Abstract
Genetically encoded voltage indicators (GEVIs) are a promising technology for fluorescence readout of millisecond-scale neuronal dynamics. Previous GEVIs had insufficient signaling speed and dynamic range to resolve action potentials in live animals. We coupled fast voltage-sensing domains from a rhodopsin protein to bright fluorophores through resonance energy transfer. The resulting GEVIs are sufficiently bright and fast to report neuronal action potentials and membrane voltage dynamics in awake mice and flies, resolving fast spike trains with 0.2-millisecond timing precision at spike detection error rates orders of magnitude better than previous GEVIs. In vivo imaging revealed sensory-evoked responses, including somatic spiking, dendritic dynamics, and intracellular voltage propagation. These results empower in vivo optical studies of neuronal electrophysiology and coding and motivate further advancements in high-speed microscopy.
To dissect the mechanisms of high-speed neuronal information processing in the live brain, neuroscientists need to track cellular and subcellular electrophysiological activity with millisecond-scale resolution in identified neuron types. Genetically encoded fluorescent Ca2+ indicators report isolated, individual action potentials from many cell types in live animals (1, 2). However, Ca2+ indicators’ slow kinetics (~50 to 1000 ms) precludes high-fidelity studies of fast-spiking cell types, determinations of spike waveforms, resolution of individual spikes in fast spike trains, and precise estimates of spike timing. Moreover, the magnitude of Ca2+ influx in response to an action potential varies across cell types and even within individual cells (1, 2). In vivo Ca2+ imaging also poorly tracks subthreshold or dendritic voltage dynamics, due to insensitivity to hyperpolarizations and confounds from synaptic Ca2+ influx. Organic voltage-sensitive dyes typically have much faster kinetics than Ca2+ indicators but are generally highly phototoxic, allow neither genetically targeted delivery nor long-term imaging studies of single cells, and have been incapable of reporting single spikes in the live mammalian brain (3).
GEVIs combine genetic targeting and optical readout of transmembrane voltage (3, 4), and in principle can sense spikes and subthreshold dynamics. Nevertheless, to date, GEVIs have lacked the capabilities to detect individual action potentials and fast spike trains in live animals (3, 4). Past efforts fused fluorescent proteins to voltage-sensitive domains (VSDs) from voltage-sensitive phosphatases (5–9) or used Archaerhodopsin (Arch), which is both a fast VSD and a dim fluorophore (10). Although Arch variants work well in cultured neurons, the intense illumination required (1 to 10 W · mm−2) plus the consequent heating, autofluorescence, and photodamage have precluded imaging studies in intact tissue over wide fields of view (10).
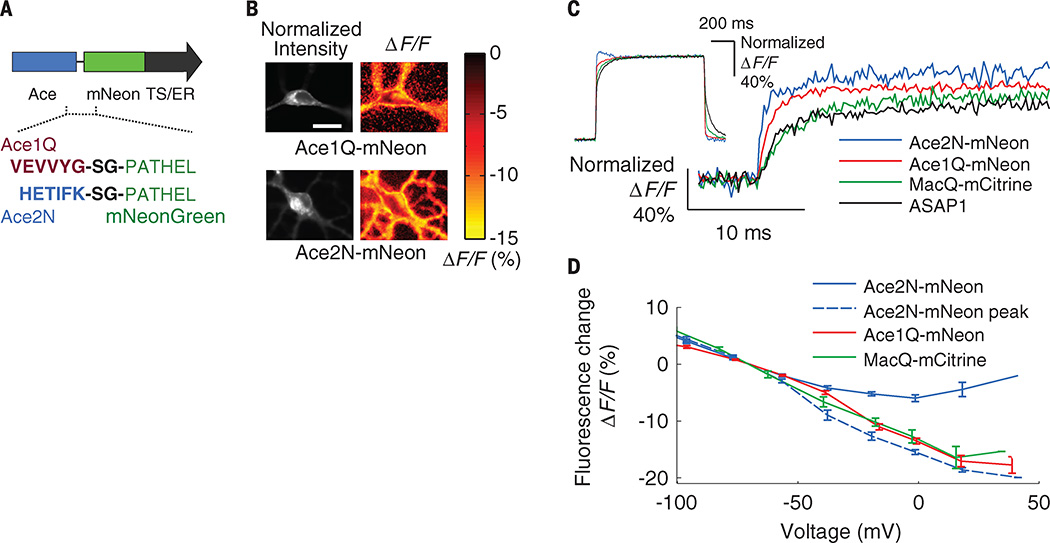
Here, we present fast GEVIs (<1-ms response) that fuse the Acetabularia acetabulum rhodopsin (Ace) (11) and mNeonGreen (12) fluorescent protein to enable voltage-sensitive fluorescence resonance energy transfer (FRET) (Fig. 1A and table S1). We previously introduced this “FRET-opsin” configuration (13, 14), which combines the fast kinetics of a rhodopsin VSD with a bright fluorophore and provides high-fidelity membrane potential and spike train readouts at illumination levels ~50 to 100 times lower than those used with Arch indicators. A FRET-opsin indicator based on Leptosphaeria maculans (Mac) rhodopsin and yellow fluorescent mCitrine reported fast neural spiking in brain slices and Purkinje neurons’ dendritic activation in live mice (13). These results had suggested that optical recordings of action potentials and dendritic voltage dynamics in live animals might be attainable. Ace-mNeon indicators now enable high-fidelity imaging of individual spikes and fast spike trains in live mice and flies due to their faster kinetics and superior brightness compared with all prior GEVIs. Ace is about six times as fast as Mac, and mNeonGreen has a ~50% higher extinction coefficient than mCitrine and nearly threefold better photostability (12). We created Ace mutants (Ace1Q and Ace2N) with an inactivated proton pump; these have blue-shifted absorption spectra compared with Mac and Arch (11, 13), yielding superior FRET acceptors when paired with green or yellow emitters (figs. S1 and S2). When used together with protein trafficking signals, the fusions provide high FRET efficiency and minimal protein aggregation in live neurons (Fig. 1, A and B), key attributes of a FRET indicator (13, 14).
