Abstract
Different combinations of histone modifications have been proposed to signal distinct gene regulatory functions, but are poorly addressed by existing technologies. Here, we apply high-throughput single-molecule imaging to decode combinatorial modifications on millions of individual nucleosomes from pluripotent stem cells and lineage-committed cells. We identify definitively bivalent nucleosomes with concomitant repressive and activating marks, as well as other combinatorial modification states whose prevalence varies with developmental potency. We show that genetic and chemical perturbations of chromatin enzymes both preferentially affect nucleosomes harboring specific modification states. Finally, we combine this proteomic platform with single-molecule DNA sequencing technology to simultaneously determine the modification states and genomic positions of individual nucleosomes. This single-molecule technology has the potential to address fundamental questions in chromatin biology and epigenetic regulation.
The activity of genes and regulatory elements is modulated by their cell type-specific chromatin organization. The fundamental building block of chromatin is the nucleosome. Nucleosomal histones are chemically modified at many amino acid positions (1, 2), which has led to the hypothesis that combinatorial marks specify distinct regulatory outcomes (the ‘histone code’) (3). However, our understanding of the histone code and other models (4) has been constrained by our limited ability to detect, quantify and map combinatorially-modified nucleosomes. Chromatin immunoprecipitation (ChIP), a predominant method in chromatin biology, can identify the genomic location of a specific modification, but cannot effectively distinguish whether coincident marks co-exist on the same nucleosome or originate from different alleles or cells. Mass spectrometry can only compare marks if they are adjacent on the same histone peptide and does not address genomic location (5). Although alternative approaches (6), such as flowing nucleosomes through nanochannels (7), show promise, they have limited throughput and/or do not provide genomic information.
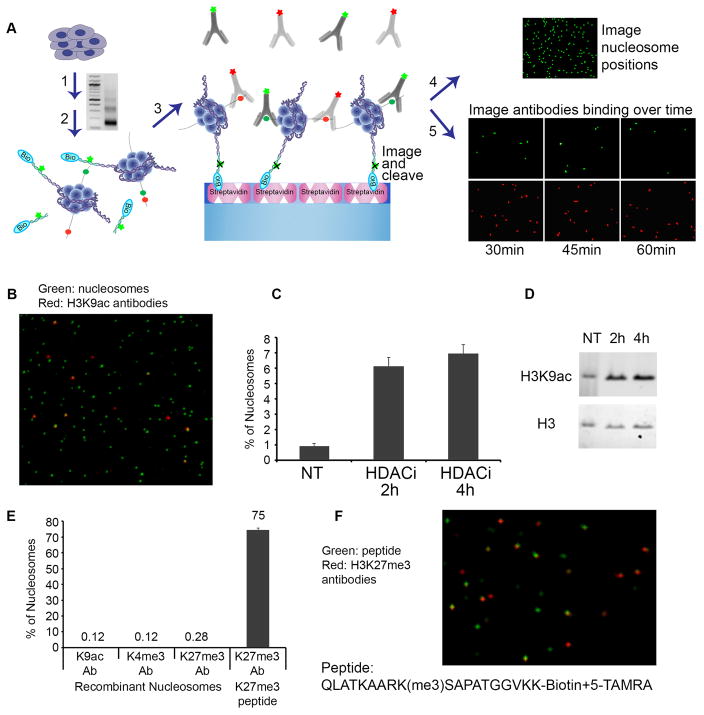
Here we address these limitations by establishing a single-molecule-based assay for investigating combinatorial histone modifications (Fig. 1A, S1, S2, S3). We begin by isolating mono-nucleosomes from cells and ligating fluorescent, biotinylated oligonucleotide adaptors to their free DNA ends. We capture the ligated nucleosomes in a spatially distributed manner on PEG-streptavidin coated slides, and incubate with fluorescently-labeled antibodies to histone modifications. We then use total internal reflection (TIRF) microscopy to record the position and modification state of each nucleosome. As proof of principle, we captured adaptor-ligated mono-nucleosomes from HEK293 cells, and incubated with antibody to histone H3 lysine 9 acetylation (H3K9ac). We used a TIRF microscope to simultaneously detect nucleosomes by their fluorescent adaptors (Alexa555, green), and to distinguish the subset with H3K9ac by the antibody label (Alexa647, red) (Fig. 1B).
