Abstract
In human and nonhuman primates, the amygdala is known to play critical roles in emotional and social behavior. Anatomically, individual amygdaloid nuclei are connected with many neural systems that are either differentially expanded or conserved over the course of primate evolution. To address amygdala evolution in humans and our closest living relatives, the apes, we used design-based stereological methods to obtain neuron counts for the amygdala and each of four major amygdaloid nuclei (the lateral, basal, accessory basal, and central nuclei) in humans, all great ape species, lesser apes, and one monkey species. Our goal was to determine whether there were significant differences in the number or percent of neurons distributed to individual nuclei among species. Additionally, regression analyses were performed on independent contrast data to determine whether any individual species deviated from allometric trends. There were two major findings. In humans, the lateral nucleus contained the highest number of neurons in the amygdala, whereas in apes the basal nucleus contained the highest number of neurons. Additionally, the human lateral nucleus contained 59% more neurons than predicted by allometric regressions on nonhuman primate data. Based on the largest sample ever analyzed in a comparative study of the hominoid amygdala, our findings suggest that an emphasis on the lateral nucleus is the main characteristic of amygdala specialization over the course of human evolution.
INDEXING TERMS: amygdala, comparative neuroanatomy, human evolution, lateral nucleus, ape, stereology
The amygdala is comprised of numerous discrete nuclei with distinct cytoarchitecture, chemoarchitecture, and patterns of connectivity with other brain regions (Freese and Amaral, 2009). Given its integrative function, there is a high degree of intranuclear connectivity within the amygdala (Pitkänen and Amaral, 1998; Barton et al., 2003; Freese and Amaral, 2009). Extrinsically, specific nuclei communicate with diverse neural systems such as the autonomic nervous system, the striatopallidal system, and neocortical sensory regions (Price et al., 1987; Stefanacci and Amaral, 2002; Heimer and Van Hoesen, 2006). Thus, the amygdala is strategically positioned to bridge higher order sensory information from the neocortex with brainstem and subcortical structures that facilitate the production of adaptive physiological and motor responses (Price et al., 1987; Heimer et al., 1999; Freese and Amaral, 2009). Across mammals, the amygdala has been shown to modulate emotional responses to external stimuli, especially fear-producing stimuli (MacLean, 1949; LeDoux, 2007). In human and nonhuman primates, the amygdala has been characterized as a detector of salience, ambiguity, value, and threat (Amaral et al., 2003; Bechara et al., 2003; Adolphs, 2010; Morrison and Salzman, 2010), and it has also been associated with social behavior and social affiliation (Brothers, 1990; Adolphs, 2003; Bickart et al., 2010).
Although the gross anatomical structure of the amygdala is similar across primate species (Fig. 1) (Price et al., 1987; Heimer et al., 1999; Schumann and Amaral, 2005; Barger et al., 2007; Carlo et al., 2010), its internal organization has been shown to vary across species both qualitatively (Pitkänen and Kemppainen, 2002) and quantitatively (Stephan et al., 1987; Barger et al., 2007). In earlier comparative analyses of primates, Stephan and colleagues (1987) determined that a gross subcomponent of the amygdala, which included its basolateral division (i.e., its lateral, basal, and accessory basal nuclei) and some of its superficial cortical nuclei, increased at substantially greater rates relative to overall brain size than the rest of the amygdala, which primarily included the dorsalmost set of cortical nuclei and the central nucleus (Stephan and Andy, 1977). Barton and Aggleton (2000) extended these analyses to show that the basolateral division, in particular, is larger in humans than predicted by allometry, and that it correlates with 1) social group size and 2) parvocellular visual pathway size.
Figure 1.
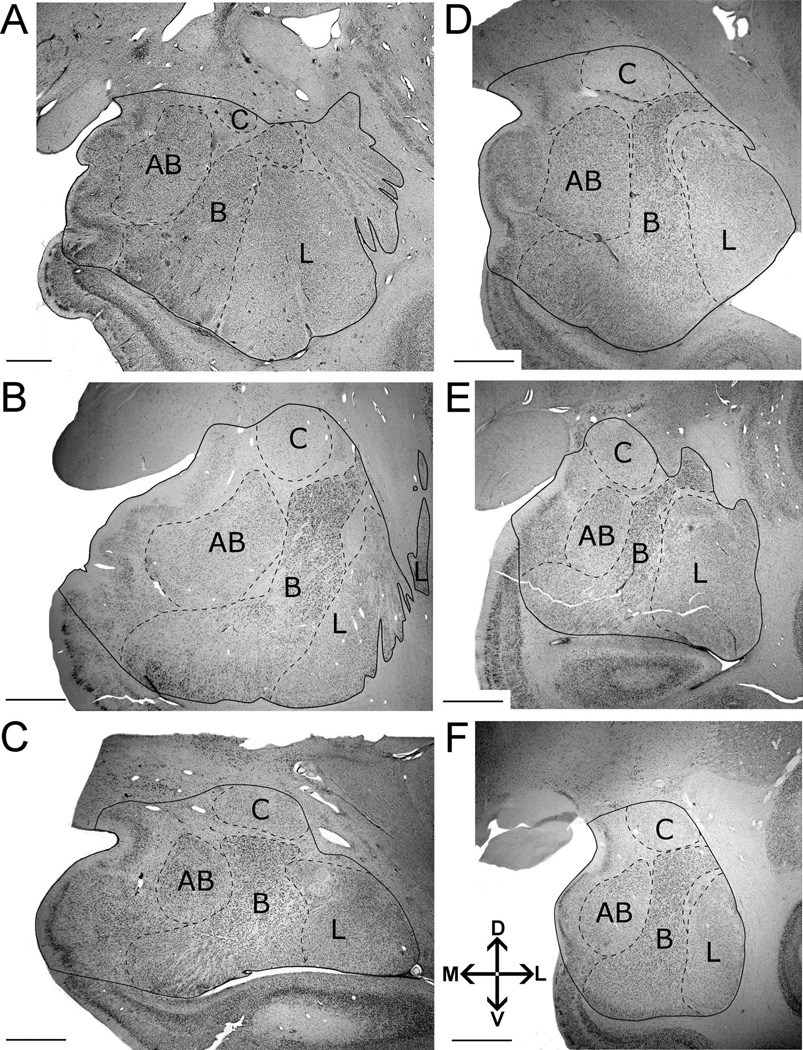
Delineation of left amygdala and nuclei included in this analysis illustrating the consistency of borders across species. Images were taken from midrostrocaudal levels in the following primates: A: human. B: gorilla. C: orangutan. D: chimpanzee. E: gibbon. F: long tailed macaque. Abbreviations: AB, accessory basal nucleus; B, basal nucleus; C, central nucleus; L, lateral nucleus. Other amygdaloid nuclei are not represented in this comparative figure, but are highlighted in Figure 2. The human image (A) is modified from Schumann and Amaral (2005). Images follow radiological conventions. Scale bar = 2 mm in A–F.
We have recently investigated these early findings in more detail, anatomically, by targeting the evolution of discrete nuclei in the primate amygdala. Across Old World and New World monkey species, we established that the volumes and numbers of neurons in the lateral, basal, and accessory basal nuclei generally increase at the same rate as the volume and number of neurons in the whole amygdala. In contrast, increases in the volume and number of neurons in the central nucleus are hypometric, i.e., they do not keep up with increases in the whole amygdala (Carlo et al., 2010). In humans and apes, we have found that as brain size increases, amygdala volume expands at similar rates as the whole basolateral division (Barger et al., 2007). To date, no comparable quantitative information is available for the central nucleus in apes.
Moreover, our previous volumetric data indicate that, in its internal organization, the human amygdala exhibits specializations that are unique to our species (Barger et al., 2007). Specifically, the human lateral nucleus is significantly larger than predicted for an ape of human brain size. Consequently, the lateral nucleus is the largest nucleus in the human amygdala (Schumann and Amaral, 2005; Barger et al., 2007), whereas the basal nucleus is the largest nucleus in the ape amygdala. Thus, human amygdala evolution is not necessarily characterized by passive increases in volume associated with increases in overall brain size, but rather by evolutionary reorganization (Holloway, 1968) of its component nuclei, perhaps as a response to selection pressures in human evolution (Semendeferi et al., 2010). However, the number of neurons in the ape amygdala has never been investigated, leaving open questions about the relationship between increases in volumes and neuronal populations in the evolution of large-brained primate species.
The goal of this study was to determine whether the number of neurons in the amygdala and in each of four major amygdaloid nuclei (lateral, basal, accessory basal, and central nuclei) differ between humans and our closest living relatives, the apes. This study comprises the largest sample ever used in a comparative analysis of the hominoid amygdala (35 specimens total). In addition to humans, the sample includes all of the large, or “great”, ape species (chimpanzees, bonobos, gorillas, and orangutans), as well as representatives of the more distantly related smaller, or “lesser” apes, (gibbons). The present study builds on our previous comparative study of amygdala volumes (Barger et al., 2007) in the following ways: First, we used assumption-free stereological methods to estimate neuron numbers. Second, we counted neurons in the central nucleus to test the hypothesis that the central nucleus might be more conserved across hominoids than the basolateral nuclei. Third, we included a macaque monkey species in the sample to provide a phylogenetic outgroup. Based on our volumetric findings, we predicted that the number of neurons in the basolateral nuclei would increase at greater rates across primate species than in the central nucleus. Additionally, we predicted that the number of neurons in the basal nucleus would be higher in apes than in humans, whereas the number of neurons in the lateral nucleus would be disproportionately higher in human than in nonhuman primates.
MATERIALS AND METHODS
Specimens
Our sample (Table 1) comprised 35 specimens including humans (n = 11), chimpanzees (n = 5), bonobos (n = 4), gorillas (n = 5), orangutans (n = 4), gibbons (n = 3), and long-tailed macaques (n = 3). The sample includes specimens from our collective libraries (C.M.S., K.S., J.M.A., and J.A.B.), as well as nine new ape specimens processed by N.B. (Table 1).
TABLE 1.
Specimens in Sample1
| Species | Common name | Sex | Age (yr) | Hemisphere |
|---|---|---|---|---|
| Homo sapiensb | Human | M | 11 | Left |
| Homo sapiensb | Human | M | 14 | Right |
| Homo sapiensb | Human | M | 17 | Left |
| Homo sapiensb | Human | M | 18 | Left |
| Homo sapiensb | Human | M | 24 | Right |
| Homo sapiensb | Human | M | 25 | Left |
| Homo sapiensb | Human | M | 27 | Right |
| Homo sapiensb | Human | M | 27 | Left |
| Homo sapiensb | Human | M | 32 | Left |
| Homo sapiensb | Human | M | 44 | Left |
| Homo sapiensc | Human | M | 75 | Left |
| Pan troglodytesa | Common chimpanzee | F | 2 | Left |
| Pan troglodytesc | Common chimpanzee | F | 24 | Left |
| Pan troglodytesa | Common chimpanzee | F | 27 | Left |
| Pan troglodytesa | Common chimpanzee | F | 42 | Left |
| Pan troglodytesc | Common chimpanzee | F | Adult | Left |
| Pan paniscusc | Bonobo | F | 2 | Left |
| Pan paniscusc | Bonobo | F | 11 | Left |
| Pan paniscusd | Bonobo | F | 25 | Left |
| Pan paniscusd | Bonobo | M | Adult | Right |
| Gorilla gorilla gorillaa | Western lowland gorilla | M | 10 | Right |
| Gorilla gorilla gorillac | Western lowland gorilla | F | 20 | Left |
| Gorilla gorilla gorillaa,f | Western lowland gorilla | M | 22 | Left |
| Gorilla gorilla gorillaa | Western lowland gorilla | M | 34 | Right |
| Gorilla gorilla gorillaa | Western lowland gorilla | F | 50 | Right |
| Pongo pygmaeusc | Orangutan | M | 17 | Left |
| Pongo pygmaeusa | Orangutan | F | 23 | Right |
| Pongo pygmaeusc | Orangutan | M | 34 | Left |
| Pongo pygmaeusc | Orangutan | F | Adult | Right |
| Hylobates muelleria,f | Müller’s Bornean gibbon | M | 19 | Left |
| Hylobates concolorc | White-cheeked gibbon | F | 22 | Right |
| Hylobates larc | White-handed gibbon | F | Adult | Right |
| Macaca Fascicularise | Long-tailed macaque | M | 4 | Left |
| Macaca Fascicularise | Long-tailed macaque | M | 5 | Left |
| Macaca Fascicularise | Long-tailed macaque | M | 5 | Left |
1New histological series processed by aN.B. were combined with specimens from the collections of aC.M.S., cK.S., dJ.M.A., and eJ.A.B. to yield a large sample suitable for statistical analysis. fC.C.S. and P.R.H. provided tissue for two specimens sectioned at 40 microns.