Fig. 1. Ace FRET-opsin sensors report membrane voltage with ~1-ms response times.
(A) Linker sequences bridging Ace1Q and Ace2N to mNeonGreen. Endoplasmic reticulum (ER) export sequence and Golgi export trafficking signal (TS) at the construct’s C terminus improve the sensor’s membrane localization and hence the signaling dynamic range. (B) Fluorescence signals from neurons expressing Ace1Q-mNeon or Ace2N-mNeon. (Left) Baseline fluorescence emissions from mNeonGreen. (Right) Spatial maps of the fluorescence response (ΔF/F) to a voltage step of approximately 100 mV. Areas of fluorescence and voltage response were generally colocalized. Scale bar, 20 µm. Illumination intensity, 15 mW · mm−2. (C) Step responses of the Ace sensors, ASAP1 and MacQ-mCitrine to +100 mV command voltage steps, normalized to each sensor’s maximum (or steady state) ΔF/F response to the command voltage. The initial rise of the Ace2N-mNeon sensor was more than six times as fast as that of ASAP1 and MacQ-mCitrine (table S2). (Inset) The full step response of all sensors. The trace for Ace2N-mNeon exhibits hysteresis ~40 to 200 ms after the voltage step, outside the interval shown in the main plot. Illumination intensity, 15 to 50 mW·mm−2. Data acquisition rate, 5 kHz. Inset traces were down-sampled to 250 Hz. (D) Steady-state responses of FRET-opsin sensors as a function of membrane voltage in cultured neurons (N = 10 cells per trace). Because Ace2N-mNeon exhibited hysteric responses to voltage steps (fig. S2), we also plot its peak initial responses. Illumination intensity, 15 mW · mm−2. Error bars, SEM.
We measured responses of Ace1Q-mNeon and Ace2N-mNeon to voltage depolarization steps in cultured human embryonic kidney–293T (HEK293T) cells. These sensors responded five to six times as fast as MacQ-mCitrine (13) and the ASAP1 indicator (7) (Fig. 1C and table S2). At termination of a step depolarization, Ace2N-mNeon exhibited some hysteresis during its relaxation back to baseline fluorescence levels (Fig. 1C, inset, and fig. S3). However, the steady-state response varied linearly with the transmembrane voltage within the physiological range of subthreshold potentials, and the initial peak response increased linearly between −100 mV and +30 mV (Fig. 1D). These relationships were similar to those of previous FRET-opsin sensors (Fig. 1D and fig. S3), but the faster kinetics of the Ace indicators enabled superior spike detection fidelity, even for rapid successions of spikes that would have blurred together with Ca2+ imaging (Fig. 2, A to E). During action potentials in cultured neurons, Ace2N-mNeon and Ace1Q-mNeon had peak changes in fluorescence intensity relative to baseline values (ΔF/F) of 12.0 ± 0.8% (mean ± SEM; N = 12 cells) and 8.5 ± 0.7% (N = 10 cells), respectively, while leaving the spike waveform unperturbed (Fig. 2, A and B, and fig. S4). Photobleaching rates were those of the constituent mNeonGreen, which is as photostable as enhanced green fluorescent protein (12) and comparable to those of other brightly fluorescent voltage indicators (table S2).
