The GRAS protein family is illustrious. The GRAS domain is plant specific, named for the first three proteins found to contain it: GIBBERELLIC ACID INSENSITIVE (GAI), REPRESSOR of GAI, and SCARECROW. In addition to these founding members, which function in gibberellin signaling and root patterning, the GRAS family includes DELLA proteins—important in gibberellin, jasmonate, and light signaling—as well as Nodulation Signaling Pathway proteins, which regulate nodulation in legumes (reviewed in Sun et al., 2012). GRAS family proteins have been reported in almost 300 plant species and generally appear to influence plant growth, development, and responses to environment via transcriptional regulation. Whereas their N-terminal regions vary, members of this family are characterized by a C-terminal region harboring the GRAS domain. In a landmark article, Li et al. (2016) report the crystal structure of the GRAS domain, thereby providing long-awaited insight into potential mechanisms underlying the widespread importance of GRAS domain proteins.
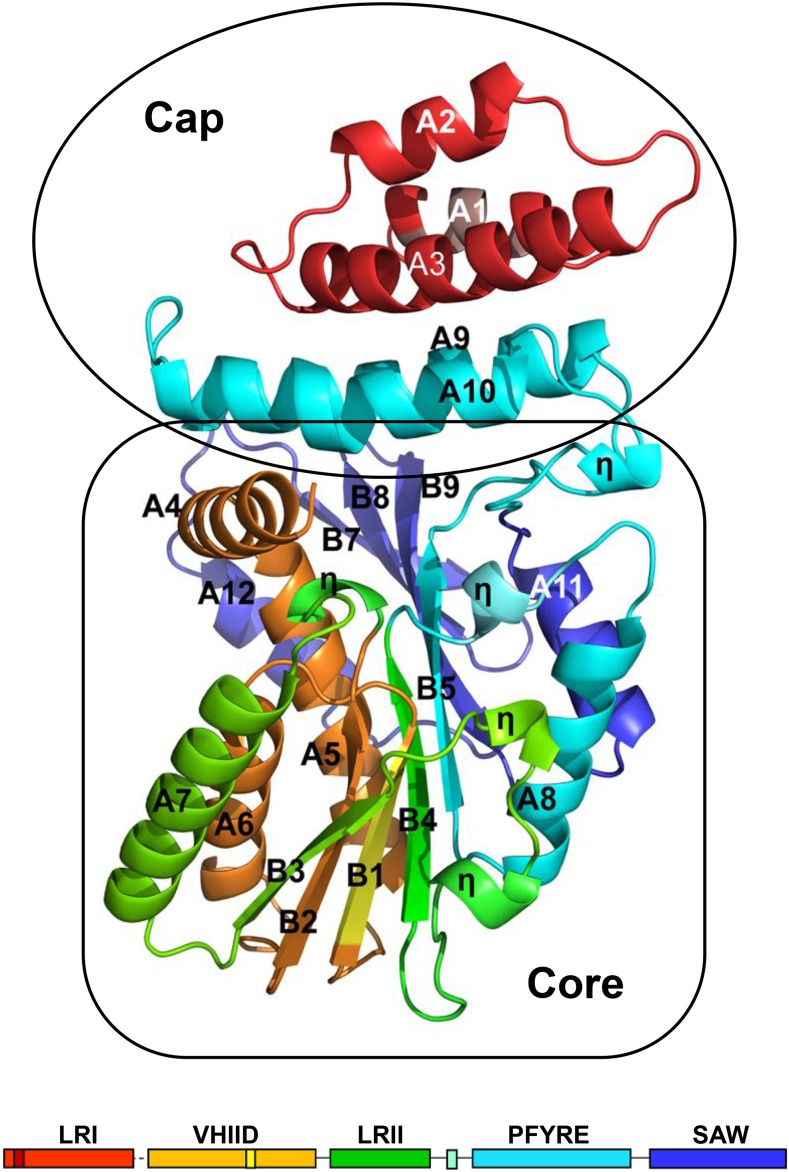
Li et al. crystalized the GRAS domain of rice (Oryza sativa) SCARECROW-LIKE7 (SCL7). The crystal structure showed that the domain forms a core region topped with a cap structure (see figure). GRAS domains comprise five conserved subdomains: LRI, VHIID, LRII, PFYRE, and SAW. The cap of the GRAS domain structure was formed of α-helices of the LRI subdomain with two from the PFYRE subdomain. The remaining subdomains made up the core of the structure. The region between the helices of the LRI cap and the VHIID core was missing, suggesting that there could be movement in this area. Interestingly, the core structure of the GRAS domain consisted of a β-sheet of sandwiched between α-helices, resembling a Rossmann fold. This finding lends support to the idea that GRAS proteins should be considered part of the Rossmann fold superfamily of methyltransferases despite lacking residues for methyltransferase activity (Zhang et al., 2012).
Crystal structure of the GRAS domain. The five subdomains of the Os-SCL7 GRAS domain are colored according to the schematic below the structure (adapted from Figure 1 of Li et al. [2016]).
The Os-SCL7 GRAS domain structure provides important insights into the potential roles of the various GRAS subdomains. The LRI subdomain contains a characteristic leucine-rich repeat, which formed a leucine zipper in the structure, and a nuclear localization signal, which was placed on the surface in the structure. In the VHIID subdomain, the conserved IHIVD motif formed a β-strand that appeared to be important for stabilizing the overall structure of the GRAS domain. The leucine repeats of the LRII subdomain were positioned so that they might interact with other proteins. Both the PFYRE and SAW subdomains appeared to contribute to the maintaining the structure of the domain.
Importantly, Os-SCL7 was crystallized as a dimer, with a groove formed between the cap regions. This groove contained two positive patches and was of a size appropriate for binding DNA. When the authors modeled DNA docking in the groove of the dimer, they found tight interactions. They tested this binding experimentally, showing that the Os-SCL7 GRAS domain could bind DNA in electrophoretic mobility shift assays. Furthermore, this binding was diminished upon mutation of the Os-SCL7 residues that showed interaction with DNA in the docking model.
This elucidation of the crystal structure of the GRAS domain addresses questions surrounding the origins of this domain as well as revealing important residues for protein-protein interactions (including dimerization) and showing that the GRAS domain can bind DNA directly.
Footnotes
Articles can be viewed without a subscription.
References
- Li S., Zhao Y., Zhao Z., Wu X., Sun L., Liu Q., Wu Y. (2016). Crystal structure of the GRAS domain of SCARECROW-LIKE7 in Oryza sativa. Plant Cell 28: 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Jones W.T., Rikkerink E.H. (2012). GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling. Biochem. J. 442: 1–12. [DOI] [PubMed] [Google Scholar]
- Zhang D., Iyer L.M., Aravind L. (2012). Bacterial GRAS domain proteins throw new light on gibberellic acid response mechanisms. Bioinformatics 28: 2407–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]