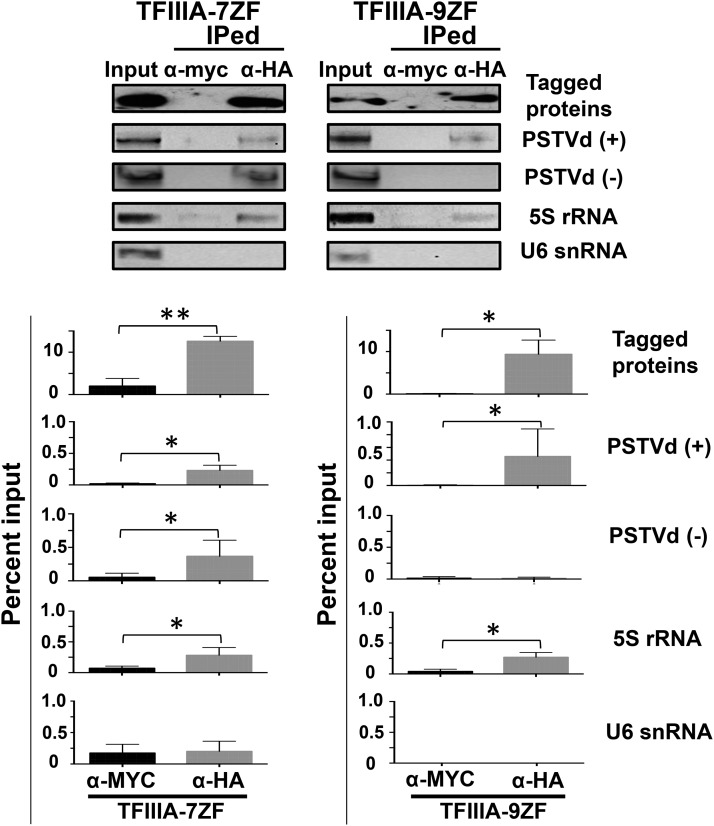
To determine whether the two TFIIIA isoforms interact with PSTVd in vivo, we used Agrobacterium tumefaciens-mediated transient expression to produce HA-tagged proteins in PSTVd-infected N. benthamiana leaves. Anti-HA antibodies were used to immunoprecipitate the recombinant proteins, and RT-PCR was used to test for presence of PSTVd in the immunoprecipitated fraction. Our results from three independent experiments showed the presence of both PSTVd strands in the immunoprecipitated fractions of TFIIIA-7ZF but only the (+)-PSTVd in the TFIIIA-9ZF fraction (Figure 3), consistent with the in vitro binding data (Figure 2). An additional validation for the pull-down was provided by the presence of 5S rRNA in the immunoprecipitated pellet. The interaction between TFIIIA-9ZF and 5S rRNA is well documented in Xenopus laevis, yeast, human, and Arabidopsis (Szymański et al., 2003; Mathieu et al., 2003). Because previous work showed that the first three zinc finger domains of TFIIIA-9ZF are dispensable for the interaction of TFIIIA with 5S rRNA (Theunissen et al., 1992; Lu et al., 2003; Lee et al., 2006), we predicted and confirmed the interaction of N. benthamiana TFIIIA-7ZF with 5S rRNA. U6 snRNA, whose biogenesis is TFIIIA independent (White, 2011; Mroczek and Dziembowski, 2013), was included as a negative control. As expected, U6 snRNA, while present in the input, was absent from the immunoprecipitated pellet.
Figure 3.
RNA Immunoprecipitation Assays to Demonstrate the Interaction between TFIIIA and PSTVd in Vivo.
The cell lysates were incubated with anti-HA antibodies (α-HA) to detect HA-tagged TFIIIA-7ZF and -9ZF proteins or with anti-myc antibodies (α-myc) as a negative control for specificity. The immunoprecipitated fractions were subject to PSTVd detection by RT-PCR. 5S rRNA was used as a positive control while U6 snRNA was used as a negative control. The presence of both the (+)- and (−)-PSTVd RNAs was examined in the immunoprecipitate. The quantitation and the statistical analyses are shown at the bottom panel. Briefly, the signals from the input lysates served as a reference representing 1% for RNA and 10% for protein for standardization. The band intensity in negative (α-myc) and positive (α- HA) treatments was quantitated and normalized to the signals of inputs to obtain their percentage values. Three biological repeats were used for the two-tailed t test (**P < 0.01 and *P < 0.05).
Taken together, these data indicate that both forms of N. benthamiana TFIIIA interact with the (+)-strand PSTVd in vitro and in vivo, but only the shorter variant is capable of binding to the (−)-strand PSTVd under the conditions we tested. These findings, which indicate clear differences between the two forms of TFIIIA in their interactions with PSTVd, have functional implications.
TFIIIA-7ZF Is Uniquely Involved in PSTVd Replication
To test the potential role of TFIIIA in PSTVd replication, we investigated the consequences of downregulation and overexpression of TFIIIA. The alternatively retained intron, which distinguishes the TFIIIA-7ZF and TFIIIA-9ZF mRNAs, has four regions in the N. benthamiana genome (http://solgenomics.net) that show sequence similarity, with at least two of the four regions mapping to annotated cDNAs. Thus, we recognized that any attempts to specifically downregulate the two TFIIIA variants will engender off-target effects and interfere with interpretation of the outcomes; we also entertained the possibility that the two isoforms could lead to decreased PSTVd replication through independent routes, again complicating analysis of whether transcription was specifically affected. Therefore, we adopted a two-pronged strategy: examine the consequences from antisense-mediated downregulation of both isoforms and ascertain the effects from overexpression of each variant.
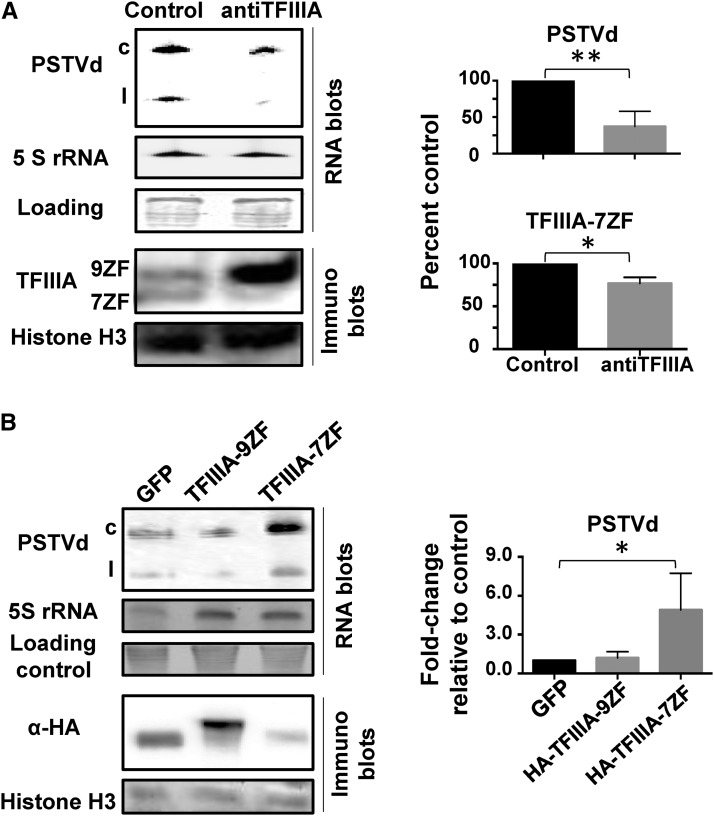
Through agroinfiltration, we could examine PSTVd replication when expressing a TFIIIA antisense construct in PSTVd-infected N. benthamiana plants. As shown in Figure 4A, antisense-mediated suppression of TFIIIA expression led to a drastic reduction in PSTVd accumulation in infected plants in repeated experiments. When we analyzed the levels of endogenous TFIIIA proteins, we observed decreased TFIIIA-7ZF and increased TFIIIA-9ZF (Figure 4A). We do not understand the basis for the increased TFIIIA-9ZF levels, but similar counterintuitive results have been reported in other systems (Jason et al., 2007; Carrieri et al., 2012). Our observation shows that a decrease in PSTVd accumulation is attributable to either lower levels of TFIIIA-7ZF and/or higher levels of TFIIIA-9ZF. However, when we transiently overexpressed HA-TFIIIA-7ZF, HA-TFIIIA-9ZF, or HA-GFP in PSTVd-infected N. benthamiana plants, only overexpression of HA-tagged TFIIIA-7ZF significantly increased PSTVd accumulation; neither TFIIIA-9ZF nor GFP overexpression had any detectable effect. This observation supports the notion that the reduction in levels of TFIIIA-7ZF is associated with a lower accumulation of PSTVd. Collectively, these data provide compelling evidence that TFIIIA-7ZF, but not TFIIIA-9ZF, plays a unique role in promoting PSTVd replication in planta.
Figure 4.