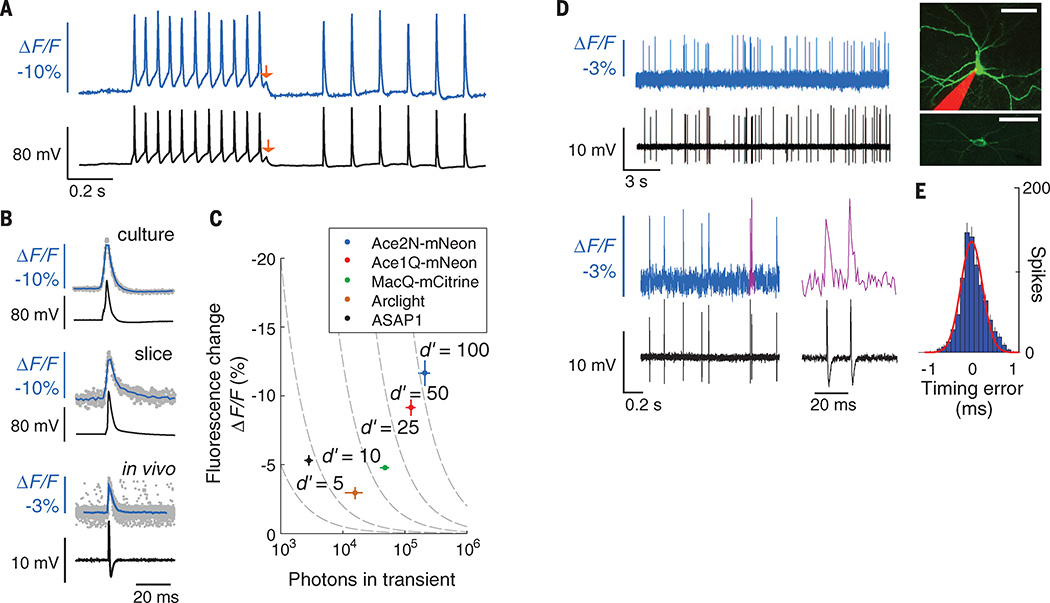
Fig. 2. Ace sensors provide about threefold to tenfold better spike detection fidelity (d′) than previous GEVIs.
(A) Fluorescence signals from cultured neurons expressing Ace2N-mNeon (blue trace) had sharp peaks closely matching action potentials in concurrent electrophysiological recordings (black trace). Red arrows mark a ~5 mV depolarization apparent in both traces. (B) Optical and electrical waveforms of single action potentials in example neurons expressing Ace2N-mNeon in cell culture (top), Ace2N-mNeon in mouse brain slice (middle), and Ace2N-4AA-mNeon in anesthetized mouse brain (bottom). Data (gray points) acquired optically from different spikes were temporally aligned to the corresponding peaks in the electrical traces. Mean waveforms of the optical data (blue traces; averaged over N = 30 spikes; resampled to 2 kHz) are aligned with the mean electrical waveforms (black traces; whole-cell patch recordings for cells in culture and brain slice; loose patch recording for live mouse). (C) Peak ΔF/F responses to action potentials, as a function of the total number of photons detected per spike in cultured neurons. Isocontours (dashed lines) of spike-detection fidelity, d′, were determined from measured brightness and optical waveforms, as in (B). Error bars, SEM. (D) Concurrent optical (colored) and juxtacellular electrical (black) recordings in an anesthetized mouse from V1 cells expressing Ace2N-4AA-mNeon. Magenta trace (lower right) shows the two magenta spikes in adjacent trace. Across 837 spikes, the electrical and optical traces were in perfect accord. (Inset) Two different neocortical neurons imaged by two-photon microscopy. A maximum projection of a dual-color image stack (top) acquired in a live mouse shows an Ace2N-4AA-mNeon–labeled cell and a pipette, filled with red dye, that recorded somatic electrical activity. An image acquired in a brain slice (bottom) after in vivo experimentation shows the membrane localization of Ace2N-4AA-mNeon. Scale bars, 40 µm. (E) Histogram of timing errors for spikes detected optically as in (D), using the electrical trace to provide the actual spike time (N = 837 spikes from three cells). Red line, Gaussian fit. Error bars, SD, estimated as counting errors. Illumination: 15, 25, and 25 mW · mm−2, respectively, for studies in culture, brain slice, and live mice. Image acquisition rates: 440 Hz, 440 Hz, and 1000 Hz, respectively.
To quantitatively compare multiple indicators, we used a metric of spike detection fidelity, d′, from signal detection theory (9, 15). This metric accounts for an indicator’s optical waveform in response to a spike and thus captures rates of correct and false spike detection better than other metrics, such as signal-to-noise ratio (SNR) (16). Modest improvements in d′ can have huge benefits for experimentation because false detection rates decline faster than exponentially with increases in d′ (9, 15). Compared to MacQ-mCitrine, the superior brightness and peak ΔF/F responses of the Ace indicators increased d′ by factors of 2.2 to 3.0, with d′ values of 100 ± 12 (Ace2N-mNeon, mean ± SEM) and 70 ± 8 (Ace1Q-mNeon) in cultured neurons (Fig. 2C). Our calculations predicted that this should be sufficient to image spikes >0.4 ms in duration, not only in brain slices but also in live animals (fig. S5).