Human and ape brains were extracted within 24 hours of the individual’s natural death and were free of neuropathologies. Brains were subsequently immersion-fixed in either 10% formalin, Bodian solution, or 4% paraformaldehyde. For each collection, specimens were either paraffin-embedded and sectioned (K.S. collection) or stored at 4°C in a solution of phosphate buffered saline (pH 7.4) and 0.01% sodium azide (C.C.S., C.M.S., J.A., J.A.B., J.M.A., and P.R.H. collections) prior to tissue processing. Macaque brains were perfused with a 4% paraformaldehyde solution and subsequently submerged in a sucrose solution for cryoprotection (Buckwalter et al., 2008). Our sample included individuals spanning developmental periods from juvenile to adulthood. We did not anticipate that the inclusion of younger individuals would substantially influence our results, because it is broadly held that neurons in the amygdala complete migration by birth (Schumann et al., 2011). Although postnatal neurogenesis has been evidenced in the adult primate amygdala (Bernier et al., 2002), we found that, within each species, neuron numbers in juveniles fell close to or overlapped adult values and that age was not significantly correlated with neuron number. Little is known about the effect of aging on amygdala neuron number in humans, but magnetic resonance imaging data suggest that limbic structures, including the amygdala, are largely preserved into the eighth decade of life (Grieve et al., 2005). Stereological analyses of amygdala aging have only been performed in rats and indicate that neuron numbers are relatively similar in adult and aged mice (von Bohlen und Halbach and Unsicker, 2002; Rubinow and Juraska, 2009). Thus, the inclusion of juvenile and aged individuals should not substantially influence estimated mean neuron numbers within species.
Tissue processing
For this study, we produced nine new series of sections from ape brain tissue including three chimpanzees, four gorillas, one orangutan, and one gibbon (Table 1). Either an entire hemisphere or a 3–4-cm anterior temporal lobe block was prepared for cryosectioning by submerging and saturating the tissue in increasing grades of a sucrose and PBS (10%, 20%, and 30%). The block was then serially sectioned at 50 µm, except for one gibbon and one gorilla specimen, which were cut at 40 µm (Table 1). Every 10th section was mounted and stained for Nissl substance with thionin.
Processing parameters for series drawn from existing libraries were as follows. Ten human brains (C.M.S) were cryoprotected, sectioned at 50 µm, and stained for Nissl substance with thionin (Schumann and Amaral, 2005). Eleven ape and human specimens (K.S.) were paraffin-embedded, sectioned at 20 µm, and stained for Nissl substance with a modification of the Gallyas silver stain (Merker, 1983; Semendeferi et al., 1998). Two bonobo brains (J.M.A) were cryoprotected, sectioned at 100 µm, and stained for Nissl substance with Cresyl Violet (Allman et al., 2010). Three long-tailed macaque brains (J.A.B.) were cryoprotected, sectioned at 50 µm, and stained for Nissl substance with thionin (Buckwalter et al., 2008). All brains were sectioned in the coronal plane. We followed standard stereological procedures to estimate neuron counts, which are robust against variation in section thickness and processing techniques.
Anatomical delineation
The amygdala is a roughly ovoid structure located in the anteromedial temporal lobe (Fig. 1), containing at least 13 distinct nuclei in primates (Price et al., 1987). The anatomical borders of the primate amygdala and its nuclei can be reliably defined across species in Nissl-stained material (Price et al., 1987; Heimer et al., 1999; Schumann and Amaral, 2005; Barger et al., 2007; Carlo et al., 2010). In particular, the nuclei chosen for this analysis exhibit boundaries that are clear in Nissl preparations and are easily distinguishable across all species analyzed (Fig. 1). Borders for the hominoid amygdala and the lateral, basal, accessory basal, and central nuclei were defined using anatomical descriptions of the macaque (Price et al., 1987) and human amygdala (Heimer et al., 1999; Schumann and Amaral, 2005). Although each nucleus can be further parcellated into discrete subdivisions, all nuclear subdivisions do not show consistent chemoarchitectonic homologies between macaques and humans (Pitkänen and Kemppainen, 2002) and could not be reliably defined in great apes without considerable further study. One investigator (N.B.) hand-traced the boundaries of the amygdala and the lateral, basal, accessory basal, and central nuclei in serial sections under 1× and 2× objectives (N.A. 0.4 and 0.06, respectively) of a Nikon Eclipse 80i (Melville, NY) microscope with the StereoInvestigator software suite (MicroBrightField, Williston, VT). The anatomical borders for each region were identified using the following criteria.
Amygdala
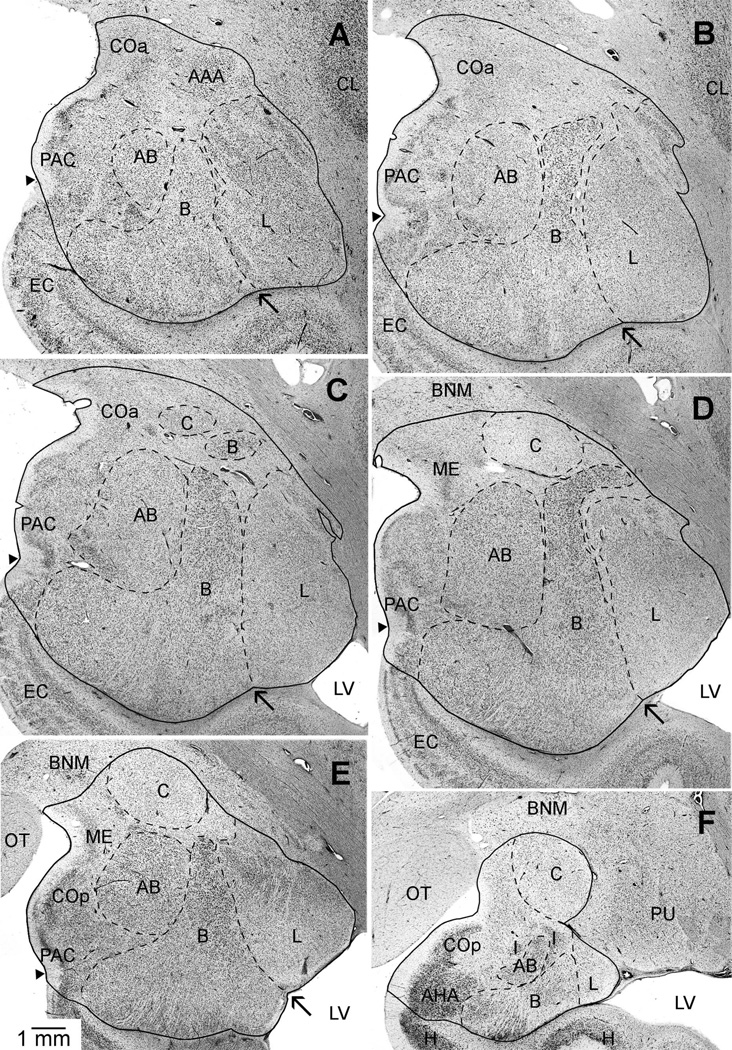
The rostral pole of the amygdala was marked by the first appearance of the basolateral nuclei (Schumann and Amaral, 2005). The external capsule borders the amygdala dorsolaterally, especially at rostral levels. The putamen also borders the amygdala dorsolaterally in caudal sections and can be differentiated from the amygdala by differences in cell structure, density, and organization (Fig. 2F). Dorsomedially, the amygdala is bounded by the substantia innominata, marked by the presence of the basal nucleus of Meynert (Fig. 2D–F). Ventromedially, the semiannular sulcus separates (Figs. 2A–E) the entorhinal cortex from the amygdala and can generally be used as a reliable landmark in addition to cytoarchitecture (Amaral et al., 1987; Sorvari et al., 1995; Insausti et al., 1995; Schumann and Amaral, 2005). At caudal levels, the lateral ventricle and hippocampus form the amygdala’s ventrolateral borders (Figs. 2E,F), whereas, at rostral levels, temporal lobe white matter forms the ventral border (Fig. 2A,B). Within the amygdala, the longitudinal association fiber bundles (Price et al., 1987), also referred to as the meduallary laminae (Heimer et al., 1999), generally mark the boundaries between the major nuclei.
Figure 2.
A–F: A series of brightfield photomicrographs illustrating the boundaries of the amygdala, lateral, basal, accessory basal, and central nuclei in coronal sections of the left hemisphere of a chimpanzee. Images are from rostral (A,B), midrostrocaudal (C,D), and caudal (E,F) positions in the amygdala. Arrows point to the “notch” that separates the ventral borders of the lateral and basal nuclei. Small arrow-heads indicate the position of the semiannular sulcus used to mark the division between the cortical amygdaloid nuclei and the adjacent entorhinal cortex (anterior) or hippocampus (posterior). Abbreviations: AAA, anterior amygdaloid area; AB, accessory basal nucleus; AHA, amygdalohippocampal area; B, basal nucleus; BNM, basal nucleus of Meynert; C, central nucleus; CL, claustrum; COa, anterior cortical nucleus; COp, posterior cortical nucleus; EC, entorhinal cortex; H, hippocampus; I, intercalated nuclei; L, lateral nucleus; LV, lateral ventricle; ME, medial nucleus; OT, optic tract; PAC, periamygdaloid cortex; PU, putamen. Images follow radiological conventions. Scale bar = 1 mm in E (applies to A–F).
Lateral nucleus
The lateral nucleus is the most laterally positioned nucleus of the amygdala and has been divided into four subdivisions in macaques and two in humans (Pitkänen and Kemppainen, 2002). Its lateral, dorsal, and ventral borders are consistent with those of the lateral amygdala. Rostrally and dorsally, the lateral nucleus is in close proximity to the ventral claustrum, which is distinguished by larger, more darkly staining cells. The medial border of the lateral nucleus is defined by the lateral medullary lamina. Cells in this region are smaller and more compact than cells in the adjacent basal nucleus. The ventral aspect of the lateral medullary lamina often terminates above the ventralmost extent of the lateral and basal nuclei, creating a notch (see arrows in Fig. 2A–E). This feature may be used as an additional landmark to distinguish between the two nuclei at ventral levels where the lamina is less prominent. Caudally, the comparatively larger cells of the lateral nucleus distinguish it from the dorsally adjacent putamen.
Basal nucleus
The basal nucleus is separated from the lateral, accessory basal, central, and intercalated nuclei by the medullary laminae. The human and nonhuman basal amygdala has been divided into three subdivisions: a large-celled “magnocellular” division, which is located dorsally, a small-celled parvicelluar division, which comprises the rostral and ventral portions of the nucleus, and an intermediate division located between the two (Price et al., 1987; Sorvari et al., 1995). The basal nucleus contains the largest cells in the amygdala and is situated between the accessory basal and lateral nucleus (Figs. 1A–F, 2B–E). The lateral medullary lamina divides the lateral aspect of the basal nucleus from the lateral nucleus. The intermediate medullary lamina divides the medial aspect of the basal nucleus from the accessory basal nucleus (Fig. 2). The basal and accessory basal nuclei are further distinguished from one another by differences in cell size. Thus, the presence of such large cells in the basal nucleus can generally be used to distinguish it from the medial aspect of the lateral nucleus and the ventrolateral aspect of the accessory basal nucleus.
Accessory basal nucleus
The intermediate medullary lamina and the large cells of the basal nucleus distinguish the lateral border of the accessory basal nucleus. The medial border is demarcated by the medial medullary lamina, which divides the accessory basal nucleus from the superficial cortical nuclei. Our definitions of the accessory basal nucleus included three recognized subdivisions (Price et al., 1987; Sorvari et al., 1995; Freese and Amaral, 2009; but see also de Olmos, 2004, for a different delineation scheme). The small-celled parvicellular division is located rostrally and laterally. The large-celled magnocellular division is positioned dorsally and runs from midrostrocaudal levels to the caudal extent of the nucleus. The ventromedial division comprises a small, compact, grouping of large sized, darkly stained cells on the ventromedial aspect of the nucleus. It runs for only a short extent through midrostrocaudal levels of the nucleus (Fig. 2B–D) and shows a slightly different histochemical profile than the immediately adjacent parvicellular division. For example, parvalbumin levels in this division are intermediate between those in the magnocellular and parvicellular divisions (Sorvari et al., 1995; Ichinohe and Rockland, 2005).