Fig. 1. Single-molecule detection of post-translational modifications on nucleosomes.
(A) Experimental scheme: (1) Nucleosomes from cells are prepared by Micrococcal Nuclease (MNase) digestion. Gel depicts nucleosomal DNA fragments of expected lengths; (2) Free DNA ends are ligated to fluorescent, biotinylated oligonucleotide adaptors; (3) Adaptor-ligated mono-nucleosomes are purified on a glycerol gradient and captured on PEG-streptavidin coated slides. (4) Nucleosome positions on the surface are imaged by TIRF microscopy, and then the fluorophore is cleaved from the adaptor. (5) Attached nucleosomes are incubated with fluorescently-labeled antibodies to histone modifications. Time-lapse images detect repeated binding and dissociation events and are integrated to score modified nucleosomes. (B-D) HEK293 cells were treated with HDAC inhibitor. (B) Single-Molecule detection of labeled nucleosomes (Alexa555, green) bound by labeled H3K9ac antibodies (Alexa647, red). (C) Proportion of nucleosomes marked by H3K9ac under each condition is determined by single-molecule counting. (D) Western blot confirms increased H3K9ac in treated cells. (E–F) Recombinant unmodified nucleosomes and H3K27me3-modified peptide were probed with the indicated antibodies. (F) Single-molecule detection of labeled H3K27me3 peptide (TAMRA, green) with labeled H3K27me3 antibodies (Alexa647, red) at a single time point.
We imaged millions of nucleosomes and decoded their modification state. We found that while the nucleosome positions were fixed, the H3K9ac antibodies repeatedly bind and dissociate at a specific subset of these nucleosome positions (Fig. S2). Summation of individual binding events over time revealed that ~1% of the nucleosomes are marked by H3K9ac (Fig. 1C). In contrast, when we treated cells with histone deacetylase inhibitors (HDACi), the fraction of acetylated nucleosomes increased to 7% (Fig. 1C, 1D). When we repeated the analysis with recombinant unmodified nucleosomes, just 0.1% of nucleosomes scored (Fig. 1E, S4). We also fluorescently labeled antibodies to H3 lysine 4 tri-methylation (H3K4me3), lysine 27 tri-methylation (H3K27me3), lysine 27 di-methylation (H3K27me2) and lysine 27 acetylation (H3K27ac), and confirmed their specificities by imaging unmodified recombinant nucleosomes as well as marked peptides, and by probing arrays of modified peptides (Fig. 1E, 1F, S4, S5).
In embryonic stem cells (ESCs), developmental gene promoters appear to be concomitantly marked by repressive (H3K27me3) and activating (H3K4me3) histone modifications (8, 9). This ‘bivalent’ chromatin state has been suggested to poise these genes for alternate fates, but remains controversial (10). Sequential ChIP and IP – mass spectrometry provide evidence for coexistence of the opposing marks (8, 11), but cannot identify definitely an individual bivalent nucleosome. We leveraged our single-molecule platform to quantify directly the co-existence of these key marks on nucleosomes derived from pluripotent ESCs, from ESCs differentiated to embryoid bodies (EBs), or from fully committed lung fibroblasts.