Studies in mouse brain slices and live mice validated these predictions (Fig. 2, B to E, and Fig. 3, A to F). To facilitate spike detection, we sparsely expressed the GEVIs using the SAD-ΔG viral vector, which efficiently transduces neurons through axons (17). This labeling approach reduced background fluorescence from out-of-focal-plane or nonspecifically labeled sources. Previous studies with this virus have shown that there is minimal toxicity in neurons up to 10 days after infection (17). We imaged thick brain slices 3 to 5 days after viral injection and found that Ace2N-mNeon reported action potentials with ΔF/F = 6.5 ± 1.5% and d′ = 32 ± 5 (mean ± SEM; n = 5 cells) (Fig. 2B). Neurons expressing the GEVI had statistically indistinguishable spike waveforms compared with nonfluorescent neurons in the same slices (fig. S4).
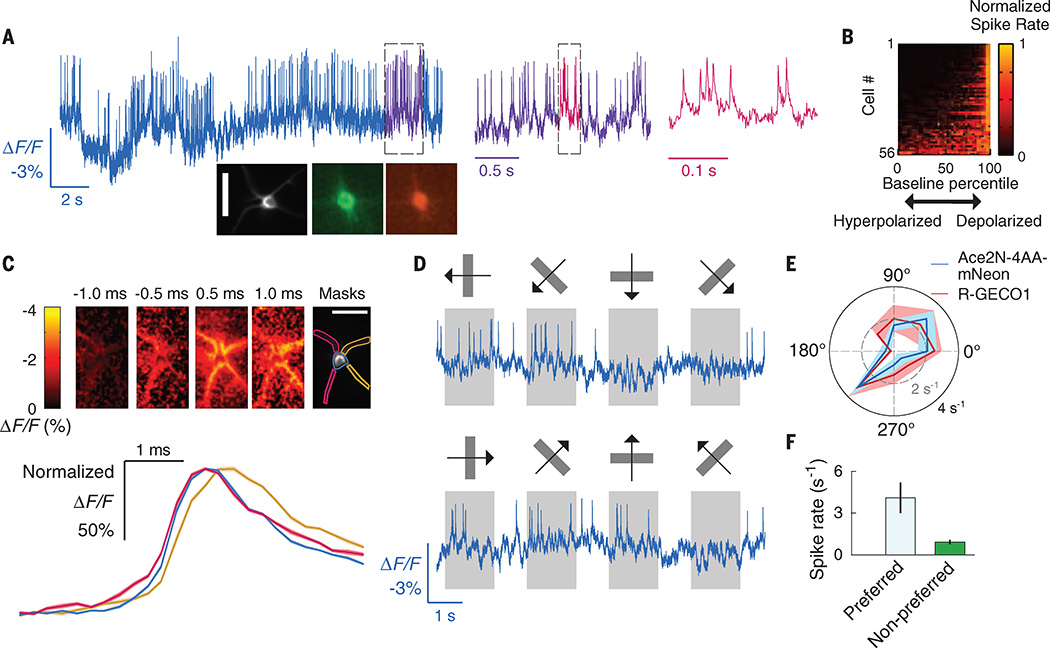
Fig. 3. Imaging single action potentials and subthreshold membrane voltage fluctuations in layer 2/3 visual cortical neurons of awake mice.
(A) Optical voltage trace acquired in an awake mouse, showing spiking by a V1 layer 2/3 neuron expressing Ace2N-4AA-mNeon via the SAD-Ace2N-4AA-mNeon-RGECO1 virus. Dashed boxes indicate intervals shown at successively expanded time scales. (Insets) Image acquired in vivo of the cell that provided the optical trace and a pair of fluorescence images taken in a fixed tissue slice of an identically labeled V1 neuron from the same preparation expressing Ace2N-4AA-mNeon (green) and RGECO1 (red). Scale bar, 40 µm. (B) Periods of reduced spiking arose when Ace2N-4AA-mNeon reported a hyperpolarization. To track each cell’s membrane voltage apart from its spikes, we applied a median filter (50-ms window) to the trace. For nonparametric comparisons, we matched each spike to the voltage at which the spike occurred, quantified as a percentile of the cell’s full range of membrane voltages over the full recording. For each cell, the plot shows the spike rate (normalized to each cell’s peak rate) at voltages across all percentiles. (C) (Top) Spike-triggered average image frames, for the cell in (A), showing the mean dendritic activation before and after firing an action potential. Times are relative to the spike peak at the soma. The left two dendrites activated before the right two dendrites. We calculated mean time traces for the soma and the left and right dendrite pairs using the spatial masks shown. (Bottom) Mean fluorescence time courses (ΔF/F) for each of the three masks, normalized to the same maximum. The traces confirm the left-to-right activation pattern; the right dendrites exhibit a voltage peak 0.25 to 0.5 ms after the left dendrites. The ΔF/F image series and time traces were sampled in 0.25-ms time bins, using data from 1900 spikes mutually aligned to their peaks. Shaded regions on the time traces denote SEM and are barely discernible. (D) Example optical traces from a cortical V1→LM neuron in an awake mouse, showing visually evoked responses to moving gratings (orientations and motion directions marked above each trace). Spiking responses to the cell’s preferred grating orientation (second column) differed from the activity suppression in response to the orthogonal orientation (fourth column). (E) Mean spike rates of the cell in (D), determined from the optical voltage trace in response to gratings moving in different orientations (10 trials per stimulus). Solid black and dashed gray circles indicate spike rates of 4 s−1 and 2 s−1, respectively. Ca2+ imaging using RGECO1 in the same cell yielded similar orientation tuning. Evoked Ca2+ signals were integrated over 1 s and normalized so that Ca2+ and spiking responses to the preferred orientation (225°) were plotted at the same radius on the polar plot. Shaded areas indicate SEM. (F) Mean ± SEM spike rates in response to gratings at the preferred orientation were higher than to those oriented orthogonally (P < 0.01 for each of seven cells; permutation test; 105 permutations). Frame acquisition rate: 1 kHz for voltage imaging and 20 Hz for Ca2+ imaging. Illumination intensity: 20 and 10 mW · mm−2, respectively, for voltage and Ca2+ imaging. Labeled neurons were ~150 µm below the brain surface.