Central nucleus
The central nucleus is encapsulated and separated from the substantia innominata, dorsally, and the basolateral nuclei, ventrally, by fiber bundles (Fig. 2C–F). This feature, as well as its smaller, more lightly staining and less densely packed cells, distinguishes it from the superior aspects of the adjacent basal and accessory basal nuclei and the ventromedial surface of the putamen (Fig. 2F). It lies caudal to the anterior amygdaloid area (Fig. 2A), which contains more darkly staining and diffuse neuronal populations than the central nucleus. Throughout much of its caudal extent, the central nucleus is often nestled between a few of the distinct, small, and darkly staining intercalated amygdaloid nuclei, which flank the white matter fibers surrounding the nucleus on its ventral border and further clarify its position (Fig. 2F). There are two recognized subdivisions of the central nucleus, a lateral and medial division, which are separated by fiber bundles (Price et al., 1987; Sorvari et al., 1995).
Data collection
Neuron numbers were estimated by using the optical disector probe in combination with fractionator sampling (West, 1993) in the StereoInvestigator software suite (MBF Bioscience, Williston, VT). For the majority of specimens, stereological analyses were performed by using a Dell workstation that received live video from an Optronics MicroFire color video camera (East Muskogee, OK) attached to a Nikon Eclipse 80i microscope equipped with a Ludl MAC5000 stage (Hawthorn, NY) and a Heidenhain z-axis encoder (Plymouth, MN). Sections were viewed through a 100× oil objective (NA 1.4) under Köhler illumination. The disector frame size was 60 × 60 µm, with a height of 9 µm, which yielded an average of one to three neurons per counting frame across species. Section thickness was measured at every site. Section thicknesses varied from 11 to 17 µm. To determine whether guard zones were necessary, we performed z-axis counts on paraffin-embedded and cryosectioned tissue (Andersen and Gundersen, 1999; Gardella et al., 2003; Carlo and Stevens, 2011). Both processing techniques yielded sections with fewer neurons at the margins of the tissue than in the center, indicating that tissue processing may have produced artifacts that impacted the distribution of neurons in the z-axis (Andersen and Gundersen, 1999; Gardella et al., 2003). To ensure that these artifacts at the margin of the tissue did not influence our counts, we applied guard zones of 1–3 µm, depending on section thickness.
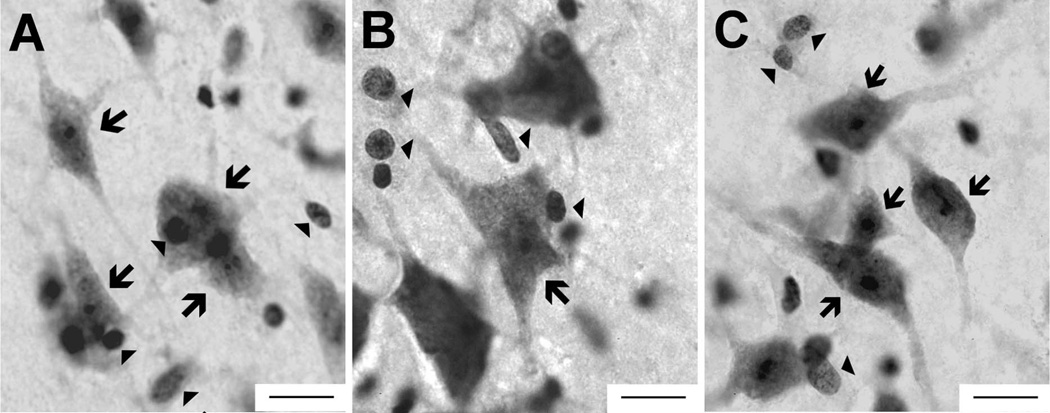
In many cases, every available section was sampled, but when the sample interval included more than one section, the starting section in the interval was chosen at random and subsequent sections were sampled at fixed intervals, as is standard procedure (West, 1993). The distance between sampled sections ranged between 0.4 and 1.2 mm, reflecting the diverse array of brain sizes in the sample. Due to these brain size differences and also to differences in volume across the nuclei, several different grid sizes were utilized for each nucleus in each species (nonhuman primates: Table 2; humans: Schumann and Amaral, 2005). A neuron was counted only if its nucleus first came into view within the counting frame or intersected the lines of inclusion located on the frame’s top and right sides, but not the lines of exclusion to the bottom and left (Schumann and Amaral, 2005). A cell was marked as a neuron if it exhibited a large, clear, lightly stained nucleus, containing a single, distinct nucleolus, surrounded by darkly stained clumps of Nissl substance covering the remainder of the neuronal perikarya extending to the proximal portions of the dendritic processes (Fig. 3, arrows). Because the nuclei of the amygdala are generally regularly shaped, we report coefficient of error values by using m = 1 rather than m = 0, the latter of which is more appropriate for irregularly shaped structures (Gundersen et al., 1999). In no case did the coefficient of error (Gundersen et al., 199, m = 1) exceed 8% for any region analyzed, indicating that the precision of stereological estimates was high. Thus sampling variance is unlikely to contribute more than 50% to observed group variance, a measure suggested to balance sampling precision and efficiency (West et al., 1991).
TABLE 2.
Grid Sizes Used in Each Species
| Nonhuman Species |
Grid area (µm2) |
Neurons counted (Average) |
Sections sampled (Average) |
|
|---|---|---|---|---|
| Amygdala | Chimpanzee | 1,7002–2,3002 | 223 | 11 |
| Bonobo | 2,3002 | 181 | 12 | |
| Gorilla | 2,4002 | 218 | 10 | |
| Orangutan | 2,4002 | 208 | 12 | |
| Gibbon | 2,000–2,4002 | 216 | 10 | |
| Macaque | 1,7002 | 288 | 10 | |
| Lateral | Chimpanzee | 1,0002–1,2002 | 220 | 10 |
| Bonobo | 1,0002–1,2002 | 179 | 11 | |
| Gorilla | 1,0002–1,2002 | 217 | 9 | |
| Orangutan | 1,0002–1,2002 | 243 | 11 | |
| Gibbon | 1,0002–1,2002 | 211 | 10 | |
| Macaque | 8002 | 378 | 9 | |
| Basal | Chimpanzee | 1,2002–1,5002 | 205 | 10 |
| Bonobo | 1,2002–1,5002 | 173 | 11 | |
| Gorilla | 1,3002 | 235 | 9 | |
| Orangutan | 1,2002–1,6002 | 164 | 10 | |
| Gibbon | 1,2002 | 150 | 10 | |
| Macaque | 9002 | 236 | 9 | |
| Accessory basal |
Chimpanzee | 8002 | 187 | 10 |
| Bonobo | 8002 | 171 | 11 | |
| Gorilla | 8002–1,0002 | 164 | 9 | |
| Orangutan | 7002–1,0002 | 173 | 10 | |
| Gibbon | 7002 | 175 | 10 | |
| Macaque | 6002 | 278 | 10 | |
| Central | Chimpanzee | 5002 | 202 | 10 |
| Bonobo | 5002 | 190 | 11 | |
| Gorilla | 5002 | 187 | 9 | |
| Orangutan | 5002 | 209 | 10 | |
| Gibbon | 4002 | 173 | 9 | |
| Macaque | 6002 | 178 | 10 |
Figure 3.
Tissue from the chimpanzee (A) lateral nucleus, (B) basal nucleus, and (C) central nucleus as viewed through a 100× objective, the magnification used for data collection. Morphological features of neurons (arrows) and glia (arrowheads) can be distinguished at this magnification. Scale bar = 15 µm in A–C.
As in our previous analysis (Schumann and Amaral, 2005), postprocessing section thickness was measured at each stereologic probe site so that mean measured section thickness could be used to estimate the disector’s thickness sampling fraction when neuron counts were calculated. Alternatively, the use of number-weighted section thickness has been advocated to estimate neuron numbers when considerable deformation is present in the z-axis (Dorph-Petersen et al., 2001). Thus, we tested whether our choice of thickness measure would significantly influence our estimates. For each nucleus within each taxonomic group, estimates calculated with number-weighted thicknesses varied less than 3% on average from values calculated with mean measured section thickness. These differences were not statistically significant when they were assessed within individual species or across the entire sample (Student’s t-test: P > 0.05 for the amygdala and all nuclei).
We quantified data from one hemisphere in each specimen (Table 1) to maximize sample size. There was no influence of laterality on amygdala volume in our previous volumetric analysis (Barger et al., 2007) using many of the same specimens. Data for 10 of the 11 human amygdala were collected by C.M.S. (Schumann and Amaral, 2005). In an interobserver reliability test performed on 2 of the 10 human specimens, N.B. produced neuron counts that were more than 95% concordant with previously published data (Schumann and Amaral, 2005), confirming that data from the two analyses could be reliably combined.
Data analysis
Data from all structures passed the Shapiro–Wilk test for normality in all species; however, we opted to use nonparametric analyses when possible, as most distributions exhibited evidence of skewness and deviation for mesokurtosis likely due to the small intraspecific sample sizes. In addition to raw neuron numbers, we calculated the percent of total amygdala neurons contained in each amygdaloid nucleus to factor out the influence of total amygdala neuron number on interspecific comparisons. This measure was defined as the quotient of the neuron number in a nucleus divided by total amygdala neuron number (e.g., central neuron number/amygdala neuron number). Both raw neuron counts and percentage data were subjected to a Kruskal–Wallis test to determine whether means differed significantly across species. If significant variation was present, we further explored differences between individual species post hoc using the Mann–Whitney U test (SPSS 17, SPSS, Chicago, IL).
We performed allometric regressions in two conditions: 1) with humans included to assess trends across primates; and 2) with humans excluded to determine whether observed human values were significantly greater than predicted by nonhuman primate values. To investigate allometric trends, species mean log-transformed data were entered into the phylogenetic independence contrasts program PDAP (Garland et al., 1992) in Mesquite 2.74 (Maddison and Maddison, 2010). Phylogenetic branch lengths (Purvis, 1995) were log transformed so that standardized contrasts did not correlate with their standard deviations (Garland et al., 1992). The number of neurons in each nucleus was regressed against the total number of amygdala neurons minus the neuron number in that nucleus to eliminate statistical artifacts that results from regressing a structure against itself. Regression equations and confidence intervals obtained from PDAP were mapped back into the original data space, representing contemporary species data, for subsequent analysis. We chose to include all nonhuman primate species in the interest of increased statistical power. Although the macaque mean data point may be regarded as a possible statistical outlier that may influence the results of our analysis, the slopes of regression lines drawn through non-macaques fit well within the 95% confidence intervals of lines drawn through all species.
We tested for significantly positive or negative residuals to determine whether changes in neuron distribution reflected adherence to allometric trends across primates or derived features deviating from these trends. We also use this metric because of the tendency for PDAP to produce inflated prediction intervals (Midford et al., 2003). The value of each species’s mean residual was subjected to a Student’s one-sample t-test to determine whether residuals significantly deviated from 0. For more intuitive interpretation, we provide and report percent residuals for each species, which were calculated from untransformed values by using the following formula: (observed − predicted value)/predicted value.
Photomicrograph production
Images were taken on either a Nikon Eclipse 80i microscope at 1× (Fig. 2) or 100× (Fig. 3) magnification or a Leica MZ6 stereomicroscope at 0.63× magnification (Fig. 1) with an Optronics MicroFire camera and the program Picture Frame 2.3 (Optronics, Inc, East Muskogee, OK). The entire chimpanzee amygdala is too large to be captured at 1× thus component images were montaged in Adobe Photoshop Elements 5.0 (Adobe Systems, San Jose, CA) to produce each panel in Figure 2. To ensure that published images best approximated the clarity and contrast of slides as viewed under the microscope, brightness, contrast, and sharpness were manipulated in all images by using GIMP 2.6.2 (http://www.gimp.org/) and Adobe Photoshop Elements 5.0. Boundaries for published images were drawn with GIMP 2.6.2.