Effects of TFIIIA on PSTVd Accumulation in N. benthamiana Plants.
(A) Downregulating expression of TFIIIA resulted in reduced accumulation of PSTVd as well as 5S rRNA, shown by RNA gel blotting. Immunoblots of leaf extracts showed reduced levels of TFIIIA-7ZF as well as increased level of TFIIIA-9ZF upon antisense suppression of TFIIIA. Histone H3 served as a loading control for protein gel blots, while ethidium bromide staining of rRNAs served as a loading control for RNA gel blots.
(B) Increased replication of PSTVd upon ectopic expression of TFIIIA-7ZF but not of TFIIIA-9ZF.
The quantification and normalization for the experiments are shown on the right of each gel panel. Gel loading controls were used as a standard for normalization. The band intensities were quantitated and normalized to the corresponding standards to obtain their percentage or fold values. Three biological repeats were used for the statistical analyses (**P < 0.01 and *P < 0.05). c, circular PSTVd, l, linear PSTVd.
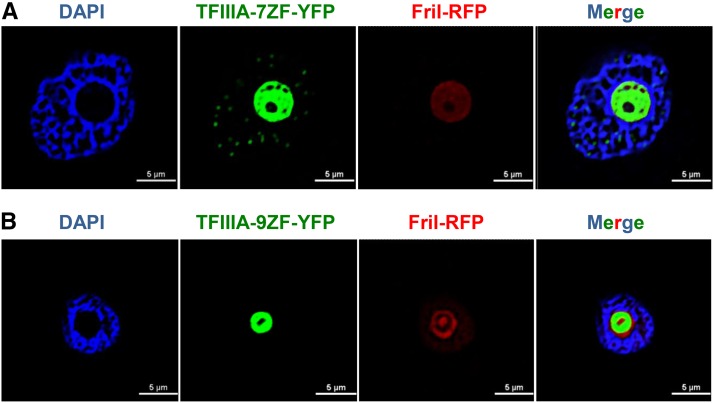
TFIIIA-7ZF Is Localized in the Nucleoplasm and Nucleolus in Contrast to the Nucleolar Localization of TFIIIA-9ZF
Subcellular localization is critical for protein function. We therefore determined the localization patterns of N. benthamiana TFIIIA-7ZF and -9ZF. We transiently expressed these proteins as YFP fusions in N. benthamiana leaves via agroinfiltration. Before observing the subcellular localization of the recombinant proteins, both TFIIIA-7ZF-YFP/HA and TFIIIA-9ZF-YFP/HA were tested for effects on PSTVd accumulation (Supplemental Figure 7) and PSTVd in vivo binding capacity (Supplemental Figure 8). Indeed, these fusions are wild type like in their behavior.
Fibrillarin is a nucleolar marker in plants and other organisms (Beven et al., 1996; Sirri et al., 2008), and we coexpressed the Arabidopsis fibrillarin fused with RFP to identify the nucleoli in N. benthamiana leaf cells. We further used 4′,6-diamidino-2-phenylindole (DAPI) staining to identify the nucleoplasm. In this transient expression system, TFIIIA-7ZF-YFP/HA localized in the nucleoplasm as well as the nucleolus (Figure 5A), whereas TFIIIA-9ZF-YFP/HA localized primarily in the nucleolus (Figure 5B). In particular, TFIIIA-7ZF-YFP/HA was present as distinct dots, reminiscent of nuclear speckles (green dots in Figure 5A) in nucleoplasm, which resemble Pol II transcription sites (Lamond and Spector, 2003). Examination of serial Z-sections of confocal images reproducibly verified these distinct localization patterns in 30 cells examined from each transformation (Supplemental Figure 9). Because PSTVd replication takes place in the nucleoplasm (Qi and Ding, 2003), where Pol II resides (Lamond and Spector, 2003; Pontes et al., 2006), localization of TFIIIA-7ZF in the nucleoplasm is consistent with its role in PSTVd replication catalyzed by Pol II. In contrast, the exclusive nucleolar localization of TFIIIA-9ZF might account for its inability to play a role in PSTVd replication.
Figure 5.
Subcellular Localization of TFIIIA-7ZF and TFIIIA-9ZF in N. benthamiana Leaves Infected by PSTVd.
TFIIIA-YFP is shown in green, Arabidopsis FriI-RFP is in red, and the nucleoplasm is shown in blue by DAPI staining.
(A) Localization of TFIIIA-7ZF in the nucleoplasm (speckled green dots) as well as in the nucleolus.
(B) Localization of TFIIIA-9ZF exclusively in the nucleolus. To enable better visualization of the speckle-like structures in the TFIIIA-7ZF panels relative to TFIIIA-9ZF, the contrast for all images was slightly enhanced but uniformly in all. The original confocal images are presented in Supplemental Figure 9.
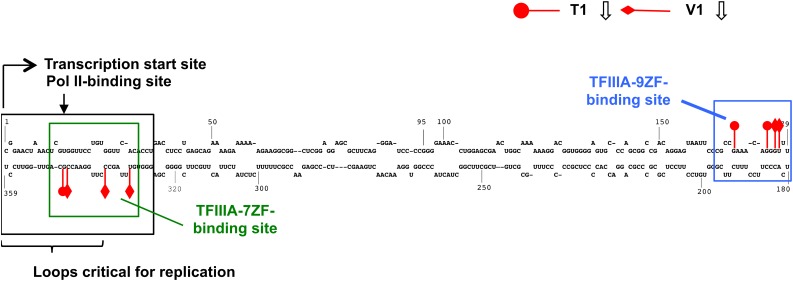
TFIIIA-7ZF Interacts with the Left-Terminal Region of PSTVd and TFIIIA-9ZF Interacts with the Right-Terminal Region
To gain insights into the role of TFIIIA-7ZF in PSTVd replication, we next employed RNase footprinting assays to identify the binding site(s) in PSTVd for both proteins. We used RNase V1, which specifically cleaves Watson-Crick base-paired RNA stems, and RNase T1, which cleaves 3′ of single-stranded G residues (Nichols and Yue, 2008). Rather than the more traditional approach of using radiolabeled RNAs and denaturing polyacrylamide gel electrophoresis, we employed for our footprinting studies 5′-fluor-labeled PSTVd RNA and capillary electrophoresis coupled to laser-induced fluorescence detection (Sobczak and Krzyzosiak, 2002). Comparison of RNase V1 and T1 cleavage sites on PSTVd demonstrated that they were mostly mutually exclusive, consistent with the fact that V1 and T1 cleave double- and single-stranded RNA, respectively. Quantitative comparison of V1 and T1 cleavage patterns of PSTVd alone or TFIIIA-7ZF-bound PSTVd reproducibly identified the left-terminal region of PSTVd, spanning nucleotides 331 to 347 (and covering loops 3 to 5), as the site protected by TFIIIA-7ZF against RNase cleavage in three independent experiments (Figure 6; Supplemental Figure 10). Furthermore, TFIIIA-7ZF binding protected mostly the lower strand of nucleotides in this region. By contrast, the TFIIIA-9ZF binding site on PSTVd was mapped to the right-terminal region from G168 to U177 (Figure 6; Supplemental Figure 11). These mutually exclusive binding sites suggest different roles for the two variants. The right-terminal region, where TFIIIA-9ZF binds, plays a role in systemic trafficking of PSTVd rather than replication (Zhong et al., 2008). The left-terminal region, where TFIIIA-7ZF binds, is critical for initiation of PSTVd transcription (Kolonko et al., 2006), replication (Zhong et al., 2008), and Pol II binding in vitro (Bojić et al., 2012) (Figure 6). Thus, the nonoverlapping PSTVd binding sites of the TFIIIA variants, together with their distinct subcellular locales, provide strong support to the idea that TFIIIA-7ZF is a genuine transcription factor that aids in the RNA-templated RNA transcription by Pol II in planta.
Figure 6.
Summary of RNase Protection (Footprinting) Assay Data Depicting the Binding Sites of TFIIIA-7ZF and TFIIIA-9ZF in the PSTVd Genome.
Pol II Interacts with TFIIIA Variants
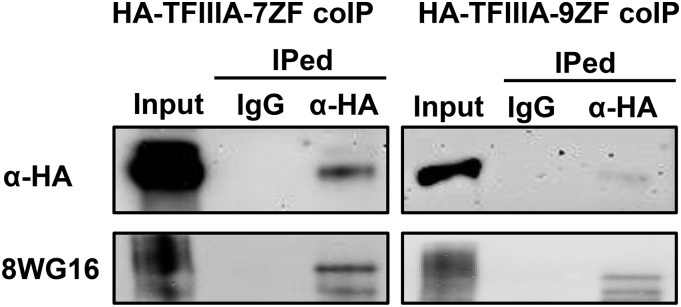
If TFIIIA-7ZF functions as a transcription factor for Pol II-mediated PSTVd transcription, it should interact with Pol II in vivo. To test this idea, we performed coimmunoprecipitation experiments. After using agroinfiltration to express TFIIIA-7ZF-YFP/HA in N. benthamiana leaves, we immunoprecipitated TFIIIA-7ZF-YFP/HA with anti-HA resin and detected the presence of Pol II largest subunit with the mouse monoclonal antibody 8WG16 (two independent biological replicates). The presence of Pol II and TFIIIA-7ZF in the immunoprecipitated pellet was validated by immunoblotting (Figure 7), clearly demonstrating the interaction of Pol II and TFIIIA-7ZF in vivo. While it is beyond the scope of this study to uncover structural and mechanistic differences in the interaction of the two TFIIIA variants with Pol II and Pol III, the interaction of Pol II and TFIIIA-7ZF in vivo provides a physical basis for the cellular outcomes for PSTVd replication that we observed.