We next imaged Ace2N-expressing neurons in the mouse visual cortex, using an indicator variant (Ace2N-4AA-mNeon) with four extra amino acids in the linker domain (table S1). This variant yielded improved expression and superior visualization of individual cells’ dynamics in live mice (Figs. 2D and 3A, inset). Because extending the linker left the VSD unchanged, the voltage-dependent kinetics were unaffected, although the increased distance between the FRET pairs reduced the peak ΔF/F response to neural spikes (fig. S6 and table S2). In anesthetized mice (Fig. 2, D and E) and awake mice (Fig. 3), we readily observed spiking waveforms in labeled neurons in cortical layer 2/3, with interspike intervals as brief as ~10 ms—i.e., an interval briefer by a factor of 20 than the signal integration time of the fast Ca2+ indicator GCaMP6f (1).
Ace2N-4AA-mNeon also provided a high level of spike detection fidelity (d′ = 16 ± 1.5; mean ± SEM; N = 56 layer 2/3 neurons, ~150 µm below the brain surface; N = 20 awake mice), at which the frequency of spike detection errors is vastly less than with previous GEVIs. Imaging at a 1-kHz frame rate at a d′ value of 16 corresponds to a mean rate of <10−7 detection errors per day (15). Consistent with this, during simultaneous optical and loose-patch electrical recordings in the same neocortical neurons of anesthetized mice (Fig. 2D), the spike trains attained using Ace2N-4AA-mNeon and those from the electrical recordings were in perfect concordance (837 total spikes detected in dual recordings; N = 3 mice). Highlighting the importance of Ace2N’s ~threefold increase in d′ over the best previous GEVIs, in vivo imaging with a value threefold lower (d′ = 5.3) leads to an unacceptable four spike-detection errors per second. The nearly vanishing error rate predicted at d′ = 16 is unattainable over hours under true experimental conditions due to the indicator’s finite photostability and nonstationary noise sources in the live brain other than photon shot noise. Nevertheless, this calculation underscores the huge effect of modest rises in d′, in that Ace2N has an error rate that is orders of magnitude lower than previous GEVIs and is the first GEVI with acceptable error rates in the detection of single action potentials in live mammals.
Ace2N-4AA-mNeon also provided exquisite spike-timing accuracy, as determined from the dual optical and electrical recordings in live mice (Fig. 2E). Relative to the spike times in the electrical traces, the spike times in the optical traces differed by only 0.24 ± 0.01 ms (SEM; N = 837 spikes) (16). This degree of spike-timing accuracy approached the theoretical physical limits, 0.14 ± 0.03 ms, set by quantum mechanical photon shot noise (9, 15, 16).
We also examined in awake mice whether individual cells’ ongoing fluctuations in baseline Ace2N-4AA-mNeon emissions reflect subthreshold membrane voltage dynamics. If so, periods of optically reported membrane hyperpolarization should have reduced spike rates, and periods of reported depolarization should have elevated firing rates. This prediction proved correct for all 56 layer 2/3 neurons imaged in awake mice (Fig. 3B). As determined optically, each neuron fired spikes from baseline voltage levels that were significantly higher than its median resting potential (P < 0.05 for all 56 cells; Wilcoxon rank-sum test). As a population, the 56 cells were far more likely to fire spikes when the optically reported membrane voltages were above median resting levels (P < 10−15; Binomial test). Ace2N-4AA-mNeon also revealed submillisecond-scale features of dendritic voltage dynamics during spike generation (Fig. 3C and movie S1). Thus, Ace2N reports aspects of individual cells’ subthreshold, dendritic, and high-speed membrane potential dynamics in the live brain.