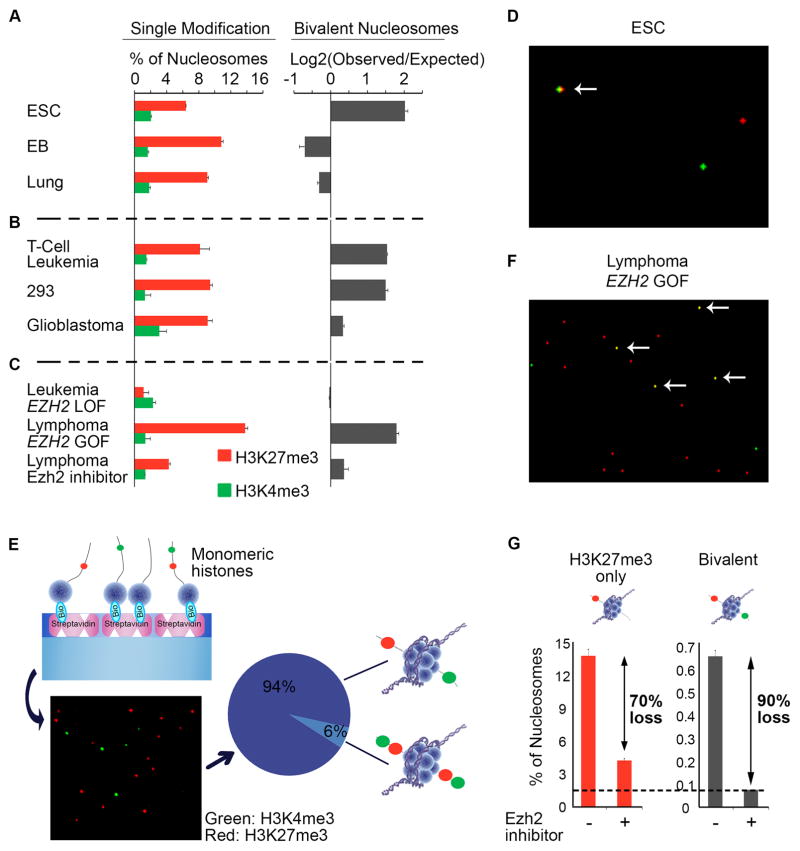
We captured nucleosomes and determined their positions as above, chemically cleaved their fluorophores, and then incubated them simultaneously with H3K4me3 (green) and H3K27me3 (red) antibodies (Fig. S6, S7). TIRF imaging revealed that ~6.5% of nucleosomes from ESCs carry H3K27me3 (Fig. 2A, S8, S9). This fraction is somewhat higher in EBs and lung fibroblasts, consistent with prior reports that this repressive mark expands during differentiation (2). The fraction of nucleosomes marked by H3K4me3 is relatively constant (ESCs: ~2%; EBs: ~1.6%; lung: ~1.6%). Single-molecule counting revealed that 0.5% of nucleosomes in ESCs carry both marks and thus are truly bivalent (Fig. 2A, 2D, S8). In contrast, in both EBs and fibroblasts, bivalent nucleosomes are much less prevalent and actually depleted relative to random expectation (Fig. 2A).
Fig. 2. Single-molecule imaging of symmetric and asymmetric bivalent nucleosomes.
(A) We queried modifications on nucleosomes from pluripotent ESCs, EBs and lung fibroblasts (lung). Left: Colored bars indicate proportions of nucleosomes with H3K27me3 (red) or H3K4me3 (green). Right: Black bars indicate relative over- or under-representation of bivalent nucleosomes. Results are presented as the Log2 ratio of the observed proportion of bivalent nucleosomes divided by the expected random association of these two marks (random = fraction H3K27me3 * fraction H3K4me3; Fig. S10). (B) Nucleosomes from a T-cell acute lymphoblastic leukemia line, HEK293 cells and glioblastoma stem cells were decoded as in (A). (C) Nucleosomes from an acute leukemia line with a loss-of-function EZH2 mutation, a lymphoma line with a gain-of-function EZH2 mutation, and the lymphoma cells treated with EZH2 inhibitor GSK126. (D) Magnified TIRF image overlay reveals three nucleosomes, one with H3K27me3 (red), one with H3K4me3 (green), and one with concomitant bivalent modifications (arrow). (E) Image depicts H3K27me3 and H3K4me3 antibody binding to individual histones isolated from ESCs. (F–G) The lymphoma cell line with gain-of-function EZH2 was treated with GSK126 for 3 days. (F) Nucleosomes were decoded for H3K27me3 and H3K4me3. Shown is image for pre-treated samples, with arrows highlighting bivalent nucleosomes. (G) Plot depicts proportions of H3K4me3-negative (left, H3K27me3 only) and H3K4me3-positive (right, bivalent) nucleosomes that carry H3K27me3. Bivalent nucleosomes are more likely to lose H3K27me3 following treatment with GSK126.