Figure 7.
Coimmunoprecipitation of TFIIIA-7ZF and TFIIIA-9ZF with Pol II.
YFP/HA-dual-tagged TFIIIA-7ZF and TFIIIA-9ZF served as baits. Nonspecific mouse IgG-conjugated resin served as a negative control.
Surprisingly, when we applied the same procedures to test any possible interaction between Pol II and TFIIIA-9ZF-YFP/HA (three biological replicates), we obtained a similar result as that of TFIIIA-7ZF-YFP/HA (Figure 7). TFIIIA-9ZF has been well established to participate in Pol III-catalyzed transcription of 5S rDNA, and it has no other known function as a transcription factor. However, previous studies have shown that Pol II and Pol III share several transcription factors (Meyers and Sharp, 1993; Raha et al., 2010). Thus, though the presence of TFIIIA-9ZF in the nucleoplasm is negligible as assessed by confocal microscopy (Figure 5), we cannot rule out that a small amount of TFIIIA-9ZF interacted with Pol II. A more reasonable explanation is that TFIIIA-9ZF interacts with Pol II post-cell lysis, which has been observed in the interactions between Argonaute proteins and small RNAs (Riley et al., 2012). These two possibilities await clarification by future experiments. Nevertheless, our data clearly demonstrate a physical basis for the interaction of Pol II and TFIIIA-9ZF.
TFIIIA-7ZF Enhances Pol II-Catalyzed in Vitro Transcription of the PSTVd RNA Genome
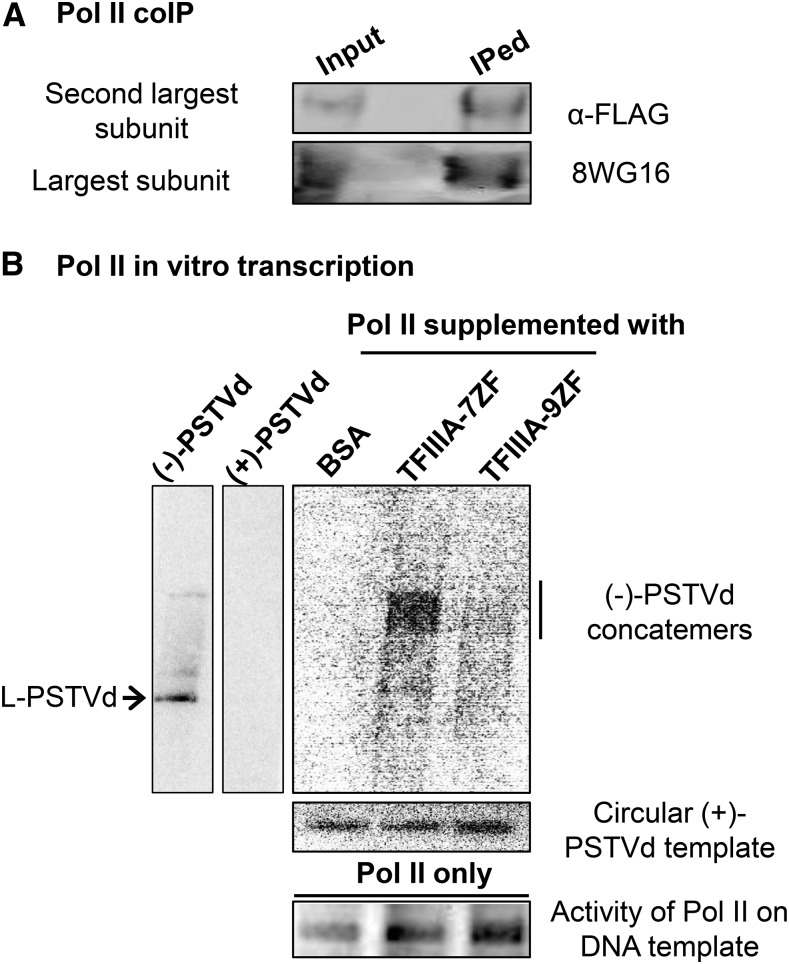
To obtain direct evidence to support the involvement of TFIIIA-7ZF in Pol II-catalyzed transcription of an RNA template, we tested the activity of partially purified Pol II to transcribe a PSTVd circular genome in vitro with or without supplementation of TFIIIA variants. We first followed an established protocol (Ream et al., 2009) to purify Pol II using an affinity approach that exploits a FLAG tag on the second largest subunit of Pol II from Arabidopsis and validated the presence of Pol II by detection of both second largest subunit (using anti-FLAG serum) and the largest subunit (8WG16 serum). As shown in Figure 8, both the largest and second largest subunits were present in the immunoprecipitated pellet, suggesting the presence of an active Pol II holoenzyme. We then set up an in vitro transcription using partially purified Pol II and PSTVd circular genome (Rackwitz et al., 1981).
Figure 8.
TFIIIA-Mediated Increase in in Vitro Transcription by Pol II.
(A) Immunoprecipitation of the FLAG-tagged second largest subunit of Pol II from a partially purified Pol II holoenzyme.
(B) Ability of partially purified Pol II holoenzyme to transcribe circular (+)-PSTVd in the presence of BSA, TFIIIA-7ZF, or -9ZF. The position of the linear unit-length PSTVd and the concatemers are indicated by and an arrow and a bar, respectively. The transcription products were detected by using a 32P-labeled strand-specific probe. The specificity of the probe is shown by the detection of T3 phage polymerase generated (+)-PSTVd (40 ng) and (−)-PSTVd (10 ng) in vitro transcripts.
Based on calculations informed by the results from our EMSA analysis, we supplemented these reactions with amounts of TFIIIA-7ZF and -9ZF that permit complete formation of PSTVd RNA-TFIIIA RNP complexes. BSA was used as a negative control. After a 4-h incubation at 28°C, the reactions were treated with proteinase K and were directly subject to denaturing PAGE. A 32P-labeled probe complementary to the (−)-PSTVd was used for detection of transcription products. The probe showed high specificity in recognizing (−)-PSTVd with no cross-reactivity to (+)-PSTVd (Figure 8B). In vitro transcription of the PSTVd circular genome by our Pol II preparation supplemented with BSA showed barely detectable signal; in contrast, supplementing with TFIIIA-7ZF significantly enhanced the transcriptional efficiency as evident from the accumulation of (−)-PSTVd concatemer RNAs (Figure 8B). These concatemers resemble the (−)-PSTVd concatemers that accumulate in protoplasts (Qi and Ding, 2002). Interestingly, TFIIIA-9ZF also weakly enhanced in vitro transcription but clearly with a much reduced efficiency as compared with that of TFIIIA-7ZF (Figure 8B). These results were reproducible in at least two trials.
We used two controls to ensure that the enhanced Pol II transcription of PSTVd in the presence of TFIIIA-7ZF was not due to unequal loading of either RNA templates or Pol II complexes (Figure 8B). While one aliquot (3 μL) of each reaction was used to verify the lack of variability in circular (+)-PSTVd templates, another (1 μL) was used to transcribe a CaMV 35S promoter-containing plasmid to compare the level of Pol II activities in each reaction. Results from these control experiments show near equal loading of Pol II holoenzyme and the circular (+)-PSTVd template (Figure 8B) and confirm that the enhanced transcription of PSTVd RNA by Pol II upon addition of TFIIIA-7ZF is not due to technical artifacts.
DISCUSSION
The rolling circle replication of the plant viroid PSTVd involves transcription, cleavage, and ligation (Supplemental Figure 1B). When Nohales et al. (2012) demonstrated that tomato (Solanum lycopersicum) DNA ligase 1 catalyzes the ligation step of PSTVd, they pointed out how PSTVd and other viroid relatives engender efficient replication by switching the template and substrate specificity of a DdRP to RdRP and of a DNA to an RNA ligase. Here, our key findings help shed further light on the modus operandi of this noncoding RNA pathogen. Foremost, we provide firm evidence for association of PSTVd and Pol II in N. benthamiana. Second, we show that TFIIIA-7ZF, a shorter splice variant of the full-length TFIIIA-9ZF, aids PSTVd transcription in planta likely due to its role in Pol II transcription. Consistent with this hypothesis, we demonstrate that TFIIIA-7ZF binds to the left-terminal region of PSTVd, which has already been implicated in transcription initiation (Kolonko et al., 2006; Zhong et al., 2008; Bojić et al., 2012) and interaction with Pol II in vivo. Furthermore, TFIIIA-7ZF directly enhances a reconstituted Pol II transcription of circular (+)-PSTVd template in vitro. Because TFIIIA-7ZF does not have any sequence signature associated with a ribonuclease, we can exclude the possibility of it being involved in cleavage of the PSTVd transcripts, and there is not much support to invoke its role in ligation especially given the earlier findings on the tomato DNA ligase (Nohales et al., 2012).