RESULTS
Neuron numbers
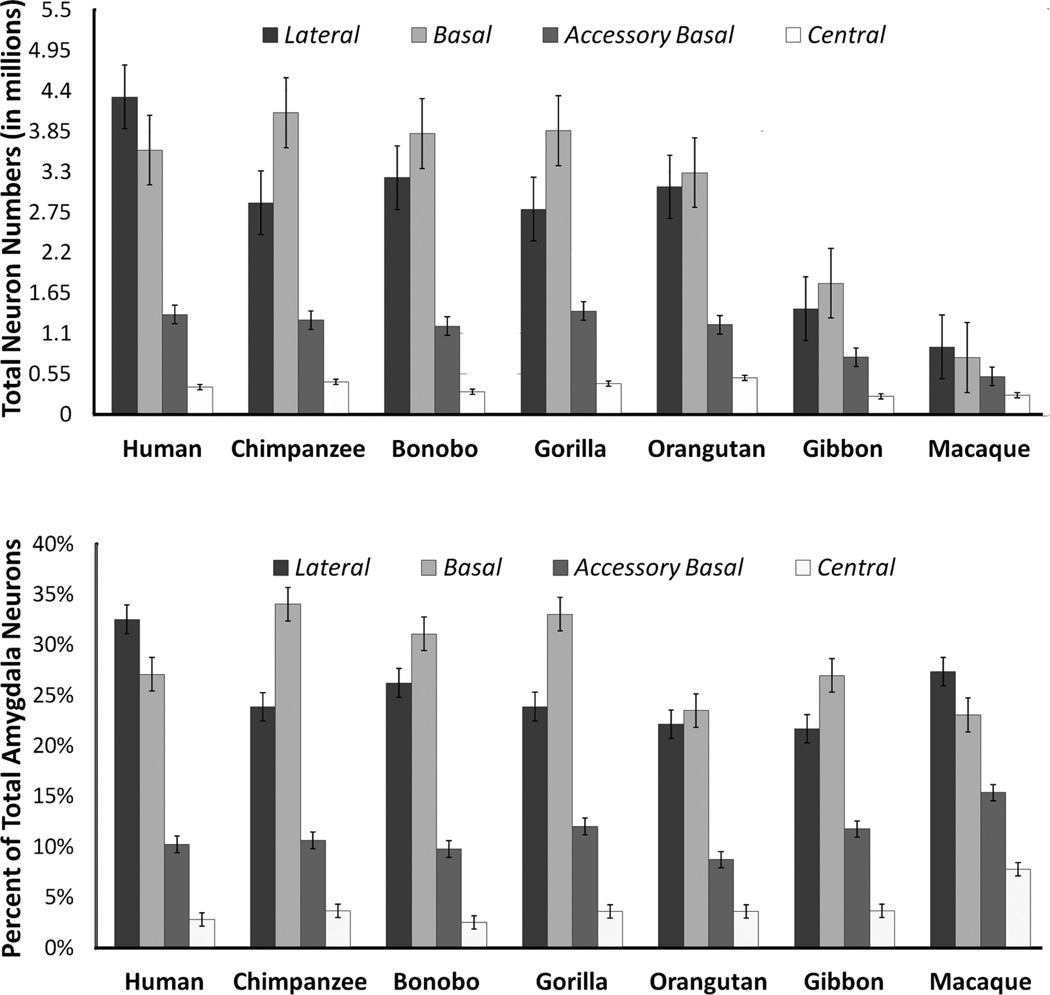
The total number of neurons in the amygdala of humans and great ape species (hominids) overlapped with one another. All hominid amygdala exhibited approximately 12–14 million neurons. The absolute number of amygdala neurons in non-hominids was generally less than in hominids. Specifically, the amygdala of lesser apes, the gibbons, contained nearly half this number (6.6 million) and the amygdala of the long-tailed macaques, roughly a fourth (3.4 million; Table 3 and Fig. 4). This observation was statistically supported by the Kruskal–Wallis analysis, which detected significant differences between species in the mean number of neurons in the amygdala and in most nuclei (Amygdala: H(6) = 14.10, P = 0.029; Lateral: H(6) = 24.20, P < 0.000, Basal: H(6) = 16.08, P = 0.013; Central: H(6) = 20.44, P = 0.002). Differences in the accessory basal nucleus approached significance (H(6) = 11.32, P = 0.079).
TABLE 3.
Average Neuron Number × 106 and (Standard Deviation) for Each Nucleus in Each Species1
| ROI | Human (Hu) |
Chimpanzee (Ch) |
Bonobo (Bo) |
Gorilla (Go) |
Orangutan (Or) |
Gibbon (Gi) |
Macaque (Ma) |
Post hoc comparisons |
|---|---|---|---|---|---|---|---|---|
| Amygdala | 13.27 (3.70) | 12.05 (5.53) | 12.28 (2.92) | 11.68 (2.37) | 13.99 (2.97) | 6.61 (2.90) | 3.35 (0.24) | Ma < Hu, Ch, Bo, Go, Or Gi < Hu, Bo, Or, Go |
| Lateral nucleus | 4.32 (1.11) | 2.87 (0.95) | 3.22 (0.60) | 2.79 (0.33) | 3.09 (0.38) | 1.43 (0.32) | 0.92 (0.02) | Ma < Hu, Ch, Bo, Go, Or Gi < Hu, Ch, Bo, Go, Or Hu > Ch, Bo, Go, Or, Gi, Ma |
| Basal nucleus | 3.59 (1.29) | 4.10 (1.44) | 3.82 (1.37) | 3.86 (1.27) | 3.29 (1.02) | 1.78 (0.57) | 0.77 (0.06) | Ma < Hu, Ch, Bo, Go, Or Gi < Hu, Ch, Bo, Or, Go |
| Accessory basal nucleus |
1.36 (0.32) | 1.28 (0.62) | 1.20 (0.50) | 1.40 (0.34) | 1.22 (0.23) | 0.78 (0.50) | 0.52 (0.12) | ANOVA, ns |
| Central nucleus | 0.37 (0.09) | 0.44 (0.02) | 0.31 (0.08) | 0.42 (0.13) | 0.50 (0.09) | 0.24 (0.06) | 0.26 (0.02) | Ma < Hu, Ch, Go, Or Gi < Hu, Ch, Go, Or Bo < Ch, Or Hu < Ch, Or |
The final column illustrates post hoc differences that were significant at P < 0.05 in boldface or that approached significance at P < 0.08 in italics.
Figure 4.
Histograms indicating the average number of neurons (× 106) in the amygdala and four nuclei (top) and the average percent of total amygdala neurons distributed to the lateral, basal, and accessory basal nuclei across species (bottom) (n = 35). Error bars represent standard error.
Post hoc comparisons confirmed that species differences in the number of neurons in each amygdaloid nucleus were largely split between large-brained hominids and smaller brained non-hominids (Table 3). That is, the amygdaloid nuclei of great apes and humans generally contained more neurons than those of gibbons and macaques.
Neuron numbers in individual nuclei stood out significantly in only two species. In humans, the lateral nucleus contained significantly more neurons (4.32 million) than all other primates analyzed (Fig. 4). Additionally, the human central nucleus contained significantly fewer neurons (0.37 million) than chimpanzee (0.44 million) and orangutan (0.50 million) central nuclei (Table 3 and Fig. 4). The bonobo central nucleus also contained significantly fewer neurons (0.31 million) than the central nuclei of chimpanzees and orangutans, but not significantly fewer than humans (Table 3 and Fig. 4). The average number of neurons in the gorilla central nucleus (0.42 million) was also greater than in bonobos or humans, but this difference did not reach significance.
Nuclei as percent of total amygdala neurons
There were significant species differences in the percent of total amygdala neurons distributed to each nucleus (Kruskal–Wallis: Lateral: H(6) = 20.52, P = 0.002; Basal: H(6) = 19.13, P = 0.004; Accessory basal: H(6) = 16.43, P = 0.012; Central: H(6) = 13.72, P = 0.033). Species’ mean values and the results of post hoc analysis are presented in Table 4, and species mean values are presented graphically in Figure 4.
TABLE 4.
Neuron Number in Each Nucleus as a % of Total Amygdala Neurons1
| ROI | Human (Hu) |
Chimpanzee (Ch) |
Bonobo (Bo) |
Gorilla (Go) |
Orangutan (Or) |
Gibbon (Gi) |
Macaque (Ma) |
Post hoc comparisons |
|---|---|---|---|---|---|---|---|---|
| Lateral nucleus | 32.5 | 23.9 | 26.2 | 23.9 | 22.1 | 21.7 | 27.4 | Hu > Ch, Bo, Go, Or, Gi, Ma |
| Basal nucleus | 27.1 | 34.0 | 31.1 | 33.0 | 23.5 | 27.0 | 23.0 | Ma < Ch, Bo, Go Or < Ch, Bo, Go Hu < Ch, Bo, Go |
| Accessory basal nucleus | 10.2 | 10.6 | 9.8 | 12.0 | 8.7 | 11.8 | 15.4 | Go > Ch, Bo, Or Or < Ch, Bo, Go, Hu, Gi, Ma Ma > Hu, Ch, Bo, Or |
| Central nucleus | 2.8 | 3.7 | 2.5 | 3.6 | 3.6 | 3.7 | 7.8 | Ma > Hu, Ch, Bo, Go, Or, Gi Bo < Ch, Go |
The final column illustrates post hoc differences that were significant at P < 0.05 in bold or that approached significance at P < 0.08 in italics.
Post hoc tests indicated that human amygdala contained a significantly greater percentage of neurons in the lateral nucleus than great apes. At 32.5%, the percentage of neurons in the human lateral nucleus was the largest of any nucleus analyzed in the human amygdala.
Ranging from 23.5 to 34%, the percentage of neurons in the basal nucleus of all ape species was the largest of any nuclei analyzed in the ape amygdala. Among the apes, the percentage of neurons in the orangutan basal nucleus (23.5%) was significantly smaller than in the other great apes (31.1–34%). Orangutans also had a significantly smaller percentage of neurons in the accessory basal nucleus (8.7%) than other apes (9.8–12%). Gorilla amygdala contained proportionately more neurons in the accessory basal nucleus than other great apes.
In long-tailed macaques, like humans, the largest percent of amygdala neurons was located in the lateral nucleus (Fig. 4). Nonetheless, the average percentage of neurons in macaque lateral nuclei (27.4%) was significantly smaller than in human lateral nuclei (32.5%). The long-tailed macaque amygdala contained a significantly greater percentage of accessory basal (15.4%) and central neurons (7.8%) than most other species.
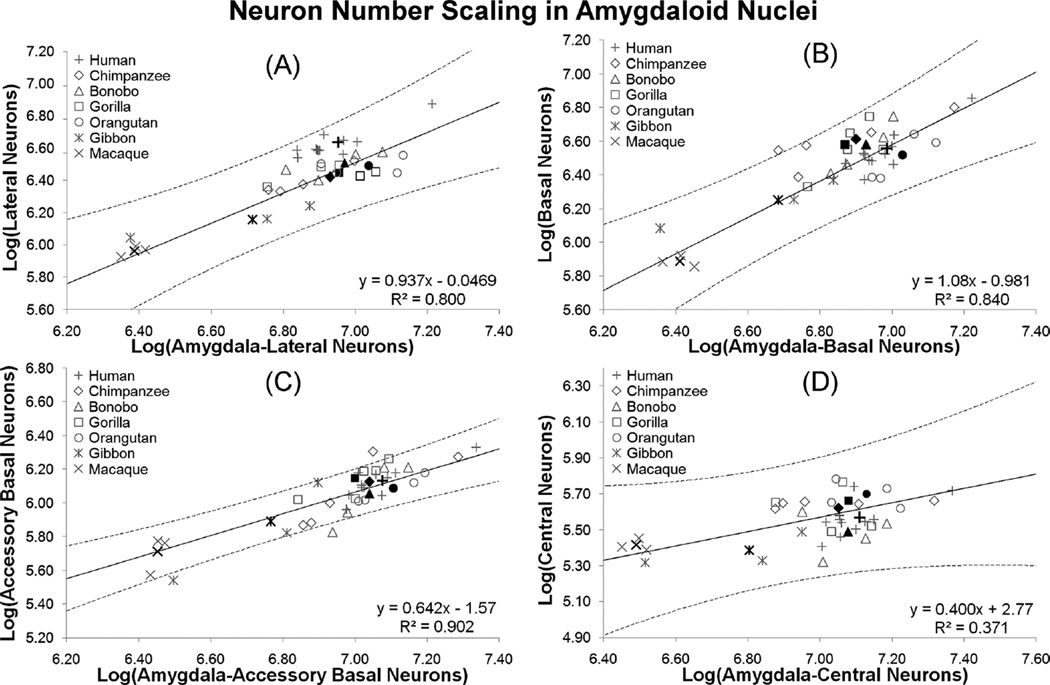
Allometric analysis
The lateral nucleus fell very slightly below isometry with respect to number of neurons in the rest of the amygdala (b = 0.937 ± 0.539 (95% CI), R2 = 0.800, P < 0.01; Fig. 5A). Neuron numbers in the basal nucleus scaled with positive allometry (b = 1.08 ± 0.542 (95% CI), R2 = 0.840, P < 0.01; Fig. 5B). Because regressions for both the basal and lateral nuclei contain a slope of 1 in the 95% confidence interval, it is possible that both nuclei scale with isometry. Neurons in the accessory basal nucleus scaled considerably more negatively (b = 0.642 ± 0.243 (95% CI), R2 = 0.902, P < 0.01; Fig. 5C). The slope for central nucleus neuron numbers was low, but did not correlate significantly with total amygdala numbers (b = 0.400 ± 0.599 (95% CI), R2 = 0.371, P = 0.147; Fig. 5D).
Figure 5.