We next studied visually evoked spiking responses. To illustrate targeting of specific subclasses of visual cortical neurons, we expressed Ace2N-4AA-mNeon selectively in cells of the primary visual area (V1) that sent axons to the lateromedial cortical area (LM) (denoted V1→LM neurons), one of 15 cortical areas in mice that receive inputs from V1 (16, 18). We presented moving grating visual stimuli to awake mice and imaged the cells’ evoked responses. Generally, layer 2/3 V1→LM neurons’ evoked spiking was preferentially elicited by gratings of one orientation (4.1 ± 1.1 spikes · s−1; mean ± SEM) versus the response (or suppression of activity) to gratings of the orthogonal orientation (1.0 ± 0.2 spikes · s−1) (Fig. 3, D to F) (N = 7 neurons from 5 mice; P < 10−3; permutation test for each cell; 105 permutations). The mean orientation selectivity index (0.82 ± 0.03; mean ± SEM), as determined from Ace2N-4AA-mNeon fluorescence traces, fit well with values determined by Ca2+ imaging in the same neurons (Fig. 3E) and with previous studies (16, 19). There were also light-sensitive V1→LM neurons that responded at the onset or offset of visual illumination regardless of the grating orientation (fig. S8).
We also studied sensory-driven neural dynamics in live fruit flies, in which electrical recordings are often challenging. Beyond in vivo imaging of fast somatic spike trains, GEVIs have the potential to reveal fast voltage dynamics in genetically identified dendrites and axons. To illustrate, we imaged fast dynamics in the olfactory system and first examined local neurons (LNs) of the olfactory glomeruli (20), in which we used the R55D11-GAL4 fly driver line (21) to express an Ace2N variant with two extra amino acids in the linker domain (Ace2N-2AA-mNeon) (Fig. 4A, fig. S6, and table S2). The baseline spike rate in the absence of odor stimulation was 7.4 ± 2.8 s−1 (N = 8 neurons from 8 flies), comparable to electrophysiological measurements (20). In response to odor stimuli, we observed fast, evoked spike trains in single trials at firing rates significantly above baseline (P < 10−3; Wilcoxon signed rank test) (Fig. 4B). The peak odor-evoked spike rate was 65 ± 7 s−1; i.e., the individual spikes fired about an order of magnitude faster than Ca2+ imaging can resolve.
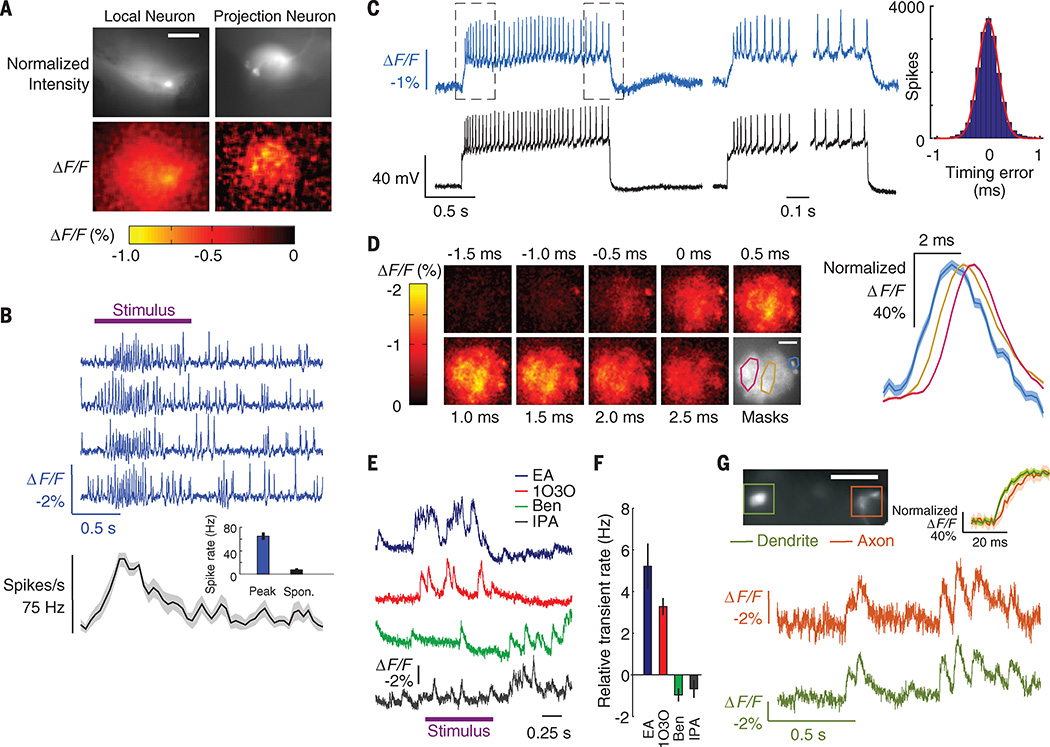
Fig. 4. Odor-evoked spiking, dendritic dynamics, and voltage propagation delays in fly olfactory neurons.