It is instructive to place our findings in the context of other proteins that are known to influence either PSTVd infectivity or the specificity of DdRPs. Previous yeast three-hybrid studies showed that tomato VirP1 binds to the right-terminal region of (+)-PSTVd (Gozmanova et al., 2003; Maniataki et al., 2003), and VirP1 was indeed shown to be critical for viroid infectivity. Because the right-terminal region is not important for PSTVd replication based on genetic evidence (Zhong et al., 2008) and it is not the Pol II-binding site, VirP1 may not be involved in replication. Indeed, though VirP1 appears to be important for PSTVd infection, there is no evidence for its involvement in replication (Kalantidis et al., 2007); instead, hypotheses center around its possible involvement in systemic spread of PSTVd (Maniataki et al., 2003) or in RNA-mediated DNA methylation (Kalantidis et al., 2007). δ70, the “housekeeping” sigma factor involved in the transcription of most genes in growing bacterial cells (Gruber and Gross, 2003), is necessary for 6S RNA template transcription in E. coli (Wassarman and Saecker, 2006). Thus, this protein aids transcription of both DNA and RNA templates by a DdRP, though the mechanism by which it accomplishes a switch in substrate specificity remains unclear. HDV encodes two proteins, small and large delta antigens (HDAg-S and HDAg-L) that are involved in HDV transcription/replication and packaging, respectively (Tseng and Lai, 2009). HDAg-L is encoded by a read-through of mRNA encoding the HDAg-S, thereby differing from the latter by having 19 extra amino acids in the C terminus. Many HDAg- and HDV RNA-interacting host factors have been identified with functions in different aspects of the HDV life cycle, but none have been demonstrated to have a role as an RNA binding transcription factor for replication (Greco-Stewart and Pelchat, 2010).
The biogenesis of TFIIIA-7ZF and its role in PSTVd replication are remarkable. By alternative splicing-mediated excision of the first two zinc fingers, a Pol III-specific transcription factor for DNA-templated transcription (TFIIIA-9ZF) is converted into a Pol II-associated transcription factor for RNA-templated transcription (TFIIIA-7ZF). Given that the endogenous function of TFIIIA-7ZF is currently unknown, the role of this protein in other novel DNA- or RNA-templated transcription activities remains to be addressed. Our data clearly support the notion that TFIIIA-7ZF has acquired distinct functions compared with the canonical TFIIIA-9ZF given its distinct subcellular localization, nonoverlapping binding sites on PSTVd, different substrate binding preferences, and variable capacity in facilitating the accumulation of PSTVd. Moreover, the spatiotemporal expression of TFIIIA-7ZF (Layat et al., 2012) suggests developmental control, as exemplified by its notable absence from seeds.
The role of TFIIIA-9ZF in PSTVd biogenesis remains unclear. TFIIIA-9ZF can interact with Pol II and facilitate Pol II transcribing (+)-PSTVd circular RNA template in vitro, albeit with a reduced efficiency compared with TFIIIA-7ZF. However, the strict nucleolar subcellular locale of TFIIIA-9ZF makes its interaction with Pol II less likely. Consistent with this expectation, TFIIIA-9ZF overexpression had no detectable effect on PSTVd accumulation in planta. Moreover, TFIIIA-9ZF cannot recognize (−)-PSTVd, which rules out its role as a transcription factor for transcribing (−)-PSTVd template. On the other hand, binding of nucleolar-localized TFIIIA-9ZF to the (+)-PSTVd RNA prompts the idea that it might aid nucleolar transport and/or exit of the (+)-PSTVd RNA generated during replication.
Our findings, which indicate that eukaryotic cells utilize dedicated transcription factors to assist DdRPs to perform RNA-templated RNA synthesis, should motivate a broader search for such proteins. Equally profitable will be structural and mechanistic studies that explore the molecular underpinnings for the substrate specificity switch and processivity gains when a specific transcription factor alters the function of Pol II. In this regard, establishing a robust in vitro transcription system with PSTVd, Pol II, and TFIIIA-7ZF provides a valuable starting point to test the contribution of auxiliary protein factors. Additionally, given that viroids cause diseases of agronomic significance in many crops (Kovalskaya and Hammond, 2014), chemical or genetic strategies that change the ratio of the two spliced forms of TFIIIA merit consideration as a protective strategy.
METHODS
Plant Materials and Growth Conditions
All plants were grown in a growth chamber controlled at 14-h light (Sylvania fluorescent bulb tubes; F96T12HO), 27°C/10-h dark, 24°C cycles. PSTVd infection was achieved by inoculation of 200 ng of (+)-PSTVd in vitro transcripts onto four true leaf stage seedlings of Nicotiana benthamiana. PSTVd infection (20 d postinoculation) was verified by RNA gel blotting using (−)-PSTVd riboprobes. Details regarding RNA purification and RNA gel blotting are described below.
Molecular Cloning
Total RNA of N. benthamiana leaves and Arabidopsis thaliana flowers was extracted using RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s instructions. Reverse transcription was performed using SuperScript III transcriptase (Life Technologies) and the supplied oligo(dT) primer. To express desired cDNAs or gene fragments in planta or in Escherichia coli, gene-specific primers (see Supplemental Table 1 for primer details) were used for PCR amplification using FideliTaq polymerase (USB). The PCR products were inserted into pCR8/GW/TOPO vector (Life Technologies) to generate entry clones for easy access to recombination-based cloning. The CaMV 35S promoter construct for Pol II in vitro transcription was PCR amplified using pRS300 plasmid and the primers (35Sf and 35Sr) described in Supplemental Table 1. The PCR product was recombined to pDONR207 vector using BP clonase (Life Technologies). The sequence-verified entry clones were subject to recombination reactions to generate destination expression clones using LR clonase II (Life Technologies; see Supplemental Table 1 for destination vectors). All constructs were verified by DNA sequencing at the OSU Plant-Microbe Genomics Facility.
Recombinant Protein Purification
Recombinant TFIIIA proteins with an intein-chitin binding domain (CBD) tag were overexpressed using the E. coli BL21(DE3) Rosetta strain (EMD Millipore). Bacterial cultures were grown to A600 ∼0.6, before adding 0.4 mM IPTG to induce protein expression. After induction, bacterial cultures were grown at 20°C for 12 to 16 h before they were harvested. Bacterial cells from a 1.6-liter culture were pelleted by centrifugation at 6000g for 20 min and resuspended in a lysis buffer containing 20 mM Tris-HCl (pH 8.5), 400 mM NaCl, 20 µM PMSF, 5 µM ZnSO4, and 0.1% (v/v) Triton X-100. Sonication with a Bioruptor (Diagenode) was then used to lyse the cells, followed by centrifugation at 15,000g for 1.5 h at 4°C. The supernatant (cell lysate) was collected and incubated for 1 h with 2 mL of 50% slurry of chitin resin (New England Biolabs) before loading onto an empty EconoPac gravity-flow column (14 cm length, 1.5 cm internal diameter; Bio-Rad Laboratories). After washing with 100 mL of the lysis buffer, incubation for 18 h at 4°C in a cleavage buffer (20 mM Tris-HCl, pH 8.5, 500 mM NaCl, 50 mM DTT, and 5 µM ZnSO4) was used to trigger on-column cleavage of the intein-CBD tag according to the instructions in the IMPACT kit (New England Biolabs). Fractions containing tagless proteins were dialyzed against 10 mM Tris-HCl, pH 7.5, 20 mM KCl, 5 µM ZnSO4, and 1 mM DTT. Protein concentrations were estimated by Coomassie Brilliant Blue staining of an SDS-PAGE gel using BSA of known concentrations as reference standards. Proteins were concentrated with an Amicon protein concentrator (EMD Millipore) to ∼20 µM before being stored in 50-μL aliquots at −80°C. Proteins used for RNA footprinting assays were dialyzed against 10 mM Tris-HCl (pH 7.5), 20 mM KCl, 200 mM NaCl, 5 µM ZnSO4, and 1 mM DTT and then concentrated to ∼50 µM as described above.