A bivalent nucleosome could reflect symmetric co-occurrence of H3K4me3 and H3K27me3 on the same histone tail, or asymmetric marking on opposite tails. To address this, we extracted individual histone molecules from ESCs, biotinylated them, spatially distributed them on our surface and detected H3K4me3 and H3K27me3 modified tails (Fig. 2E). We found that 0.4% of H3K27me3-modified tails also carry H3K4me3. For comparison, just 0.04% of H3K27me3-modified tails scored for H3K27ac, a combination that is chemically prohibited (Fig. S8). The 10-fold excess in detection of bivalent H3 tails relative to background suggests that symmetric bivalent nucleosomes do exist in ESCs. Nonetheless, we estimate that 94% of bivalent nucleosomes are asymmetric, while just 6% are modified on the same tail (Fig. S8).
Genomic loci marked by bivalent chromatin in ESCs are frequently deregulated in cancer cells (12). Moreover, the Polycomb and trithorax complexes that catalyze H3K27me3 and H3K4me3 are often mutated in cancer (12). We therefore investigated these modifications in cancer cells (Fig. 2B, 2C, S8). We detected bivalent nucleosomes in three cell lines that lack known Polycomb or trithorax mutations, T-cell acute leukemia (DND-41), embryonic kidney (HEK293), and glioblastoma (GSC8), at levels higher than in our differentiated models (EB, lung), but lower than ESCs. When we next examined a leukemia line (SKM-1) with a loss-of-function (LOF) mutation of the PRC2 subunit EZH2 (13), we observed very few H3K27me3-marked nucleosomes (~1%; Fig. 2C). We also examined a lymphoma cell line (Karpas422) with a gain-of-function (GOF) EZH2 mutation that increases its catalytic activity (14). We detected H3K27me3 on ~15% of nucleosomes. Bivalent nucleosomes are also prevalent in the lymphoma cells, with roughly half of all H3K4me3-marked nucleosomes carrying H3K27me3 (Fig. 2C, 2F, S8). The proportion of bivalent nucleosomes is ~4-fold greater than expected from random overlap, suggesting that the mutant EZH2 preferentially catalyzes H3K27me3 on nucleosomes that are marked by H3K4me3. This is consistent with the increased H3K27me3 over active promoters seen in EZH2 GOF lymphomas (15). When we treated these lymphoma cells with an EZH2 inhibitor (16), H3K27me3 was preferentially lost from bivalent nucleosomes (Fig. 2G). This may reflect increased nucleosome turnover and/or preferential demethylation in such regions.