Independent contrasts regression plotting the log of total amygdala neuron number against the log of the neuron numbers in (A) the lateral nucleus, (B) the basal nucleus, (C) the accessory basal nucleus, and (D) the central nucleus with all species included in each regression. Individual data points are plotted as open, gray markers and species mean values are plotted as closed, black markers.
Human departures from allometry
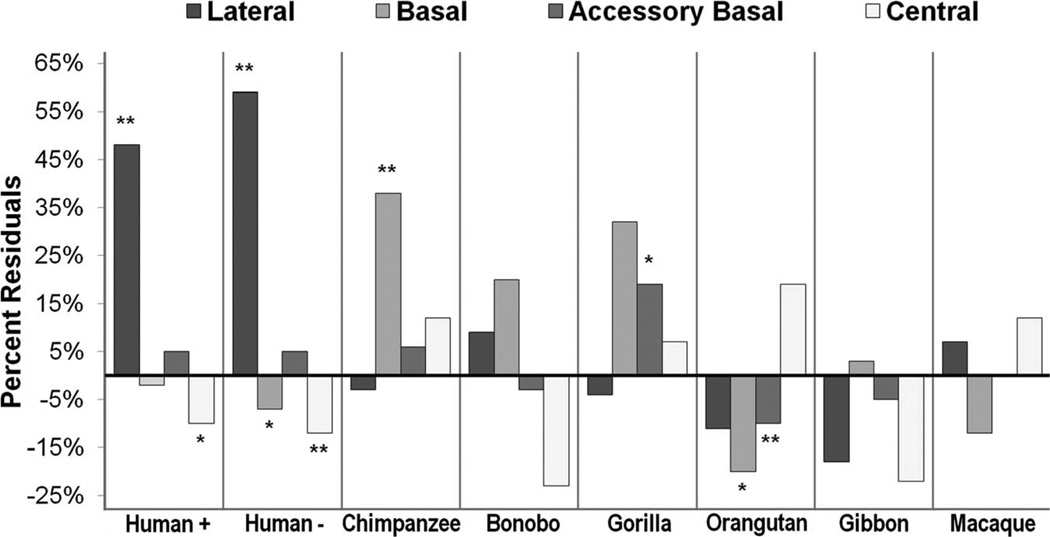
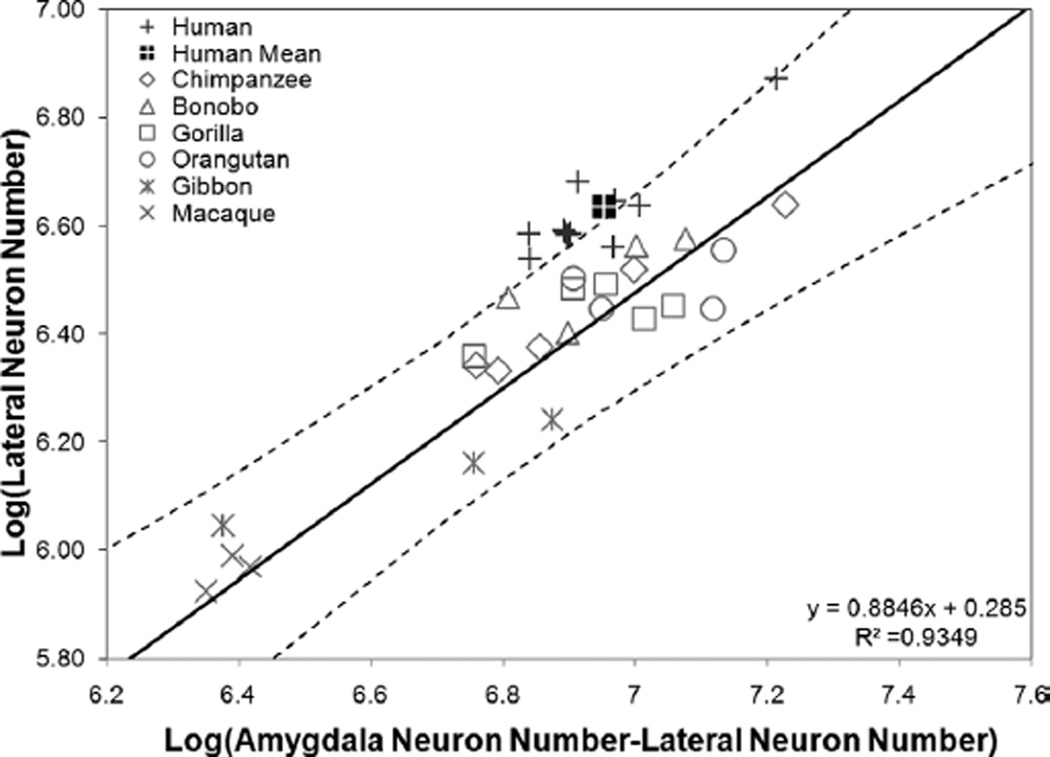
Human residuals for the lateral nucleus were significantly positive whether humans were included (residual = 0.174, P < 0.000) or excluded (residual = 0.202, P < 0.000) from the prediction equation (Table 5 and Fig. 6). When humans were excluded from the regression, the percent residual for observed human values was 59% (Table 5 and Fig. 6). Additionally, human data points largely fell outside of the 95% prediction interval when they were excluded from the analysis, and the human mean clearly fell outside of this range (Fig. 7). Human residuals for the central nucleus fell 12% below predicted values when humans were excluded (residual = −0.061; P = 0.028; Table 5 and Fig. 6), but the regression equation did not reach significance (b = 0.423; R2 = 0.390; P = 0.185). Neuron numbers in the human basal nucleus were nearly significantly smaller than expected when humans were excluded from the regression (residual = −0.037, P = 0.067), but the magnitude of this deviation was low, approximately 7% (Table 5 and Fig. 6).
TABLE 5.
Percent Residuals Derived From Log–Log Independent Contrast Regressions1
| Human+ | Human− | Chimpanzee | Bonobo | Gorilla | Orangutan | Gibbon | Macaque | |
|---|---|---|---|---|---|---|---|---|
| Lateral nucleus | 48 | 59 | −3 | 9 | −4 | −11 | −18 | 7 |
| Basal nucleus | −2 | −7 | 38 | 20 | 32 | −20 | 3 | −12 |
| Accessory basal nucleus | 5 | 5 | 6 | −3 | 19 | −10 | −5 | 0 |
| Central nucleus | −10 | −12 | 12 | −23 | 7 | 19 | −22 | 12 |
Residuals that were significant at P < 0.05 are in bold and that approached significance at P < 0.08 are in italics.
Human+, percent residual with all species included; Human−, percent residual excluding human data from the regression.
Figure 6.
Average percent residuals from regression equations in each nucleus for each species. Starred bars represent values that were statistically significant (**) or close to statistically significant (*) from a residual of 0. Human+, percent residual with all species included; Human−, percent residual excluding human data from the regression.
Figure 7.
Independent contrasts regression plotting the log of amygdala neuron number against the log of neuron numbers in the lateral nucleus with humans excluded from the regression (n = 24).
Allometric departures in nonhuman primates
Results are presented in Table 5 and graphically in Figure 6. Chimpanzees exhibited significant positive residuals for basal nucleus neuron number (residual = 0.16, P = 0.033). In contrast, orangutan mean residual neuron numbers for the basal nucleus were nearly significantly smaller than predicted by regressions drawn through other primates (residual = −0.11, P = 0.059). Because human residuals were low for this nucleus and may have a negative influence on the regression line, we also tested orangutan residuals in a regression that excluded human data points for the basal nucleus. In this case, the number of neurons in the orangutan basal nucleus was significantly smaller than predicted for a nonhuman primate with a similar number of total amygdala neurons (residual = −0.13, P = 0.035). Orangutans’ residuals were significantly negative for the accessory basal nucleus, as well (residual = −0.046, P = 0.035). Alternatively, gorillas’ mean accessory basal neuron number residual was positive and approached significance (residual = 0.071, P = 0.080). When humans are excluded from the lateral nucleus regression, bonobo residuals for this nucleus are nearly significantly positive (residual = 0.05, P = 0.053).
Summary
Absolute neuron numbers in the amygdala and most nuclei generally overlapped in humans and great apes and were greater in these species than in gibbons and macaques. In one of the few deviations from this general observation, the human lateral nucleus contained significantly more neurons than the lateral nucleus of any other species in the analysis. When the numbers of neurons in each nucleus were considered as a proportion of total amygdala neurons, neuron numbers in the lateral nucleus were greatest in humans as well. Accordingly, humans exhibit 59% more neurons than predicted by allometric regression lines drawn through other primates. Together, the data provide robust evidence that a greater proportion of amygdala neurons are distributed to the lateral nucleus in humans when compared with our closest relatives.
The amygdala in apes contained a higher percentage of neurons in the basal nucleus than macaques and humans, and the human basal nucleus contained slightly fewer neurons than predicted by trends across nonhuman species. The chimpanzee basal nucleus contained more neurons than predicted, whereas gorillas distributed more neurons to the accessory basal nucleus. Neuron numbers in the basal and accessory basal nuclei are smaller in orangutans than predicted by trends across other nonhuman primates.
DISCUSSION
The goal of the present analysis was to examine the distribution of neurons in the amygdala of humans and apes. We quantified the number of neurons in the amygdala and its lateral, basal, accessory basal, and central nuclei in 24 nonhuman primate specimens representing all great ape species, gibbons, and macaques (Table 3). We found that the human amygdala is not simply an evolutionarily “scaled-up” version of an ape amygdala. The human amygdala contained significantly and proportionately more neurons in the lateral nucleus than the ape amygdala (Fig. 4). This number was greater than expected based on trends across apes and macaques (Fig. 7). In contrast, neuronal populations in the ape amygdala were highest in the basal nucleus. The data indicate that, after the human lineage split from the last common ancestor we shared with great apes, a shift in amygdala organization occurred that resulted in increased neural populations in the lateral nucleus.
Evolutionary scaling of amygdaloid nuclei across species
The percentage and number of neurons found in each amygdaloid nucleus varied across species, with most of this variation accounted for by allometric scaling expectations. Because each nucleus exhibited a different scaling rate (Fig. 5), an increase in amygdala neuron number will have different, but largely predictable, consequences for the percentage of neurons distributed to any particular nucleus. In our sample, basal nucleus neuron numbers increased at a slightly greater rate than total amygdala neuron number (slope = 1.1). Thus, increases in total amygdala neuron number will lead to an increasingly larger percentage of neurons being distributed to the basal nucleus. In the lateral nucleus, neuron number scaled with slight negative allometry (slope = 0.9), nearly keeping up with changes in total amygdala neuron number. As confidence intervals for the regression of both the lateral and basal nuclei contain a slope of 1, it cannot be discounted that neuron numbers in both nuclei scale isometrically with total amygdala neuron number. The accessory basal nucleus, in contrast, exhibited clear negative allometry with a slope of 0.6 (and upper confidence limit of 0.9), suggesting that neurons in this nucleus will only double for every tripling of total amygdala neuron number on average. Increases in central nucleus neuron populations did not show a strong relationship with total amygdala neuron number. The regression data suggest a trend for neurons in the central nucleus to double for every fivefold increase in total amygdala neuron number, although larger samples are needed to determine whether this relationship is significant.
Evolutionary specializations in hominoid amygdala
Human amygdala
The human lateral nucleus contained a disproportionately large number of neurons compared with other primates, especially the great apes. The human amygdala contained significantly more neurons in the lateral nucleus, both absolutely and proportionately, than was the case in apes (Tables 3 and 4), and this number was greater than expected based on trends across apes and macaques (Table 5 and Fig. 6). Neuron numbers in the human lateral nucleus were nearly 60% greater than predicted by allometric trends, a degree of magnitude rarely seen in comparative analyses of human brain evolution (Sherwood et al., 2012). For example, the volume of the human neocortex is 24% larger than expected for a primate of our brain size (Rilling and Insel, 1999), whereas the human frontal lobe, long assumed to be enlarged, is approximately the size expected for an ape of human brain size (Semendeferi et al., 2002; Semendeferi and Damasio, 2000). Increases in lateral neuron populations are perhaps balanced by decreases in neuron numbers in the central and basal nuclei, which exhibit subtle reductions in humans (Table 3 and Fig. 6).
We previously reported that the volume of the amygdala is, on average, over 3 times larger in humans than in great apes (Barger et al., 2007). In contrast, we found that amygdala neuron number did not differ between the two groups. Given that great ape and human neuron numbers also overlap in area 13, a functionally and connectively related limbic structure in the posterior orbitofrontal cortex (Semendeferi et al., 1998), neuron numbers in hominid limbic structures may be characterized by evolutionary conservation. However, area 13 is less than twice as large in humans as it is in great apes (Semendeferi et al., 1998). Given this difference, one possibility that remains to be investigated is the potential importance of neuropil expansion in the evolution of the human amygdala.