(A) (Top) Fluorescence images of a local neuron (left) and a projection neuron (right) expressing Ace2N-2AA-mNeon in the fly antenna lobe. (Bottom) Spatial maps of the fluorescence response (ΔF/F) at the peak of an odor-evoked transient. Scale bar, 40 µm. (B) (Top) Optical voltage traces (blue traces) from the local neuron in (A) reveal increased spiking during 5% benzaldehyde odor presentation (purple bar). (Bottom) Odor-evoked spike rate as a function of time, averaged over five trials (shaded area denotes SEM). (Inset) Mean peak spike rate during odor presentation was higher than baseline rates without odor [P < 10−3; Wilcoxon signed-rank test; N = 8 neurons and 8 flies (20 × UAS-Ace2N-2AA-mNeon/+; R55D11-GAL4/+)]. (C) (Left) Concurrent optical voltage and whole-cell patch electrical recordings in whole brain explants, from the same cell type as in (B). Spikes were evoked by current injection. (Middle) Paired optical and electrical traces, taken from periods enclosed by dashed boxes in the left panel. Spikes are clearly distinguished in the optical traces from both the plateau of subthreshold depolarization and each rise to spike threshold. (Right) Histogram of timing errors for the spikes detected optically, relative to the spike times in the whole-cell patch recordings (N = 18,141 spikes from 4 flies). Red line is a Gaussian fit. Standard deviation of the timing errors was 0.19 ± 0.002 ms. Error bars estimating the SD as counting errors are too small to be seen. (D) (Left) Spike-triggered average image frames for the cell in (C), showing the mean activation in the neural processes before and after firing a somatic spike, as determined from spike times in the electrical recording. Depolarization started at about −1.0 ms in the soma (located in the upper right of each image) and propagated right to left across the dendritic tree during a spike. We calculated mean time traces for the soma and two subportions of the dendrites using the spatial masks in the lower right panel (cyan line encloses the soma; red and gold lines enclose dendritic regions). Scale bar, 40 µm. (Right) ΔF/F for each of the three spatial masks, normalized to the same maximum. The traces confirm the right-to-left activation pattern; the left dendrites exhibited a voltage peak 0.5 to 0.7 ms after the right dendrites. Mean ΔF/F image series and traces were sampled in 0.25-ms bins, using data from 1300 spikes, temporally aligned to the spike peaks. Shaded regions denote SEM and are only discernible for the soma. (E) Odor-evoked, optical voltage traces from the DL3 projection neuron dendrites of (A), with significant rises in activity over baseline for some [10% ethyl acetate (EA) and 3-octanol (1O3O); P < 0.04 for both EA and 1O3O), but not other odors (10% benzaldehyde (BEN) and isopropanol (IPA); P = 1 and 0.4 for BEN and IPA, respectively; Wilcoxon signed rank tests; N = 6 flies (20 × UAS-Ace2N-2AA-mNeon/+; R26B04-GAL4/+)]. (F) Odor-evoked dendritic activation levels, relative to baseline levels. (G) (Top) Example fly expressing Ace2N-2AA-mNeon in DL3 cells in which we concurrently imaged dendrites and axons. (Bottom) Optical traces from both regions show concurrent activity. (Inset) Averaged traces aligned to onset of dendritic activity reveal a ~4-ms propagation delay between dendrites and axons (N = 30 transients; shaded regions denote SEM). Scale bar, 50 µm. Illumination, 20 mW · mm−2. Frame acquisition rate, 1 kHz. For display only, each optical trace in (B) was high-pass filtered by subtracting a median-filtered 50-ms window) version of the trace. Traces in (E) were processed the same way for display, but with a 1-s filter window.
As in mice, we performed dual optical and electrical recordings to verify the spike trains, this time using whole-cell patch-clamp electrodes to access LN neurons’ intracellular potentials in intact fly brain explants (Fig. 4C, left). Across 18,141 recorded spikes, the spike trains provided by Ace2N-2AA-mNeon perfectly matched those from the patch-clamp recordings (N=4 fly brains) and had spike-timing errors of 0.19 ± 0.002 ms (SEM; N = 18,141 spikes), close to the theoretical optimum of 0.11 ± 0.03 ms (Fig. 4C, right). Subthreshold dynamics were readily apparent in the optical traces, including plateau potentials and the rising and falling voltage waveforms surrounding action potentials. Ace2N-2AA-mNeon also reported the submillisecond-scale dynamics of spike back-propagation into the dendritic tree, revealing ~0.5- to 1.0-ms delays between the initiating voltage peak at the soma and those in the dendrites (Fig. 4D and movie S2).
We next imaged olfactory projection neurons (PNs), which generally have more selective odor tuning than LNs (22, 23). We used live flies and focused on R26B04-GAL4–labeled PN dendritic arbors (Fig. 4A), which exclusively innervate the DL3 glomerulus (21). In response to specific odors, the arbors exhibited fast, odor-evoked voltage dynamics with temporal structure at the 10- to 100-ms time scale (Fig. 4, E and F). We compared these dynamics to those seen by Ca2+ imaging using the GCaMP6f indicator (fig. S9). The odor-tuning profiles determined from the two modalities were in broad agreement, but voltage imaging with Ace2N revealed both small-amplitude responses and fast temporal signals that GCaMP6f failed to convey (fig. S9). Finally, we imaged the PN dendritic arbors and axonal boutons simultaneously. Odor-evoked voltage signals in the two structures were similar, but there was a 3.7 ± 0.4 ms (mean ± SEM; N = 3 flies) propagation delay from dendrites to axons (Fig. 4G). Neither Ca2+ imaging nor electrical recordings have been capable of revealing this type of fast, intracellular signal propagation in a live animal.