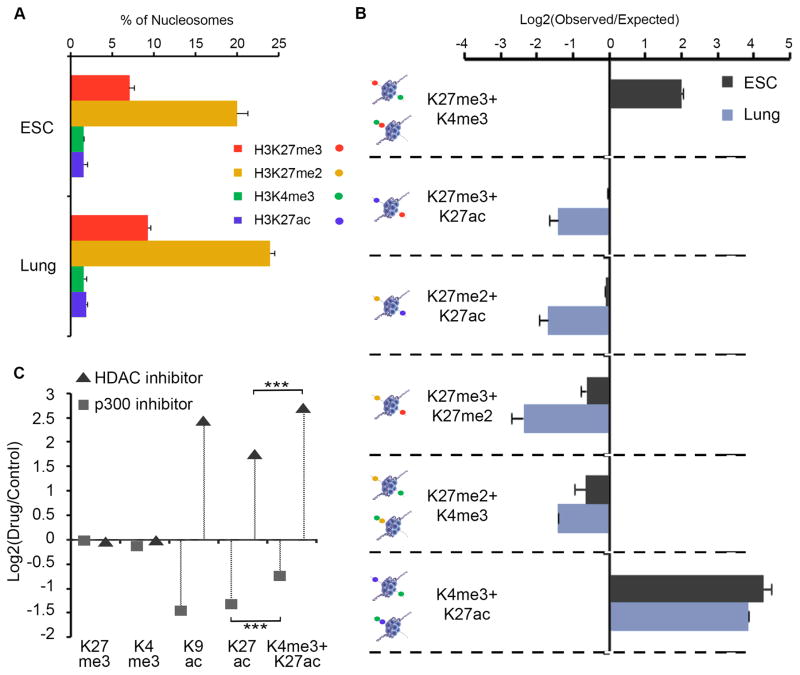
We next explored higher-order combinations of H3K27ac (marks active enhancers (2)), H3K27me2 (intergenic regions (17)), H3K4me3 and H3K27me3 (Fig. 3A, S11). Monitoring four or more histone modifications on single nucleosomes is carried out in successive steps of antibody incubation and imaging, which are separated by a wash step to remove antibodies. Single molecule counting reveals that the proportions of nucleosomes marked by each of the four modifications are similar in ESCs and lung fibroblasts, with the exception that H3K27 methylations increase modestly in the differentiated cells (Fig. 3A). When we considered these modifications in combination, however, we observed considerable differences between cell types (Fig. 3B, S11).
Fig. 3. Higher-order modification states across cellular states and inhibitor treatments.
(A) Individual nucleosomes from ESCs and lung fibroblasts decoded for H3K4me3, H3K27me3, H3K27me2 and H3K27ac, as described in Fig. 1. Bars depict fraction of nucleosomes with the indicated modification. (B) Bars indicate, for each possible modification pair, relative over- or under-representation, compared to random expectation, as in Fig. 2A. Opposing modifications are relatively more likely to co-exist in ESCs than lung fibroblasts. (C) ESCs were treated with DMSO (control), HDAC inhibitor (Sodium butyrate) or p300 inhibitor (C646). Nucleosomes were isolated and decoded for H3K27me3, H3K4me3, H3K9ac and H3K27ac. Plot shows effects of inhibitors on each single modification or the combination of H3K27ac and H3K4me3.
ESC chromatin is enriched for the bivalent combination, and for the pairwise combination of the two active marks, H3K4me3 and H3K27ac. The other pairwise combinations are present in roughly the same proportions as would be expected by chance overlap. The lung fibroblasts are enriched for the pairwise combination of active marks, like the ESCs, but not for the bivalent nucleosomes. However, the lung fibroblast chromatin is depleted for the other combinations (Fig. 3B). This includes the three pairwise states for H3K27 modifications, which are by definition asymmetric. These distinct combinatorial modification patterns likely relate to alternate chromatin environments in the respective cell types, most notably the hyper-dynamic nature of ESC chromatin (18) (Fig. S12).
We next examined how combinatorial chromatin states change upon treatment with a pan- histone deacetylase (HDAC) inhibitor or with a p300 histone acetyltransferase (HAT) inhibitor. HDAC inhibition significantly increased levels of H3K9ac and H3K27ac, while p300 inhibition had the opposite affect (Fig. 3D, S11). Neither treatment altered H3K4me3 or H3K27me3. However, the changes in histone acetylation occurred preferentially on nucleosomes with specific markings. In the case of HDAC inhibition, increased acetylation preferentially affected H3K4me3-marked nucleosomes, consistent with HDACs modulating acetylation levels at active promoters (19). In contrast, H3K4me3-marked nucleosomes were less affected by p300 inhibition, consistent with a role for this enzyme at enhancers (20).