Generation of α-TFIIIA Antibodies
To generate antibodies against N. benthamiana TFIIIA, the His6-TFIIIA-7ZF construct was cloned (Supplemental Table 1) and transformed into the E. coli BL21(DE3) Rosetta strain (EMD Millipore) for expression. The His6-tagged TFIIIA-7ZF protein was purified with the aid of a Ni-NTA resin (Qiagen) under denaturing conditions using protocols provided by the manufacturer. The purified proteins were separated by SDS-PAGE (8% [w/v] polyacrylamide) and identified by Coomassie Brilliant Blue staining. The dominant 34-kD band corresponding to TFIIIA-7ZF was excised from the gel and shipped to Cocalico Biologicals to raise polyclonal antibodies against TFIIIA-7ZF in rabbits. The antibodies were further purified from the serum following a previous report (Kim et al., 2003).
RNA Preparation in Vitro
To prepare 32P-labeled PSTVd (+)-strand and (−)-strand probes for EMSAs, SmaI-linearized pInt95-94(+) and pInt95-94(−) were used as templates for in vitro transcription with a T3 MEGAscript (Life Technologies). PSTVd transcripts were purified using the MEGAClear kit (Life Technologies). This (+)-PSTVd synthesized was also used for N. benthamiana infection. The purified PSTVd RNAs were radiolabeled using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (100 µCi/µmol; Perkin Elmer Life Sciences). Unincorporated ATP was removed using Sephadex G-25 columns (GE Healthcare Life Sciences) to obtain probes with specific activities of 2 to 4 × 106 dpm/pmol.
PSTVd circularization was performed following the method of Beaudry and Perreault (1995). Briefly, 10 μg of linear PSTVd transcripts was incubated at 37°C for 2 h in a final volume of 200 μL containing 150 units of calf intestinal alkaline phosphatase (New England Biolabs), 80 units of RNase inhibitor (New England Biolabs), and 1× calf intestinal alkaline phosphatase buffer. After phenol-chloroform extraction and ethanol precipitation, the pellet was resuspended in 10 μL DEPC water and incubated at 37°C for 2 h in a final volume of 100 μL containing 100 units of T4 polynucleotide kinase (New England Biolabs), 80 units of RNase inhibitor, 10 mM ATP, and 1× T4 polynucleotide kinase buffer. After extraction and precipitation, the pellet was resuspended in 20 μL DEPC water and incubated at 37°C for 2 h in a final volume of 200 μL containing 150 units of T4 RNA ligase (New England Biolabs), 80 units of RNase inhibitor, 10 mM ATP, and 1× T4 RNA ligase buffer. The circular transcripts were separated from linear transcripts by denaturing polyacrylamide gel electrophoresis. Circular products were detected by UV shadowing, excised, and eluted with 0.3 M NaCl at 4°C overnight. After removal of gel pieces, the products were ethanol precipitated, dried, and dissolved in DEPC water.
EMSAs
Binding reactions were performed at 25°C for 30 min and entailed incubations of 32P-labeled RNAs (20,000 dpm) with increasing concentrations of recombinant proteins (as specified in the figures). The binding assays (10 μL final volume) contained 20 mM Tris-HCl (pH 7.5), 35 mM KCl, 5 µM ZnCl2, 3.5 mM MgCl2, 10 nM yeast tRNA (Life technologies), and 10% (v/v) glycerol. Electrophoresis was performed at 4°C in 6% (w/v) polyacrylamide (29:1) gels for 2 h at 120 V using Tris borate buffer (65 mM Tris and 22.5 mM boric acid, pH 8). The gels were then fixed, dried, exposed overnight in a phosphor imager cassette, and visualized using a Personal Molecular Imager (Bio-Rad Laboratories). The intensity of each band on the gel was quantified using Quantity One software (Bio-Rad Laboratories). The binding curves were obtained by plotting the fraction of RNA bound with proteins, Ɵ (total bound RNA in all bands divided by the sum of bound and unbound RNAs) as a function of protein concentration. Origin software (OriginLab) was used to calculate the binding constants by fitting the data to the Hill equation:
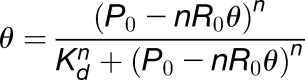 |
where R0 is the total concentration of RNA added in each lane, P0 is the total concentration of proteins added in each lane, and n is the Hill coefficient. Kd and n values listed in the figures represent the mean ± sd from three independent experiments; R2 was > 0.98 in all curve fits.
Agroinfiltration
PSTVd-infected N. benthamiana plants were infiltrated with a GV3101 strain of Agrobacterium tumefaciens harboring the appropriate DNA construct following a previously described procedure (Itaya et al., 2007). Three days postinoculation, infiltrated leaves were used for analyzing PSTVd accumulation and/or protein accumulation and subcellular localization patterns. For examining PSTVd accumulation, ∼2 g of leaves was ground in liquid nitrogen for nuclear extraction. A high SDS buffer (100 mM Tris-HCl, pH 8.8, 150 mM DTT, 30% [v/v] glycerol, 5% [w/v] SDS, and 1 mM PMSF) was used for lysis of nuclei. One-half of the lysate was used for immunoblotting, with the remainder used for RNA gel blotting after RNA extraction.
RNA Extraction and RNA Gel Blots
Total RNA from leaves (1 g fresh weight) or from immunoprecipitated pellets was isolated using Trizol reagent (Life Technologies). RNA samples were electrophoresed on 5% (w/v) polyacrylamide/8 M urea gels, transferred to Hybond-XL nylon membranes (Amersham Biosciences) using a vacuum blotting system (Amersham), and immobilized by UV cross-linking. After overnight hybridization to [α-32P]UTP-labeled riboprobes at 70°C in ULTRAhyb reagent (Life Technologies), the membranes were washed twice at 70°C in 2× SSC-0.1% (w/v) SDS (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.2× SSC-0.1% (w/v) SDS and exposed to a storage phosphor screen (Kodak). For detecting PSTVd, probes were obtained by transcribing in vitro HindIII-linearized pRZ6-2 template using a T7 MAXIscript kit (Life Technologies). For detecting 5S rRNA, probes were obtained by transcribing in vitro NcoI-linearized pGEMT-5S rRNA template using a SP6 MAXIscript kit (Life Technologies). The probes were purified by NucAway column (Life Technologies).
RNA Immunoprecipitation, Coimmunoprecipitation, and Immunoblotting
The tagged TFIIIA proteins were expressed in planta via agroinfiltration in PSTVd-infected N. benthamiana plants. At 4 d postinfiltration, 4 g of leaves were ground into a fine powder using liquid nitrogen. (Note: To demonstrate the presence of circular PSTVd RNA in the Pol II immunoprecipitates, we used 40 g of PSTVd-infected leaves.) The leaf powder was then incubated in a nuclear extraction buffer (0.4 M sucrose, 10 mM Tris-HCl, pH 8, 5 mM DTT, 10 mM NaF, and a plant protease inhibitor cocktail from Sigma-Aldrich), filtered through two layers of Miracloth (GE Healthcare), and then centrifuged at 2100g and 4°C for 10 min. The pellets were washed with buffer I (0.25 M sucrose, 10 mM Tris-HCl, pH 8, 10 mM MgCl2, 1% [v/v] Triton X-100, 5 mM DTT, 10 mM NaF, and a plant protease inhibitor cocktail [Sigma-Aldrich]) for 10 min with shaking at 4°C, followed by centrifugation at 12,000g for 10 min at 4°C.
For RNA immunoprecipitation, the pelleted nuclei were lysed by first incubating with buffer II (50 mM Tris-HCl, pH 7.9, 0.32 M sucrose, 5 mM CaCl2, 10 mM MgCl2, 10 mM NaF, 0.2% [v/v] Nonidet P-40, 20 units of Turbo DNase [Life Technologies], RNaseOUT [Life Technologies], and a plant protease inhibitor cocktail [Sigma-Aldrich]) at 4°C for 30 min. After adding NaCl to a final concentration of 0.36 M, the mixture was sonicated on the high setting in a Bioruptor (Diagenode) for 20 min, and centrifuged for 3 min at 16,100g and at 4°C. The supernatant was immediately diluted to 150 mM NaCl. The prepared nuclear lysates were divided into four parts: 2 μL served as RNA extraction input, 20 μL served as protein input, and 200 μL each was incubated with nonspecific antibody EZview Red c-myc-agarose (Sigma-Aldrich) and specific antibody EZview Red HA-agarose (Sigma-Aldrich), respectively, for 2 h with shaking at 4°C. The samples were then spun down at 600g and at 4°C, and the pellets washed four times with 1× TBS supplemented with 0.1% (v/v) Triton X-100. The agarose beads were treated with 2× SDS-PAGE loading buffer at 90°C for 5 min. One half of the eluate was used for immunoblotting to detect the presence of target proteins and the other half used to detect specific coprecipitated RNAs. For RNA detection, the RNA in the precipitates was purified via standard Trizol RNA extraction protocol (Life Technologies). RT-PCR or RNA gel blots were performed to analyze the presence of PSTVd, 5S rRNA, and U6 snRNA (see Supplemental Table 1 for primers) using the reagents described above. To normalize and quantify the results, 1% of RNA input and 10% of the protein input were used as the reference to calculate the pull-down efficiency in the final immunoprecipitated fractions.