Other hominids
Human share the phylogenetic classification of hominid with our closet living relatives, the great apes. These include chimpanzees, bonobos, gorillas, and orangutans, in order of their phylogenetic relatedness to humans. Even though neuron numbers were similar across hominids, the distribution of neurons across amygdaloid nuclei varied between humans and great apes, indicating that the human amygdala is evolutionarily reorganized in relation to great ape amygdala. High rates of allometric scaling in the basal nucleus (Fig. 5B) may explain our related finding that neuron numbers in great ape amygdala were highest in the basal nucleus absolutely and proportionately (Fig. 4)
In some cases, we found preliminary evidence that individual great ape species may exhibit neural specializations in the amygdala. The chimpanzee basal nucleus contained 38% more neurons than predicted for a species with a similar number of amygdala neurons, although the absolute number and percentage of basal nucleus neurons was not significantly greater in chimpanzees. We found that the amygdala of bonobos (or “pygmy chimpanzees”) did differ from that of common chimpanzees, and this is consistent with a recent neuroimaging study (Rilling et al., 2011). Bonobo central nuclei contained the smallest number of neurons among hominids. They had nearly significantly fewer neurons in the central nucleus than most other great apes (Table 3). Additionally, bonobo lateral nuclei contained more neurons than all nonhuman hominids, although deviations from predicted values only approached significance in allometric regressions across nonhuman primates. Given this pattern, it is tempting to speculate that, of all the apes, bonobos might come closest to approximating human amygdala organization, but a substantially higher sample size would be needed to test that hypothesis. In gorillas, the accessory basal nucleus contained a larger percentage of neurons than any other hominid species, although residuals for this nucleus only approached significance in regression analyses (Table 5).
Among the great apes, orangutans are the most distantly related to humans. Although, like other great apes, the basal nucleus of orangutans contained more neurons than any other nucleus, the orangutan basal nucleus contained approximately 10% fewer neurons than that of other great apes (Table 4) and neuron numbers in the orangutan basal nucleus were smaller than predicted when scaling rates in nonhuman primates were taken into account (i.e., when humans were excluded from the analysis). In addition, the proportion of neurons in the accessory basal nucleus of the orangutan amygdala was small compared with other primates, and neuron numbers in this nucleus were 10% fewer than predicted by allometric regressions (Tables 4, 5 and Fig. 6). This was not the case for all basolateral nuclei, as the number and percent of neurons in the orangutan lateral nucleus were close to those of other great apes and residuals were not significantly different from 0.
Other hominoids
We are using the term hominoid to refer to the larger phylogenetic classification that includes humans, great apes, and lesser apes, the gibbons and the siamang. Gibbon amygdala contained fewer neurons than human and great ape amygdala, as a whole and in each nucleus analyzed (Table 3), but the organization of the gibbon amygdala followed the pattern present in great apes. Neurons in the gibbon amygdala were distributed predominantly to the basal nucleus (Fig. 4). In no case did the number of neurons in gibbons exceed predicted values for any nucleus analyzed (Fig. 6).
Gibbon neuron numbers exhibited a high degree of individual variation, which may increase the probability that our statistical analyses would produce negative results. An important feature of our gibbon sample is that it represented three distinct species. Traditionally, the social organization of all gibbon species was thought to be the monogamous pair bond; more recent data have challenged this presumption (Malone and Fuentes, 2009). In our study, the two gibbons with the highest numbers of amygdala neurons (Fig. 5) are from two species, Hylobates lar and H. concolor, which have been reported to travel in groups of more than two individuals. H. muelleri, the gibbon species with the lowest number of amygdala neurons in this analysis (Fig. 5), has not been observed traveling in larger groups (Malone and Fuentes, 2009). Thus, it is possible that neuroanatomical variation in our sample might reflect behavioral variation among gibbon species, given that social group size has been shown to correlate with amygdala volume (Barton and Aggleton, 2000; Bickart et al., 2010). Subsequent analyses with larger samples and a broader array of gibbon species would be needed to assess this hypothesis.
Cercopithecoids
One Old World monkey species, the long-tailed macaque, was added to our sample as a phylogenetic outgroup to contrast with hominoids. The number of neurons in individual nuclei of the long-tailed macaque amygdala did not deviate significantly from predictions based on allometric regressions. Thus, it is most likely that differences between the organization of the ape and long-tailed macaque amygdala, i.e., a high percentage of amygdala neurons in the accessory basal and central nuclei, reflect the allometric relationships particular nuclei share with total amygdala neuron number rather than neural adaptations specific to this species (Table 5). A larger cercopithecoid sample would be needed to explore this finding further. Macaque values also appeared to cluster together more closely than great ape or human values. If the coefficient of variation is calculated (standard deviation/mean), macaques exhibit consistently lower values than hominoids.
Although the human amygdala clearly contained more lateral nucleus neurons than any species analyzed, we found that both human and long-tailed macaque amygdala emphasized the lateral nucleus. This does not imply, however, that the human and macaque amygdala are more similar morphometrically than the human and great ape amygdala. Despite the fact that macaques in our study do distribute more neurons to the lateral nucleus that to other nuclei, the human lateral nucleus still contains proportionately more neurons than the macaque lateral nuclus. Additionally, the macaque amygdala contains a higher percentage of neurons in the accessory basal and central nuclei than the human amygdala. Our recently published study evidences a similar amygdala organization in long-tailed macaques; however, we found that rhesus macaques have more neurons in the basal nucleus than in the lateral nucleus, akin to ape amygdala organization (Carlo et al., 2010). Finally, from a phylogenetic perspective, the last common ancestor of humans and apes would share a similar amygdala organization that differs from those of cercopithecoids. Thus, based on the law of parsimony, human-specific increases in the lateral nucleus must have occurred after humans split with our most recent last common ancestor shared with apes and would not reflect the preservation of an ancestral cercopithecoid state (presuming long-tailed macaques represent that state). It may be the case that similarities in the amygdala organization of long-tailed macaques and humans reflect evolutionary parallelism related to functional adaptations. If the distribution of neurons does reflect amygdala function in closely related species, it may be important to consider issues of species-specific variation when investigating functional aspects of the primate nervous system and when using macaque species to model human disorders.
Comparison with previous volumetric findings
Several of the findings from the present analysis are concordant with volumetric findings from our previous analysis (Barger et al., 2007). Specifically, the human lateral nucleus is significantly larger than predicted for a hominoid of our brain size, which is reflected in our findings for neuron numbers in this nucleus. We found that orangutans have significantly smaller accessory basal and basal nuclei than other great apes and this finding is also paralleled by reduced neuron numbers in both nuclei. Although the finding only approached significance, increased neuron numbers in the gorilla accessory basal nucleus would concord with our finding that volume is also increased in gorillas. In contrast, chimpanzees appear to have more neurons in the basal nucleus than predicted, but no such increase was indicated in our volumetric analysis.
Methodological considerations
Given that many of the species in our sample are endangered and tissue samples are rare, we sought to maximize sample size by combining species from a variety of laboratories and collections. Considerable debate has arisen concerning the influence of artifacts from tissue processing on stereological data collection. Counts from paraffin-embedded tissue tend to be higher than from cryosectioned tissue (Ward et al., 2008), and we found this to be the case in our sample to some degree. However, counts from paraffin-embedded tissue were not significantly different from those obtained from cryosectioned tissue, for nearly all nuclei in all species (Mann–Whitney U test: Z = −1.80, P > 0.05, two-tailed). The only exception was chimpanzee total amygdala counts. We tested whether counts from paraffin-embedded or cryosectioned chimpanzee tissue were significantly different from the combined mean to assess the potential impact of this difference. Counts from paraffin-embedded tissue did not differ significantly from the mean (one-sample T-test: t = 1.53, P = 0.26), whereas counts from cryosectioned tissue did (t = −7.94, P = 0.02), suggesting that counts from the former have a greater influence on the group mean.
Evolutionary and functional significance
Neural Connectivity and amygdala evolution
Because the basolateral nuclei are strongly connected to the neocortex (Price et al., 1987; Freese and Amaral, 2009; Stefanacci and Amaral, 2002) and the central nucleus communicates mostly with brainstem and olfactory centers (Price et al., 1987), it has been hypothesized that high rates of neocortical enlargement in primate evolution influenced the more expansive development of the basolateral division, whereas conservation of the autonomic and olfactory systems resulted in the relative stabilization of other nuclei (Stephan et al., 1987; Barton and Aggleton, 2000; Carlo et al., 2010). Barton et al. (2003) tested this hypothesis, finding that increases in neocortical volume are correlated with increases in the volume of the corticobasolateral amygdala (the lateral, basal, accessory basal, and more ventral cortical nuclei), but not the centromedial amygdala (the central nucleus, the anterior amygdaloid area, and the more dorsal cortical nuclei). This link between neocortical enlargement and basolateral volume might be a response to increased processing demands from the neocortex, as the number of neurons in the basolateral nuclei rise concomitantly (Carlo et al., 2010). Buttressing claims that subcomponents of the amygdala evolve in a mosaic fashion (Stephan et al., 1987; Barton and Aggleton, 2000), our data provide further cellular evidence for evolutionary reorganization in the primate amygdala, which occurs largely as a result of variation in the scaling patterns of individual nuclei.
In terms of cellular increase across hominoids, the basal nucleus appears to increase at the fastest rates as the total number of neurons in the amygdala increases. In primates, neocortex hyperscales with brain size, occupying increasingly larger proportions of total brain volume as brain size increases (Stephan and Andy, 1969; Rilling and Insel, 1999). Because the basal nucleus is the primary source of output to the neocortex, (Freese and Amaral, 2009), it may be the case that the processing needs of the basal nucleus increase as brain, and, correspondingly, amygdala size increases.
A low allometric coefficient indicates that central nucleus neuron populations do not keep up with changes in total amygdaloid neuron numbers. In fact, we found only a weak relationship between increases in central neuron numbers and neuron numbers in the entire amygdala. Previous analyses suggest that the central nucleus, with its heavy projections to autonomic regions, is remarkably conserved across primates in terms of volume and neuron number scaling (Stephan et al., 1987; Carlo et al., 2010). This may reflect the fact that its major targets, hypothalamic and brainstem nuclei, are themselves quite conserved (Price et al., 1987; Stephan et al., 1987; Carlo et al., 2010).
In relation to great apes, the number of neurons in the human lateral nucleus was increased; this may also reflect its connectivity. Specifically, the lateral nucleus, as the primary recipient of cortical input, evaluates multimodal information about stimulus characteristics arriving predominantly from temporal lobe association cortices (Stefanacci and Amaral, 2002; LeDoux, 2007; Freese and Amaral, 2009). The human temporal cortex is 23% larger than predicted based on trends in other primates, and the temporal lobe is the only major lobe that is known to be differentially expanded in humans in relative to apes (Rilling and Seligman, 2002). This elaboration of the temporal lobe includes increase not only in the temporal cortex but also in the subcortical white matter, which may have evolutionary and/or developmental consequences for the lateral nucleus (Rilling and Seligman, 2002; Schenker et al., 2005). It is conceivable that increased processing demands arising from the expanded temporal cortex may engender a disproportionate increase in the size of neuronal populations in the lateral nucleus. The fact that coordinated changes between the temporal cortex and amygdaloid nuclei are present only in humans suggests that these structures may have co-evolved as an integrated functional network as the human lineage split from our last common ancestor with great apes.
Social behavior and amygdala evolution
Many attempts have been made to explain the link between the conspicuously large size of the human brain and human behavioral complexity. An increasingly influential proposition has been the “social intelligence hypothesis,” which asserts that complex primate cognition has arisen in the social, rather than material, environment (Jolly, 1966; Humphrey, 1976; Dunbar, 1993; Byrne, 1996; Herrmann et al., 2007). It has been hypothesized that advanced cognitive capacities in primates arose in response to the demands of navigating complex and dynamic social environments that require an understanding of and adherence to somewhat arbitrary social rules, constraints, and conventions (Humphrey, 1976).
As the complexity of the social environment increases, cognitive systems dedicated to interpreting the identities, communicative signals, intentions, and minds of social partners may become increasingly taxed (Jolly, 1966; Humphrey, 1976; Dunbar, 1993; Byrne,1996). Given the amygdala’s role in social vigilance, its evolution may also be affected by these pressures. In gregarious, social mammals, like primates, the amygdala may be particularly involved in processing the emotional salience of stimuli that mark the relationships and the communicative intent of conspecifics as it is routinely engaged in processing emotionally communicative social signals (Sugiura et al., 2001; Yang et al., 2002; Sander et al., 2005; Adolphs, 2010). In support of this hypothesis, increases in the size of the basolateral division correlate with larger social group sizes and higher frequencies of social play across primate species (Barton and Aggleton, 2000; Lewis and Barton, 2006). In both humans and macaques, within species comparisons indicate that amygdala volume correlates with social network or social group size (Bickart et al., 2010; Kanai et al., 2011; Sallet et al., 2011). Early analyses linked measures of social complexity to neocortical elaboration in primates (Dunbar, 1995). Because neocortical expansion is linked to basolateral expansion, it is not surprising that volumetric increase in both structures appears to be correlated with similar socioecological variables.