Phosphorylation of the histone variant H2Ax (ɣH2Ax) is one of the earliest and best studied marks of DNA damage. We therefore examined ɣH2Ax levels on individual nucleosomes extracted from ESCs. We found that ~2% of the nucleosomes contain ɣH2Ax at baseline (Fig. S13). Combinatorial analysis revealed that ɣH2Ax is specifically enriched on nucleosomes with activating marks (H3K27ac and H3K4me3). Treatment with HDAC inhibitors led to concomitant increases in acetylation and ɣH2Ax levels, consistent with studies documenting high baseline levels of ɣH2Ax associated with decondensed chromatin in ESCs (21).
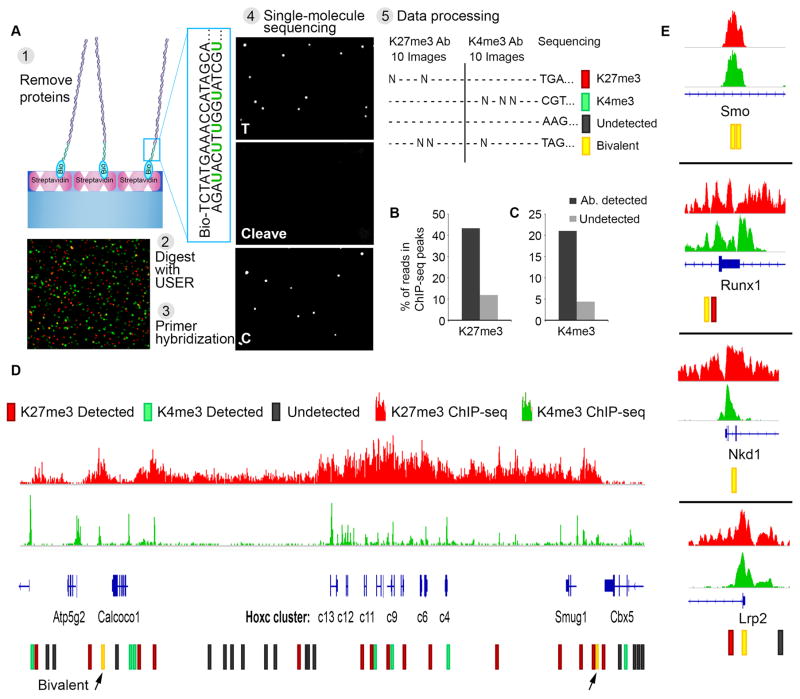
Finally, we used single-molecule sequencing technology to read the DNA associated with each individual nucleosome (Fig. 4A, S14). We captured adaptor-ligated nucleosomes from ESCs, and queried their H3K27me3 and H3K4me3 status. We then displaced the histone octamers, leaving behind double-stranded nucleosomal DNA. We enzymatically cleaved the uracil bases that were incorporated into the non-biotinylated strand of the adaptor, exposing a known sequence that was used as a priming site for single-molecule sequencing-by-synthesis (22). We used TIRF microscopy to detect repeated cycles of base addition, separated by chemical cleavage of fluorescent label and terminator.
Fig. 4. Single-molecule sequencing determines genomic positions of modified nucleosomes.
(A) Experimental scheme: (1) Nucleosomes are captured and probed for their modification state, as in Fig. 1A. Histones are evicted by increasing salt concentration. (2) The enzyme USER is applied to excise uracil bases incorporated into the non-biotinylated adaptor strand and expose a known sequence (3) Complementary primer is hybridized to the adaptor. Image shows single molecule detection of nucleosomal DNA (Alexa647, red) and primer (Alexa555, green). (4) Direct single-molecule DNA sequencing-by-synthesis (22). Images reflect two sequencing cycles: incorporation of thymine, cleavage of fluorophore and terminator, and incorporation of cytosine. (5) Data processing: for each x-y coordinate on the surface, sequence data is analyzed and integrated with the initial images scoring antibody binding and modification states of the corresponding nucleosomes. (B) Single-molecule reads were aligned to the genome. Plot indicates proportions of H3K27me3-modified nucleosome reads (‘detected’) or un-modified nucleosome reads (‘undetected’) that aligned to H3K27me3-enriched regions per conventional ChIP-seq. (C) Proportions of H3K4me3-modified nucleosome reads that aligned to H3K4me3-enriched regions per conventional ChIP-seq. (D) The HOXC gene cluster is shown along with H3K27me3 and H3K4me3 ChIP-seq tracks. Single-molecule reads that aligned to these regions are indicated, along with the modification status of the corresponding nucleosome. (E) Analogous data shown for other developmental loci for which bivalent nucleosomes were definitively identified.