For coimmunoprecipitation of Pol II and TFIIIA, we used the protocols described above with the following modifications. First, RNaseOUT was omitted. Second, micrococcal nuclease (New England Biolabs) was used instead of Turbo DNase. For Pol II RNA immunoprecipitation, mouse monoclonal antibodies (8WG16; GeneTex) were used in combination with Direct Magnetic IP/Co-IP kit (Life Technologies) following the manufacturer’s instructions. Mouse IgG (Life Technologies) was employed as a negative control.
FLAG-tagged Pol II construct was a kind gift from Craig Pikaard (Indiana University). To purify FLAG-tagged Pol II, we followed the instructions described elsewhere (Ream et al., 2009). In brief, 12 g of N. benthamiana leaves overexpressing FLAG-Pol II was collected and ground to a fine powder in liquid nitrogen, followed by extraction using 30 mL of buffer containing 20 mM Tris-HCl (pH 7.5), 300 mM NaCl, 5 mM MgCl2, 5 mM DTT, 1mM PMSF, 5 mM NaF, 5% (v/v) glycerol, and 1% (w/v) plant protease inhibitor cocktail (Sigma-Aldrich). The lysate was filtered through double-layered Miracloth and centrifuged at 16,000g for 15 min at 4°C. The supernatant was incubated with 60 μL anti-FLAG resin (BioLegend) at 4°C for 3 h and washed twice with 15 mL of cold wash buffer containing 20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 5 mM MgCl2, 5 mM DTT, 1mM PMSF, 5 mM NaF, and 0.5% (v/v) Nonidet P-40. Fifteen milliliters of CB100 buffer [20 mM HEPES-KOH, pH 7.9, 40 mM (NH4)2SO4, 5 mM DTT, 1 mM PMSF, and 20% (v/v) glycerol] was used to wash the resin, which was eventually resuspended in 60 μL of CB100 buffer and stored as 10 μL aliquots at −80°C.
For immunoblotting, we followed standard procedures for SDS-PAGE and semidry membrane transfer. We used the following antibodies in our studies: 8WG16 (GeneTex) at 1:1000, anti-Histone H3 (Genscript) at 1:1000, anti-TFIIIA (described above) at 1:400, anti-FLAG L5 monoclonal antibody (Life Technologies) at 1:1000, horseradish peroxidase (HRP)-conjugated anti-HA (Sigma-Aldrich) at 1:1000 dilution, HRP-conjugated anti-mouse IgG (Sigma-Aldrich) at 1:8000 dilution, HRP-conjugated anti-rat IgG (Sigma-Aldrich) at 1:4000 dilution, and HRP-conjugated protein A (Life Technologies) at 1:1000 dilution. (Note: NaF was omitted from all buffers when extracting Pol II for immunoblotting.)
Confocal Microscopy
Confocal laser scanning microscopy was used to study the subcellular localization of YFP- and RFP-tagged proteins in PSTVd-infected N. benthamiana plants. To identify nuclei, the leaf tissues were infiltrated with PBS (pH 7.4) containing 2 ng/μL DAPI. The samples were examined using a Nikon Confocal A1 with a 100× oil immersion objective lens and appropriate filters for DAPI, YFP, and RFP. The images were captured and processed by NIS-Elements software (Nikon).
In Vitro Synthesis and Fluor-Labeling of PSTVd RNA for RNase Protection Assays
To prepare carboxyfluorescein-labeled PSTVd, plasmid pInt95-94(+) cDNA was digested with SmaI (New England Biolabs) for 2 h and used as a template for in vitro transcription with the T3 MEGAscript kit (Life Technologies). We used 20 µg of DNA template in a 400-μL reaction containing 1× T3 buffer, 10 mM DTT, 0.005 units/μL inorganic pyrophosphatase (New England Biolabs), 4.8 mM 5′-azido-5′-deoxyguanosine (synthesized in house) (Brear et al., 2009; Wallace, 2013), 1.2 mM GTP, 6 mM ATP, 6 mM CTP, 6 mM UTP, and 40 μL T3 RNA polymerase (Life Technologies). After transcription, the DNA templates were digested with Turbo DNase and the RNA transcripts were purified using the MEGAClear kit (Life Technologies) for subsequent 5′ labeling with 6-carboxyfluorescein (FAM). In a 20-μL labeling mixture, 110 µg of PSTVd transcripts were incubated with 60 mM Tris-HCl (pH 7.5), 7.2 mM 5(6)-propargylamidofluorescein (synthesized in house; Wallace, 2013), 2% (v/v) acetonitrile, 25% (v/v) DMSO, 4 mM Cu(OAc)2 (pH 7.5), and 40 mM sodium ascorbate (pH 7.5) in the dark for 1 h at 25°C. The reaction was terminated by the addition of thiourea to 200 mM. To examine the integrity of fluorescein-labeled PSTVd, a small aliquot of labeling mixture was separated by 8% (w/v) polyacrylamide gel containing 7 M urea and Tris borate EDTA buffer, and assessed using Typhoon Trio Molecular Imager (GE Healthcare Life Sciences). Further purification of fluorescein-labeled PSTVd was performed using the MEGAClear kit (Life Technologies), followed by quantification using the NanoDrop ND-2000 (Thermo Scientific).
To prepare FAM-labeled PSTVd fragments that were used as size markers in RNase protection (footprinting) assays, PCR reactions were performed to amplify DNA templates from pInt95-94(+) prior to in vitro transcription. Primers are described in Supplemental Table 1 for the generation of DNA templates that were used in in vitro transcriptions to make PSTVd deletion variants corresponding to the first 80, 171, 230, 267, or 314 nucleotides, respectively. PCR products were purified using native PAGE, eluted from excised gel slices, and precipitated by ethanol. In vitro transcription and fluor labeling was performed as described above for the full-length RNA.
RNase Protection (Footprinting) Assays
PSTVd RNA was first refolded by heating a solution of FAM-labeled PSTVd (40 ng) and 400 ng yeast tRNA (Life Technologies) at 65°C for 5 min, and then slowly cooling to 25°C. The refolded RNA was incubated at 25°C for 30 min in a 10-μL solution containing 2.6 µg TFIIIA-7ZF (or 0.86 µg TFIIIA-9ZF) in 20 mM Tris-HCl (pH 7.5), 35 mM KCl, 5 µM ZnCl2, 3.5 mM MgCl2, and 10% (v/v) glycerol. The amount of TFIIIA and PSTVd added in each reaction was established based on EMSA results to allow complete binding of PSTVd to TFIIIA proteins. RNase T1 or V1 (Life Technologies) was then added to the binding mixture, which was further incubated at 25°C for 1 min (100 units of RNase T1) or 2 min (0.05 units of RNase V1). Control digestion with the refolded RNA was performed in the absence of proteins. Two hundred microliters of Ribozol RNA extraction reagent (Amresco) was added to stop the reaction, followed by mixing with 200 μL of chloroform:isoamyl alcohol (24:1) (Amresco). After centrifugation at 16,000g for 5 min, RNA from the aqueous phase was extracted and precipitated using ethanol and glycoblue (Life Technologies). After incubating at −80°C for 30 min, the RNA was pelleted by centrifugation at 16,000g for 15 min, and dissolved in nuclease-free water (Life Technologies). Digested RNA was then subject to capillary sequencing at the OSU Plant-Microbe Genomics Facility. To aid in assignment of sizes and nucleotide locations to peaks in the capillary electrophoretograms (CEs), we independently analyzed 40 ng of PSTVd fragments corresponding to 1 to 80, 1 to 171, 1 to 230, 1 to 267, or 1 to 314 nucleotides. The CE results were analyzed by GeneMapper software (Applied Biosystems).
We used the following procedures to analyze the footprinting data. First, the peak heights in CEs were normalized between free PSTVd and TFIIIA-bound PSTVd samples digested by the same RNase based on the assumption that the sum of signals, which is indicative of the total amount of input PSTVd, should be equal between two samples within one experimental repeat. Second, the difference in height of each peak between free PSTVd and PSTVd-TFIIIA samples was divided by the height of the same peak in the free PSTVd sample to generate a Q (difference quotient) value for every position. We used an arbitrary cutoff of Q > 0.1 for further analysis. Lastly, we identified positions where binding of PSTVd to TFIIIA led to a reduction in peak intensities (caused by T1- or V1-mediated cleavage) that was reproducible in at least three repeats. These positions were interpreted as protected by TFIIIA and taken to indicate the binding site. The Q values range from 0.34 to 0.87 for TFIIIA-7ZF and from 0.15 to 0.69 for TFIIIA-9ZF.