We provide preliminary evidence that two of the basolateral nuclei, the basal and accessory basal nuclei, are potentially reduced in terms of volume and neuron number in orangutans. Socially, orangutans are the most solitary of the apes, generally foraging in parties of one to two individuals (Delgado and Van Schaik, 2000). Previously, we found that orangutans also have reduced orbitofrontal cortex volumes (Semendeferi et al., 1997; Schenker et al., 2005). This region is a major target of the basal nucleus and, to a lesser degree, the accessory basal nucleus in primates (Ghashghaei and Barbas, 2002), and both structures are central to the neural circuit subserving social affiliation in primates (Adolphs, 2003). The association between small social groups and reductions in functionally related neural structures is intriguing but our sample size precludes firm conclusions on the subject.
Although anthropoid primate social systems have been argued to be some of the most complex among mammals (Shultz and Dunbar, 2007), human social systems exhibit both quantitative and qualitative distinctions from those of other anthropoids. Although the maximum size of chimpanzee and bonobo social groups have been reported of up to 150 individuals (Kano, 1992; Mitani and Amsler, 2003), human social networks, on average, exceed 120 individuals both in industrialized (Hill and Dunbar, 2003) and hunter-gatherer societies (Zhou et al., 2005). Qualitatively, humans are the only primates to form social groups comprised predominantly of non-kin of both sexes (Hill et al., 2011). The human social communicative repertoire is also extensive. The spontaneous use of spoken language unequivocally distinguishes human social communication from that of apes. Although humans share a proficiency for other communicative acts like facial or body gestures with our closest living relatives, the great apes (Parr et al., 2005; Pollick and de Waal, 2007; Pika, 2008), great apes do not use their gestures in a referential or symbolic fashion (Pika et al., 2005). In contrast, human gestures can be iconic and metaphoric, accentuating spoken language (McNeill, 1996), and can essentially replace it as in the case of sign language (Poizner et al., 1990).
Across hominid species, we have found that the human amygdala, specifically, is specialized in emphasizing the lateral nucleus. Evidence from the literature on human neuropathologies can provide some insight into the function of this nucleus. Pathology of the lateral nucleus has been observed in several human neurological disorders. Autistic adults exhibit considerable reductions in the number of neurons only in the lateral nucleus (Schumann and Amaral, 2006), and volumetric reduction of the lateral nucleus has been suggested to be a feature of Williams syndrome (Galaburda and Bellugi, 2000). Because both disorders are characterized by atypical social behavior, together they support a potential role for the lateral nucleus in the modulation of social behavior. Additionally, reductions in volume and neuron number in the lateral nucleus characterize bipolar disorder, which may underlie the difficulties that patients have in assigning emotional significance to external stimuli (Berretta et al., 2007).
Most theories of human and nonhuman primate amygdala function are drawn from the expansive array of literature on amygdala connectivity in nonhuman primates. As previously mentioned, the lateral nucleus is the primary recipient of cortical input in the amygdala and is the first stop for most cortical information, functioning as the primary “gateway” to the amygdala. Although many amygdaloid nuclei receive some cortical input, the lateral nucleus is the primary recipient of multimodal sensory information arriving from the temporal association cortices. Given the available evidence, we suggest that the volume and number of neurons in the human lateral nucleus have increased in response to a heightened need to process increased cortical input and emotional elements of the extensive human communicative repertoire and expansive human social networks.
Acknowledgments
We thank the Busch Gardens Zoo, the Henry Doorly Zoo, the Hoogle Zoo, the Milwaukee County Zoo, the Yerkes National Primate Research Center, and the Great Ape Aging Project for ape donations, Drs. Shirley Strum and Jim Moore for helpful discussions, and Kate Teffer for processing assistance.
Grant sponsor: The National Science Foundation; Grant number: BCS-0726240; Grant sponsor: The Wenner Gren Foundation for Anthropological Research (Dissertation Fieldwork Grant); Grant Sponsor: the Chancellor’s Interdisciplinary Collaboratory Fellowship, University of California, San Diego; Grant sponsor: the Kavli Institute for Brain and Mind, The University of California, San Diego; Grant sponsor: the James S. MacDonnell Foundation; Grant number: 22002078.
LITERATURE CITED
- Adolphs R. Cognitive neuroscience of human social behaviour. Nat Rev Neurosci. 2003;4:165–178. doi: 10.1038/nrn1056. [DOI] [PubMed] [Google Scholar]
- Adolphs R. What does the amygdala contribute to social cognition? Ann N Y Acad Sci. 2010;1191:42–61. doi: 10.1111/j.1749-6632.2010.05445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allman JM, Tetreault NA, Hakeem AY, Manaye KF, Semendeferi K, Erwin JM, Park S, Goubert V, Hof PR. The von Economo neurons in frontoinsular and anterior cingulate cortex in great apes and humans. Brain Struct Funct. 2010;14:1–23. doi: 10.1007/s00429-010-0254-0. [DOI] [PubMed] [Google Scholar]
- Amaral D, Insausti R, Cowan WM. The entorhinal cortex of the monkey: I. Cytoarchitectonic organization. J Comp Neurol. 1987;264:326–355. doi: 10.1002/cne.902640305. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Capitanio JP, Jourdain M, Mason WA, Mendoza SP, Prather M. The amygdala: is it an essential component of the neural network for social cognition? Neuropsychologia. 2003;41:235–240. doi: 10.1016/s0028-3932(02)00154-9. [DOI] [PubMed] [Google Scholar]
- Andersen BB, Gundersen HJ. Pronounced loss of cell nuclei and anisotropic deformation of thick sections. J Microsc. 1999;196:69–73. [PubMed] [Google Scholar]
- Barger N, Stefanacci L, Semendeferi K. A comparative volumetric analysis of the amygdaloid complex and basolateral division in the human and ape brain. Am J Phys Anthropol. 2007;134:392–403. doi: 10.1002/ajpa.20684. [DOI] [PubMed] [Google Scholar]
- Barton RA, Aggleton JP. Primate evolution and the amygdala. In: Aggleton JP, editor. The amygdala: a functional analysis. 2nd. New York: Oxford University Press; 2000. pp. 480–508. [Google Scholar]
- Barton R, Aggleton JP, Grenyer R. Evolutionary coherence of the mammalian amygdala. Proc Biol Sci. 2003;270:539–543. doi: 10.1098/rspb.2002.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechara A, Damasio H, Damasio AR. Role of the amygdala in decision-making. Ann N Y Acad Sci. 2003;985:356–369. doi: 10.1111/j.1749-6632.2003.tb07094.x. [DOI] [PubMed] [Google Scholar]
- Bernier PJ, Bedard A, Vinet J, Levesque M, Parent A. Newly generated neurons in the amygdala and adjoining cortex of adult primates. Proc Natl Acad Sci U S A. 2002;99:11464–11469. doi: 10.1073/pnas.172403999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta S, Pantazopoulos H, Lange N. Neuron numbers and volume of the amygdala in subjects diagnosed with bipolar disorder or schizophrenia. Biol Psychiatry. 2007;62:884–893. doi: 10.1016/j.biopsych.2007.04.023. [DOI] [PubMed] [Google Scholar]
- Bickart KC, Wright CI, Dautoff RJ, Dickerson BC, Barrett LF. Amygdala volume and social network size in humans. Nat Neurosci. 2010;14:163–164. doi: 10.1038/nn.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brothers L. The social brain: a project for integrating primate behavior and neurophysiology in a new domain. Concepts Neurosci. 1990;1:27–51. [Google Scholar]
- Buckwalter JA, Parvizi J, Morecraft RJ, van Hoesen GW. Thalamic projections to the posteromedial cortex in the macaque. J Comp Neurol. 2008;507:1709–1733. doi: 10.1002/cne.21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne RW. Machiavellian intelligence. Evol Anthropol. 1996;5:172–180. [Google Scholar]
- Carlo CN, Stevens CF. Analysis of differential shrinkage in frozen brain sections and its implications for the use of guard zones in stereology. J Comp Neurol. 2011;519:2803–2810. doi: 10.1002/cne.22652. [DOI] [PubMed] [Google Scholar]
- Carlo CN, Stefanacci L, Semendeferi K, Stevens CF. Comparative analyses of the neuron numbers and volumes of the amygdaloid complex in Old and New World primates. J Comp Neurol. 2010;518:1176–1198. doi: 10.1002/cne.22264. [DOI] [PubMed] [Google Scholar]
- Delgado RA, Jr, Van Schaik CP. The behavioral ecology and conservation of the orangutan (Pongo pygmaeus): a tale of two islands. Evol Anthropol. 2000;9:201–218. [Google Scholar]
- de Olmos JS. The amygdala. In: Paxinos G, Mai JK, editors. The human nervous system. New York: Elsevier; 2004. pp. 739–868. [Google Scholar]
- Dorph-Petersen K-A, Nyengaard JR, Gundersen HJ. Tissue shrinkage and unbiased stereological estimation of particle number and size. J Microsc. 2001;204:232–246. doi: 10.1046/j.1365-2818.2001.00958.x. [DOI] [PubMed] [Google Scholar]
- Dunbar RIM. Coevolution of neocortical size, group size and language in humans. Behav Brain Sci. 1993;16:681–693. [Google Scholar]
- Dunbar RIM. Neocortex size and group size in primates: a test of the hypothesis. J Hum Evol. 1995;28:287–296. [Google Scholar]
- Freese JL, Amaral DG. Neuroanatomy of the primate amygdala. In: Whalen PJ, Phelps EA, editors. The human amygdala. New York: The Guilford Press; 2009. pp. 3–42. [Google Scholar]
- Galaburda AM, Bellugi U. V. Multi-level analysis of cortical neuroanatomy in Williams syndrome. J Cogn Neurosci. 2000;12(suppl 1):74–88. doi: 10.1162/089892900561995. [DOI] [PubMed] [Google Scholar]
- Gardella D, Hatton W, Rind H, Rosen G. Differential tissue shrinkage and compression in the z-axis: implications for optical disector counting in vibratome-, plastic- and cryosections. J Neurosci Methods. 2003;124:45–59. doi: 10.1016/s0165-0270(02)00363-1. [DOI] [PubMed] [Google Scholar]
- Garland T, Harvey PH, Ives AR. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst Biol. 1992;41:18–32. [Google Scholar]
- Ghashghaei HT, Barbas H. Pathways for emotion: interactions of prefrontal and anterior temporal pathways in the amygdala of the rhesus monkey. Neuroscience. 2002;115:1261–1279. doi: 10.1016/s0306-4522(02)00446-3. [DOI] [PubMed] [Google Scholar]
- Grieve SM, Clark CR, Williams LM, Peduto AJ, Gordon E. Preservation of limbic and paralimbic structures in aging. Hum Brain Mapp. 2005;25:391–401. doi: 10.1002/hbm.20115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen HJ, Jensen EB, Kiêu K, Nielsen J. The efficiency of systematic sampling in stereology—reconsidered. J Microsc. 1999;193:199–211. doi: 10.1046/j.1365-2818.1999.00457.x. [DOI] [PubMed] [Google Scholar]
- Heimer L, Van Hoesen GW. The limbic lobe and its output channels: implications for emotional functions and adaptive behavior. Neurosci Biobehav Rev. 2006;30:126–147. doi: 10.1016/j.neubiorev.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Heimer L, de Olmos JS, Alheid GF, Pearson J, Sakamoto N, Shinoda K, Marksteiner J, Switzer RC., III . The human basal forebrain. Part II. In: Bloom FE, Björklund A, Hökfelt T, editors. The primate nervous system, Part III. Vol. 15. Amsterdam: Elsevier; 1999. pp. 57–226. [Google Scholar]
- Herrmann E, Call J, Hernández-Lloreda MV, Hare B, Tomasello M. Humans have evolved specialized skills of social cognition: the cultural intelligence hypothesis. Science. 2007;317:1360–1366. doi: 10.1126/science.1146282. [DOI] [PubMed] [Google Scholar]