Integration of TIRF images for antibody-based detection of modifications with subsequent sequencing reaction data collected on the same flowcell allowed us to coordinately determine the modification state and DNA sequence of each nucleosome (Fig. 4). More than 80% of the ~300,000 reads aligned to the mouse genome. We then compared the genomic localization of individual nucleosomes with their modification states. Of the ~26,000 reads for which the corresponding nucleosomes scored positive for H3K27me3, 45% aligned to genomic regions within H3K27me3 ChIP-seq peaks (Fig. 4B). This is consistent with the 30–50% of reads that map to enriched intervals in a typical H3K27me3 ChIP-seq experiment. For comparison, just 12% of H3K27me3-negative nucleosomes aligned to H3K27me3 ChIP-seq peaks. In a subset of these experiments, we probed the sequenced nucleosomes for both H3K27me3 and H3K4me3, yielding ~1,000 aligned reads for concomitantly marked molecules, and thus providing the first definitive localization of individual bivalent nucleosomes (Fig. 4D, 4E).
We present a critical step towards defining the nature and significance of combinatorial chromatin modifications. We use single-molecule technology to decode concurrent modifications on individual nucleosomes and sequence the associated DNA. We identify individual bivalent nucleosomes with concomitant repressive and activating marks, and map their genomic locations. We also document other combinatorial modification states whose proportions change during cellular specification, or upon treatment with epigenetic inhibitors. The single-molecule assay requires little starting material and is highly scalable given that many millions or even billions of nucleosomes may be decoded and sequenced in an automated imaging run.
Supplementary Material
Acknowledgments
We thank Itay Tirosh, Paul Blainey, Kan Xiong and Stephen Elledge for constructive discussions, Jeff Reifenberger for help with imaging technology and Mark Bray for help in image analysis. We thank Ik-Soo Kim, Karim Bouazoune, Charles Epstein and Jasleen Kaur for their experimental contributions and Sebastian Deindl, Brain Liau and Rusty Ryan for valuable experimental advice. E. S is supported by the Jane Coffin Childs fund for medical research. This research was supported by funds from the National Human Genome Research Institute, the Klarman Cell Observatory, and the Howard Hughes Medical Institute. Sequencing data is available in the supporting online material. E.S, D.J and B.E.B have filed a provisional patent covering single-molecule methodologies described in the paper.
Footnotes
References
- 1.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155:39. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sidoli S, Garcia BA. Properly reading the histone code by MS-based proteomics. Proteomics. 2015;15:2901. doi: 10.1002/pmic.201500298. [DOI] [PubMed] [Google Scholar]
- 6.Gomez D, Shankman LS, Nguyen AT, Owens GK. Detection of histone modifications at specific gene loci in single cells in histological sections. Nat Methods. 2013;10:171. doi: 10.1038/nmeth.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy PJ, et al. Single-molecule analysis of combinatorial epigenomic states in normal and tumor cells. Proc Natl Acad Sci U S A. 2013;110:7772. doi: 10.1073/pnas.1218495110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 9.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 10.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voigt P, et al. Asymmetrically modified nucleosomes. Cell. 2012;151:181. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 14.Yap DB, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beguelin W, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23:677. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari KJ, et al. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell. 2014;53:49. doi: 10.1016/j.molcel.2013.10.030. [DOI] [PubMed] [Google Scholar]
- 18.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7:540. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 19.Ram O, et al. Combinatorial Patterning of Chromatin Regulators Uncovered by Genome-wide Location Analysis in Human Cells. Cell. 2011;147:1628. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 21.Turinetto V, Giachino C. Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015;43:2489. doi: 10.1093/nar/gkv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harris TD, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.