Pol II in Vitro Transcription
The following protocols were modified based on previous reports (Rackwitz et al., 1981; Ream et al., 2009). FLAG-Pol II-bound resin from immunoprecipitation (10 μL aliquot, equivalent to 2 g of FLAG-Pol II expressing tissue) was divided into two parts. One microliter of resin was used for transcribing DNA template and 9 μL was used for transcribing circular PSTVd. For DNA-dependent RNA transcription, 10 μL of transcription reactions contained 50 mM HEPES-KOH, pH 7.9, 2 mM MnCl2, 100 mM KCl, 10% (v/v) glycerol, 10 units of RNase inhibitor, 500 μM rATP, 500 μM rGTP, 500 μM rCTP, 350 μM rUTP, 150 μM Digoxigenin-11-UTP (Roche), 5 ng/μL DNA template, and 1 μL FLAG-Pol II resin. For RNA-dependent RNA transcription, recombinant proteins (BSA, TFIIIA-7ZF, and TFIIIA-9ZF) were pretreated with 1 unit of Turbo DNase (Life technologies) for 10 min at 37°C. Circular PSTVd, protein factors, and FLAG-Pol II-bound resin was then incubated at 28°C for 15 min before NTP was added. Transcription reactions (20 μL) containing 50 mM HEPES-KOH, pH 7.9, 1 mM MnCl2, 6 mM MgCl2, 40 mM (NH4)2SO4, 10% (v/v) glycerol, 20 unit of RNase inhibitor, 500 μM rATP, 500 μM rGTP, 500 μM rCTP, 500 μM rUTP, 10 ng/μL PSTVd, 4 μM protein, and 9 μL FLAG-Pol II were performed at 28°C for 4 h. The reactions were terminated by proteinase K (Invitrogen) treatment for 15 min at 37°C, followed by incubation at 95°C for 5 min. The transcription products were separated on 5% (w/v) polyacrylamide/8 M urea gel, transferred to Hybond-XL nylon membranes (Amersham Biosciences) by Trans-Blot SD semidry electrophoretic transfer cell (Bio-Rad), and immobilized by UV cross-linking. Transcripts labeled with digoxigenin-11-UTP were detected via DIG Northern Starter Kit following the manufacturer’s instructions (Roche) using chemiluminescence substrate (Bio-Rad). Signals were obtained using myECL imager (Thermo Scientific). RNA transcripts from circular PSTVd were detected by 32P-labeled strand-specific probes for (−)-PSTVd using the abovementioned procedures.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At5g52470 (FRI), At4g21710 (NRPB2), KP824744 (5S rRNA), X51447 (U6 snRNA), KX098601 (N. benthamiana TFIIIA-7ZF), and KX098600 (N. benthamiana TFIIIA-9ZF).
Supplemental Data
Supplemental Figure 1. Schematic representation of PSTVd secondary structure and asymmetric rolling circle replication model.
Supplemental Figure 2. Schematic representation of TFIIIA mRNA splicing variants and their translational products.
Supplemental Figure 3. Demonstration of the specificity of Pol II antibody (8WG16).
Supplemental Figure 4. In vivo interactions between Pol II and circular (+)-PSTVd revealed by RNA immunoprecipitation assays.
Supplemental Figure 5. Expression of TFIIIA-9ZF and TFIIIA-7ZF in N. benthamiana.
Supplemental Figure 6. Binding curves for the in vitro interaction between TFIIIA proteins and PSTVd as determined from EMSA results.
Supplemental Figure 7. Specific enhancement of PSTVd replication with ectopic expression of TFIIIA-7ZF.
Supplemental Figure 8. In vivo interactions between TFIIIA and PSTVd revealed by RNA immunoprecipitation assays.
Supplemental Figure 9. Z-serial confocal images from whole mounts of PSTVd-infected N. benthamiana leaves that ectopically express either TFIIIA-7ZF-YFP/HA or TFIIIA-9ZFYFP/HA.
Supplemental Figure 10. Capillary electrophoretograms from three independent experiments for RNase T1 and V1 footprinting of the PSTVd-TFIIIA-7ZF complex.
Supplemental Figure 11. Capillary electrophoretograms from three independent experiments for RNase T1 and V1 footprinting of the PSTVd-TFIIIA-9ZF complex.
Supplemental Table 1. Primers, vectors, and bacterial strains used for cloning.
Supplemental Data 1. N. benthamiana TFIIIA-9ZF and TFIIIA-7ZF cDNA sequences.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Biao Ding, a caring mentor and exceptional colleague, who provided oversight for this investigation since its inception but died shortly before submission of this article. We thank Tien-Hao Chen and Lien Lai in the Gopalan laboratory and Michael Zianni in the OSU Plant-Microbe Genomics Facility for assistance with RNase protection analyses. We thank Edward Behrman (OSU) for advice and oversight of synthesis of 5′-azido-5′-deoxyguanosine and propargylamidofluorescein. We thank Anna Dobritsa (OSU) for the use of her confocal microscope, David M. Bisaro (OSU) for critical reading of the manuscript, Craig Pikaard (Indiana University) for gifts of FLAG-tagged Pol II construct and purification protocol, and Iris Meier (OSU) for a gift of pK7WFR2.0 and critical reading of the manuscript. This work was supported by grants from the U.S. National Science Foundation (IOS-1051655 and IOS-1354635 to B.D.; MCB-0843543 to V.G.), a seed grant from the Ohio State University Center for RNA Biology to B.D. and V.G., and the Natural Science Foundation of China (NSFC No. 31420103904 to Y.L. and B.D.). Y.W. was supported by an Excellence in PMBB Graduate Fellowship (OSU), A.J.W. was supported by a NSF Research Experience for Undergraduates on-site grant (DBI-1062144 to Amanda Simcox and Susan Cole), and J.W. was supported by a Pelotonia Fellowship (OSU).
AUTHOR CONTRIBUTIONS
Y.W., J.Q., V.G., and B.D. designed research. Y.W., J.Q., S.J., and A.J.W. performed research. Y.W., J.Q., V.G., Y.L., and B.D. analyzed data. Y.W., J.Q., V.G., and B.D. wrote the article.
Glossary
- DdRP
DNA-dependent RNA polymerase
- RdRP
RNA-dependent RNA polymerase
- PSTVd
Potato spindle tuber viroid
- HDV
hepatitis delta virus
- ZF
zinc finger
- EMSA
electrophoretic mobility shift assay
- DAPI
4′,6-diamidino-2-phenylindole
- HRP
horseradish peroxidase
- FAM
6-carboxyfluorescein
- CE
capillary electrophoretogram
Footnotes
Articles can be viewed without a subscription.
References
- Barbazuk W.B. (2010). A conserved alternative splicing event in plants reveals an ancient exonization of 5S rRNA that regulates TFIIIA. RNA Biol. 7: 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudry D., Perreault J.P. (1995). An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Res. 23: 3064–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beven A.F., Lee R., Razaz M., Leader D.J., Brown J.W., Shaw P.J. (1996). The organization of ribosomal RNA processing correlates with the distribution of nucleolar snRNAs. J. Cell Sci. 109: 1241–1251. [DOI] [PubMed] [Google Scholar]
- Bojić T., Beeharry Y., Zhang J., Pelchat M. (2012). Tomato RNA polymerase II interacts with the rod-like conformation of the left terminal domain of the potato spindle tuber viroid positive RNA genome. J. Gen. Virol. 93: 1591–1600. [DOI] [PubMed] [Google Scholar]
- Brear P., Freeman G.R., Shankey M.C., Trmcić M., Hodgson D.R. (2009). Aqueous methods for the preparation of 5′-substituted guanosine derivatives. Chem. Commun. (Camb.) 33: 4980–4981. [DOI] [PubMed] [Google Scholar]
- Carrieri C., et al. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491: 454–457. [DOI] [PubMed] [Google Scholar]
- Ding B. (2009). The biology of viroid-host interactions. Annu. Rev. Phytopathol. 47: 105–131. [DOI] [PubMed] [Google Scholar]
- Eiras M., Nohales M.A., Kitajima E.W., Flores R., Daròs J.A. (2011). Ribosomal protein L5 and transcription factor IIIA from Arabidopsis thaliana bind in vitro specifically Potato spindle tuber viroid RNA. Arch. Virol. 156: 529–533. [DOI] [PubMed] [Google Scholar]
- Flores R., Hernández C., Martínez de Alba A.E., Daròs J.A., Di Serio F. (2005). Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43: 117–139. [DOI] [PubMed] [Google Scholar]
- Fu Y., Bannach O., Chen H., Teune J.H., Schmitz A., Steger G., Xiong L., Barbazuk W.B. (2009). Alternative splicing of anciently exonized 5S rRNA regulates plant transcription factor TFIIIA. Genome Res. 19: 913–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozmanova M., Denti M.A., Minkov I.N., Tsagris M., Tabler M. (2003). Characterization of the RNA motif responsible for the specific interaction of potato spindle tuber viroid RNA (PSTVd) and the tomato protein Virp1. Nucleic Acids Res. 31: 5534–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco-Stewart V., Pelchat M. (2010). Interaction of host cellular proteins with components of the hepatitis delta virus. Viruses 2: 189–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber T.M., Gross C.A. (2003). Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57: 441–466. [DOI] [PubMed] [Google Scholar]