- Hill KR, Walker RS, Bozicević M, Eder J, Headland T, Hewlett B, Hurtado AM, Marlowe F, Wiessner P, Wood B. Co-residence patterns in hunter-gatherer societies show unique human social structure. Science. 2011;331:1286–1289. doi: 10.1126/science.1199071. [DOI] [PubMed] [Google Scholar]
- Hill RA, Dunbar RIM. Social network size in humans. Hum Nat. 2003;14:53–72. doi: 10.1007/s12110-003-1016-y. [DOI] [PubMed] [Google Scholar]
- Holloway R. The evolution of the primate brain: some aspects of quantitative relations. Brain Res. 1968;7:121–172. doi: 10.1016/0006-8993(68)90094-2. [DOI] [PubMed] [Google Scholar]
- Humphrey NK. growing points in ethology. Cambridge: Cambridge University Press; 1976. The social function of intellect; pp. 303–317. [Google Scholar]
- Ichinohe N, Rockland KS. Distribution of synaptic zinc in the macaque monkey amygdala. J Comp Neurol. 2005;489:135–147. doi: 10.1002/cne.20632. [DOI] [PubMed] [Google Scholar]
- Insausti R, Tunon T, Sobreviela T, Insausti A, Gonzalo L. The human entorhinal cortex: a cytoarchitectonic analysis. J Comp Neurol. 1995;355:171–198. doi: 10.1002/cne.903550203. [DOI] [PubMed] [Google Scholar]
- Jolly A. Lemur social behavior and primate intelligence. Science. 1966;153:501–506. doi: 10.1126/science.153.3735.501. [DOI] [PubMed] [Google Scholar]
- Kanai R, Bahrami B, Roylance R, Rees G. Online social network size is reflected in human brain structure. Proc R Soc Lond B Biol Sci. 2011;278:1498–1497. doi: 10.1098/rspb.2011.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano T. The last ape: pygmy chimpanzee behavior and ecology. Stanford, CA: Stanford University Press; 1992. [Google Scholar]
- LeDoux J. The amygdala. Curr Biol. 2007;17:R868–R874. doi: 10.1016/j.cub.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Lewis KP, Barton RA. Amygdala size and hypothalamus size predict social play frequency in nonhuman primates: a comparative analysis using independent contrasts. J Comp Psychol. 2006;120:31–37. doi: 10.1037/0735-7036.120.1.31. [DOI] [PubMed] [Google Scholar]
- MacLean PD. Psychosomatic disease and the visceral brain: recent developments bearing on the Papez theory of emotion. Psychosom Med. 1949;11:338–353. doi: 10.1097/00006842-194911000-00003. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.73. 2010 http://mesquiteproject.org. [Google Scholar]
- Malone N, Fuentes A. The ecology and evolution of hylobatid communities: causal and contextual factors underlying inter-and intraspecific variation. In: Whittaker D, Lappan S, editors. The gibbons. New York: Springer; 2009. pp. 241–264. [Google Scholar]
- McNeill D. Hand and mind: what gestures reveal about thought. Chicago: University of Chicago Press; 1996. [Google Scholar]
- Merker B. Silver staining of cell bodies by means of physical development. J Neurosci Methods. 1983;9:235–241. doi: 10.1016/0165-0270(83)90086-9. [DOI] [PubMed] [Google Scholar]
- Midford P, Garland T, Jr, Maddison W. PDAP package of Mesquite. 2003 http://mesquiteproject.org/pdap\_mesquite. [Google Scholar]
- Mitani JC, Amsler SJ. Social and spatial aspects of male subgrouping in a community of wild chimpanzees. Behaviour. 2003;140:869–884. [Google Scholar]
- Morrison SE, Salzman CD. Re-valuing the amygdala. Curr Opin Neurobiol. 2010;20:221–230. doi: 10.1016/j.conb.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr LA, Waller BM, Fugate J. Emotional communication in primates: implications for neurobiology. Curr Opin Neurobiol. 2005;15:716–720. doi: 10.1016/j.conb.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pika S. Gestures of apes and pre-linguistic human children: similar or different? First Lang. 2008;28:116–140. [Google Scholar]
- Pika S, Liebal K, Call J, Tomasello M. Gestural communication of apes. Gesture. 2005;5:41–56. [Google Scholar]
- Pitkänen A, Amaral DG. Organization of the intrinsic connections of the monkey amygdaloid complex: projections originating in the lateral nucleus. J Comp Neurol. 1998;398:431–58. doi: 10.1002/(sici)1096-9861(19980831)398:3<431::aid-cne9>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Pitkänen A, Kemppainen S. Comparison of the distribution of calcium-binding proteins and intrinsic connectivity in the lateral nucleus of the rat, monkey, and human amygdala. Pharmacol Biochem Behav. 2002;71:369–377. doi: 10.1016/s0091-3057(01)00690-6. [DOI] [PubMed] [Google Scholar]
- Poizner H, Klima ES, Bellugi U. What the hands reveal about the brain. Cambridge, MA: The MIT Press; 1990. [Google Scholar]
- Pollick AS, de Waal FBM. Ape gestures and language evolution. Proc Natl Acad Sci U S A. 2007;104:8184–8189. doi: 10.1073/pnas.0702624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Russchen F, Amaral DG. The limbic region: II. The amygdaloid complex. In: Björklund A, Hökfelt T, Swanson L, editors. Handbook of chemical neuroanatomy: Vol 5. Integrated systems of the CNS. Amsterdam: Elsevier; 1987. pp. 279–388. [Google Scholar]
- Purvis A. A composite estimate of primate phylogeny. Philos Trans R Soc Lond B Biol Sci. 1995;348:405–421. doi: 10.1098/rstb.1995.0078. [DOI] [PubMed] [Google Scholar]
- Rilling JK, Insel TR. The primate neocortex in comparative perspective using magnetic resonance imaging. J Hum Evol. 1999;37:191–223. doi: 10.1006/jhev.1999.0313. [DOI] [PubMed] [Google Scholar]
- Rilling JK, Seligman RA. A quantitative morphometric comparative analysis of the primate temporal lobe. J Hum Evol. 2002;42:505–533. doi: 10.1006/jhev.2001.0537. [DOI] [PubMed] [Google Scholar]
- Rilling JK, Scholz J, Preuss TM, Glasser MF, Errangi BK, Behrens TE. Differences between chimpanzees and bonobos in neural systems supporting social cognition. Soc Cogn Affect Neurosci. 2011;7:369–379. doi: 10.1093/scan/nsr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinow MJ, Juraska JM. Neuron and glia numbers in the basolateral nucleus of the amygdala from preweaning through old age in male and female rats: a stereological study. J Comp Neurol. 2009;512:717–725. doi: 10.1002/cne.21924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallet J, Mars RB, Noonan MP, Andersson JL, O’Reilly JX, Jbabdi S, Croxson PL, Jenkinson M, Miller KL, Rushworth MFS. Social network size affects neural circuits in macaques. Science. 2011;334:697–700. doi: 10.1126/science.1210027. [DOI] [PubMed] [Google Scholar]
- Sander D, Grandjean D, Pourtois G, Schwartz S, Seghier ML, Scherer KR, Vuilleumier P. Emotion and attention interactions in social cognition: brain regions involved in processing anger prosody. Neuroimage. 2005;28:848–58. doi: 10.1016/j.neuroimage.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Schenker NM, Desgouttes A-M, Semendeferi K. Neural connectivity and cortical substrates of cognition in hominoids. J Hum Evol. 2005;49:547–569. doi: 10.1016/j.jhevol.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Schumann CM, Amaral DG. Stereological estimation of the number of neurons in the human amygdaloid complex. J Comp Neurol. 2005;491:320–329. doi: 10.1002/cne.20704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann CM, Amaral DG. Stereological analysis of amygdala neuron number in autism. J Neurosci. 2006;26:7674–7679. doi: 10.1523/JNEUROSCI.1285-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann CM, Bauman MD, Amaral DG. Abnormal structure or function of the amygdala is a common component of neurodevelopmental disorders. Neuropsychologia. 2011;49:745–759. doi: 10.1016/j.neuropsychologia.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semendeferi K, Damasio H. The brain and its main anatomical subdivisions in living hominoids using magnetic resonance imaging. J Hum Evol. 2000;38:317–332. doi: 10.1006/jhev.1999.0381. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Damasio H, Frank R, Van Hoesen GW. The evolution of the frontal lobes: a volumetric analysis based on three-dimensional reconstructions of magnetic resonance scans of human and ape brains. J Hum Evol. 1997;32:375–388. doi: 10.1006/jhev.1996.0099. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Armstrong E, Schleicher A, Zilles K, Van Hoesen GW. Limbic frontal cortex in hominoids: a comparative study of area 13. Am J Phys Anthropol. 1998;106:129–155. doi: 10.1002/(SICI)1096-8644(199806)106:2<129::AID-AJPA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Lu A, Schenker N, Damasio H. Humans and great apes share a large frontal cortex. Nat Neurosci. 2002;5:272–276. doi: 10.1038/nn814. [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Barger N, Schenker N. Brain reorganization in humans and apes. In: Broadfield D, Schick KD, Toth N, Yuan M, editors. The human brain evolving: paleoneurological studies in honor of Ralph L. Holloway. Gosport, IN: Stone Age Institute Press; 2010. pp. 119–156. [Google Scholar]
- Sherwood CC, Bauernfeind AL, Bianchi S, Raghanti MA, Hof PR. Human brain evolution writ large and small. Prog Brain Res. 2012;195:237–254. doi: 10.1016/B978-0-444-53860-4.00011-8. [DOI] [PubMed] [Google Scholar]
- Shultz S, Dunbar RIM. The evolution of the social brain: anthropoid primates contrast with other vertebrates. Proc Biol Sci. 2007;274:2429–2436. doi: 10.1098/rspb.2007.0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorvari H, Soininen H, Paljärvi L, Karkola K, Pitkänen A. Distribution of parvalbumin-immunoreactive cells and fibers in the human amygdaloid complex. J Comp Neurol. 1995;360:185–212. doi: 10.1002/cne.903600202. [DOI] [PubMed] [Google Scholar]
- Stefanacci L, Amaral DG. Some observations on cortical inputs to the macaque monkey amygdala: an anterograde tracing study. J Comp Neurol. 2002;451:301–323. doi: 10.1002/cne.10339. [DOI] [PubMed] [Google Scholar]
- Stephan H, Andy O. Quantitative comparative neuroanatomy of primates: an attempt at a phylogenetic interpretation. Ann N Y Acad Sci. 1969;167:370–387. [Google Scholar]
- Stephan H, Andy O. Quantitative comparison of the amygdala in insectivores and primates. Acta Anat (Basel) 1977;98:130–153. doi: 10.1159/000144789. [DOI] [PubMed] [Google Scholar]
- Stephan H, Frahm H, Baron G. Comparison of brain structure volumes in Insectivora and Primates. VII. Amygdaloid components. J Hirnforsch. 1987;28:571. [PubMed] [Google Scholar]
- Sugiura M, Kawashima R, Nakamura K, Sato N, Nakamura A, Kato T, Hatano K, Schormann T, Zilles K, Sato K, et al. Activation reduction in anterior temporal cortices during repeated recognition of faces of personal acquaintances. NeuroImage. 2001;13:877–890. doi: 10.1006/nimg.2001.0747. [DOI] [PubMed] [Google Scholar]
- von Bohlen und Halbach O, Unsicker K. Morphological alterations in the amygdala and hippocampus of mice during ageing. Eur J Neurosci. 2002;16:2434–2440. doi: 10.1046/j.1460-9568.2002.02405.x. [DOI] [PubMed] [Google Scholar]
- Ward TS, Rosen GD, von Bartheld CS. Optical disector counting in cryosections and vibratome sections underestimates particle numbers: effects of tissue quality. Microsc Res Tech. 2008;71:60–68. doi: 10.1002/jemt.20525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MJ. New stereological methods for counting neurons. Neurobiol Aging. 1993;14:275–285. doi: 10.1016/0197-4580(93)90112-o. [DOI] [PubMed] [Google Scholar]
- West MJ, Slomianka L, Gundersen HJ. Unbiased stereological estimation of the total number of neurons in the subdivisions of the rat hippocampus using the optical fractionator. Anat Rec. 1991;231:482–497. doi: 10.1002/ar.1092310411. [DOI] [PubMed] [Google Scholar]
- Yang TT, Menon V, Eliez S, Blasey C, White CD, Reid AJ, Gotlib IH, Reiss AL. Amygdalar activation associated with positive and negative facial expressions. Neuroreport. 2002;13:1737–1741. doi: 10.1097/00001756-200210070-00009. [DOI] [PubMed] [Google Scholar]
- Zhou W-X, Sornette D, Hill RA, Dunbar RIM. Discrete hierarchical organization of social group sizes. Proc Biol Sci. 2005;272:439–444. doi: 10.1098/rspb.2004.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]