- Hammond M.C., Wachter A., Breaker R.R. (2009). A plant 5S ribosomal RNA mimic regulates alternative splicing of transcription factor IIIA pre-mRNAs. Nat. Struct. Mol. Biol. 16: 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya A., Zhong X., Bundschuh R., Qi Y., Wang Y., Takeda R., Harris A.R., Molina C., Nelson R.S., Ding B. (2007). A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J. Virol. 81: 2980–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jason T.L.H., Berg R.W., Vincent M.D., Koropatnick J. (2007). Antisense targeting of thymidylate synthase (TS) mRNA increases TS gene transcription and TS protein: effects on human tumor cell sensitivity to TS enzyme-inhibiting drugs. Gene Expr. 13: 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantidis K., Denti M.A., Tzortzakaki S., Marinou E., Tabler M., Tsagris M. (2007). Virp1 is a host protein with a major role in Potato spindle tuber viroid infection in Nicotiana plants. J. Virol. 81: 12872–12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Geng R., Somers D.E. (2003). Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl. Acad. Sci. USA 100: 4933–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A. (2005). The discovery of zinc fingers and their practical applicataions in gene regulation: A personal account. In Zinc Finger Proteins: From Atomic Contact to Cellular Function, S. Iuchi and N. Kuldell, eds (New York: Landes Bioscience/Kluwer Academic/Plenum Publishers), pp. 1–6. [Google Scholar]
- Kolonko N., Bannach O., Aschermann K., Hu K.H., Moors M., Schmitz M., Steger G., Riesner D. (2006). Transcription of potato spindle tuber viroid by RNA polymerase II starts in the left terminal loop. Virology 347: 392–404. [DOI] [PubMed] [Google Scholar]
- Kovalskaya N., Hammond R.W. (2014). Molecular biology of viroid-host interactions and disease control strategies. Plant Sci. 228: 48–60. [DOI] [PubMed] [Google Scholar]
- Lamond A.I., Spector D.L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4: 605–612. [DOI] [PubMed] [Google Scholar]
- Layat E., Cotterell S., Vaillant I., Yukawa Y., Tutois S., Tourmente S. (2012). Transcript levels, alternative splicing and proteolytic cleavage of TFIIIA control 5S rRNA accumulation during Arabidopsis thaliana development. Plant J. 71: 35–44. [DOI] [PubMed] [Google Scholar]
- Lee B.M., Xu J., Clarkson B.K., Martinez-Yamout M.A., Dyson H.J., Case D.A., Gottesfeld J.M., Wright P.E. (2006). Induced fit and “lock and key” recognition of 5S RNA by zinc fingers of transcription factor IIIA. J. Mol. Biol. 357: 275–291. [DOI] [PubMed] [Google Scholar]
- Lehmann E., Brueckner F., Cramer P. (2007). Molecular basis of RNA-dependent RNA polymerase II activity. Nature 450: 445–449. [DOI] [PubMed] [Google Scholar]
- Lu D., Searles M.A., Klug A. (2003). Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature 426: 96–100. [DOI] [PubMed] [Google Scholar]
- MacNaughton T.B., Gowans E.J., McNamara S.P., Burrell C.J. (1991). Hepatitis delta antigen is necessary for access of hepatitis delta virus RNA to the cell transcriptional machinery but is not part of the transcriptional complex. Virology 184: 387–390. [DOI] [PubMed] [Google Scholar]
- Maniataki E., Martinez de Alba A.E., Sägesser R., Tabler M., Tsagris M. (2003). Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hairpin with the host protein VirP1. RNA 9: 346–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O., Yukawa Y., Prieto J.L., Vaillant I., Sugiura M., Tourmente S. (2003). Identification and characterization of transcription factor IIIA and ribosomal protein L5 from Arabidopsis thaliana. Nucleic Acids Res. 31: 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers R.E., Sharp P.A. (1993). TATA-binding protein and associated factors in polymerase II and polymerase III transcription. Mol. Cell. Biol. 13: 7953–7960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modahl L.E., Macnaughton T.B., Zhu N., Johnson D.L., Lai M.M. (2000). RNA-Dependent replication and transcription of hepatitis delta virus RNA involve distinct cellular RNA polymerases. Mol. Cell. Biol. 20: 6030–6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroczek S., Dziembowski A. (2013). U6 RNA biogenesis and disease association. Wiley Interdiscip. Rev. RNA 4: 581–592. [DOI] [PubMed] [Google Scholar]
- Mühlbach H.P., Sänger H.L. (1979). Viroid replication is inhibited by alpha-amanitin. Nature 278: 185–188. [DOI] [PubMed] [Google Scholar]
- Nichols N.M., Yue D. (2008). Ribonucleases. Curr. Protoc. Mol. Biol. 3: 13. [DOI] [PubMed] [Google Scholar]
- Nohales M.A., Flores R., Daròs J.A. (2012). Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase. Proc. Natl. Acad. Sci. USA 109: 13805–13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O., Li C.F., Costa Nunes P., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S. (2006). The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell 126: 79–92. [DOI] [PubMed] [Google Scholar]
- Qi Y., Ding B. (2002). Replication of Potato spindle tuber viroid in cultured cells of tobacco and Nicotiana benthamiana: the role of specific nucleotides in determining replication levels for host adaptation. Virology 302: 445–456. [DOI] [PubMed] [Google Scholar]
- Qi Y., Ding B. (2003). Differential subnuclear localization of RNA strands of opposite polarity derived from an autonomously replicating viroid. Plant Cell 15: 2566–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackwitz H.R., Rohde W., Sänger H.L. (1981). DNA-dependent RNA polymerase II of plant origin transcribes viroid RNA into full-length copies. Nature 291: 297–301. [DOI] [PubMed] [Google Scholar]
- Raha D., Wang Z., Moqtaderi Z., Wu L., Zhong G., Gerstein M., Struhl K., Snyder M. (2010). Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc. Natl. Acad. Sci. USA 107: 3639–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ream T.S., Haag J.R., Wierzbicki A.T., Nicora C.D., Norbeck A.D., Zhu J.K., Hagen G., Guilfoyle T.J., Pasa-Tolić L., Pikaard C.S. (2009). Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol. Cell 33: 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley K.J., Yario T.A., Steitz J.A. (2012). Association of Argonaute proteins and microRNAs can occur after cell lysis. RNA 18: 1581–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D., Paul A., Roy A., Ghosh R., Ganguly P., Chaudhuri S. (2014). Differential acetylation of histone H3 at the regulatory region of OsDREB1b promoter facilitates chromatin remodelling and transcription activation during cold stress. PLoS One 9: e100343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler I.M., Mühlbach H.P. (1992). Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci. 84: 221–229. [Google Scholar]
- Shin M.R., Natsuume M., Matsumoto T., Hanaoka M., Imai M., Iijima K., Oka S., Adachi E., Kodama H. (2014). Sense transgene-induced post-transcriptional gene silencing in tobacco compromises the splicing of endogenous counterpart genes. PLoS One 9: e87869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V., Urcuqui-Inchima S., Roussel P., Hernandez-Verdun D. (2008). Nucleolus: the fascinating nuclear body. Histochem. Cell Biol. 129: 13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K., Krzyzosiak W.J. (2002). RNA structure analysis assisted by capillary electrophoresis. Nucleic Acids Res. 30: e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymański M., Barciszewska M.Z., Erdmann V.A., Barciszewski J. (2003). 5 S rRNA: structure and interactions. Biochem. J. 371: 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen O., Rudt F., Guddat U., Mentzel H., Pieler T. (1992). RNA and DNA binding zinc fingers in Xenopus TFIIIA. Cell 71: 679–690. [DOI] [PubMed] [Google Scholar]
- Thompson N.E., Steinberg T.H., Aronson D.B., Burgess R.R. (1989). Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J. Biol. Chem. 264: 11511–11520. [PubMed] [Google Scholar]
- Tseng C.H., Lai M.M. (2009). Hepatitis delta virus RNA replication. Viruses 1: 818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S.D., Yakovchuk P., Gilman B., Ponicsan S.L., Drullinger L.F., Kugel J.F., Goodrich J.A. (2013). RNA polymerase II acts as an RNA-dependent RNA polymerase to extend and destabilize a non-coding RNA. EMBO J. 32: 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A.J. (2013). Fluor-Labeling of RNA and Fluorescence-based Studies of Precursor-tRNA Cleavage by Escherichia coli Ribonuclease P. (Columbus, OH: The Ohio State University; ). [Google Scholar]
- Wassarman K.M., Saecker R.M. (2006). Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science 314: 1601–1603. [DOI] [PubMed] [Google Scholar]
- White R.J. (2011). Transcription by RNA polymerase III: more complex than we thought. Nat. Rev. Genet. 12: 459–463. [DOI] [PubMed] [Google Scholar]
- Zheng B., Wang Z., Li S., Yu B., Liu J.Y., Chen X. (2009). Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 23: 2850–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Archual A.J., Amin A.A., Ding B. (2008). A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell 20: 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.