By virtue of their bright emissions, fast (<1 ms) kinetics, and targeted expression, Ace2N GEVIs open a wide range of experimental avenues in optical neurophysiology. The capabilities to track fast-spiking, subthreshold, and both sensory-evoked and back-propagating dendritic voltage activity in vivo allow direct visualization of many temporal features of neural dynamics, plasticity induction, and coding that have long been inaccessible. When combined with chronic animal preparations (9), it will be possible to track these phenomena over multiple days. Ace2N sensors resolved sensory-evoked spike trains with firing rates as high as ~75 s−1 (the fastest we encountered) with ~0.2-ms timing accuracy. Whereas in vivo Ca2+ imaging usually resolves successive spikes if they are ~50 to 250 ms apart and has ~20- to 100-ms timing accuracy for isolated spikes (1, 2), Ace2N sensors directly reveal the constituent spikes in spike bursts, a widespread and important means of neural signaling (24, 25). The Ace2N sensors’ timing precision is also key to resolving long-standing debates over the temporal attributes of neural coding (26, 27).
The retrograde labeling strategy we used to express the Ace2N indicator in live mice also holds importance for future work, in that every soma we imaged had an axon at the virus injection site. This allowed us to target the subclass of V1 neurons with projections to area LM. A logical next step in the study of V1 would be systematic voltage imaging of visually evoked spiking across all 15 subclasses of V1 neurons, as categorized by their axonal projections (18). There are long-standing in vivo electrophysiological methods for identifying cells with specific axonal projections (28), but they are technically onerous to an extent that has precluded widespread and routine use. Voltage imaging is now poised to make commonplace the study of neural dynamics in live mammals in chosen anatomical projection pathways.
Until now, subthreshold neural voltage dynamics have usually been inaccessible in live animals, especially in fine neural processes in which intracellular recordings are nearly prohibitive. As shown here, Ace-mNeon reveals fast dendritic activity, including stimulus-evoked dynamics and spike back-propagation. This is especially important for studies of flies and other species in which fine neurites are vital to information processing (29). There is also rising appreciation of the importance of dendritic computation in mammalian neurons (30, 31). Ace2N should allow direct observations of such phenomena in vivo.
For our in vivo imaging studies, we used wide-field epifluorescence microscopy, 1-kHz imaging rates, and sparse fluorescence labeling. Fluorescent V1→LM neural somata were usually ~150 µm below the brain surface and separated by ~40 to 60 µm (fig. S7). This separation enabled high-fidelity tracking of neural spiking despite the lack of optical sectioning and the consequent background fluorescence. Comparable performance should also be feasible in deep brain areas by using microendoscopes to access deep tissues (9). Studies of both surface and deep neural ensembles would benefit from improved scientific-grade cameras for fast imaging (1 kHz) at cellular resolution over broader fields of view than are possible today. For comparison, Ca2+ imaging studies (10 to 20 Hz) capture specimen areas larger by a factor of 100 than those sampled here at 1 kHz (9).
In principle, microscopy modalities with optical sectioning should also allow denser fluorescence labeling patterns and deeper optical penetration through thick tissue. A challenge, however, is that nearly all extant in vivo microscopy modalities with sectioning involve laser scanning, with pixel dwell times typically ~0.1 to 2 µs for frame rates of ~10 to 20 Hz. Given the importance of detecting as many signal photons as possible with voltage imaging, conventional laser scanning is plainly insufficient at the faster frame rates used for in vivo voltage imaging (~0.5 to 1 kHz). The introduction of Ace2N indicators for in vivo voltage imaging puts a premium on the innovation of microscopes that provide optical sectioning without conventional laser scanning. The emergence of scanless forms of optical sectioning microscopy holds particular interest for the further development of in vivo voltage imaging capabilities (32, 33). Even with existing optical hardware, Ace-mNeon’s capacities for targeted imaging of cells of identified types or axonal projections, resolution of fast spike trains, accurate spike-timing estimation, and observation of subthreshold and dendritic voltage dynamics open many questions to empirical study that hitherto could not be examined in live animals.
Supplementary Material
Acknowledgments
We gratefully acknowledge research funding from the Defense Advanced Research Projects Agency, an NIH Pioneer award, an NIH BRAIN Initiative U01 grant, and the Stanford CNC Program. We thank J. Li for animal husbandry. Genetic sequence data are in the supplementary materials. We licensed mNeonGreen from Allele Biotechnology & Pharmaceuticals (San Diego), which charges $100 to $300 per laboratory for a materials transfer agreement.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/350/6266/1361/suppl/DC1
Materials and Methods
Figs. S1 to S9
Tables S1 and S2
Movies S1 and S2
References (34–40)
REFERENCES AND NOTES
- 1.Chen TW, et al. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inoue M, et al. Nat. Methods. 2015;12:64–70. doi: 10.1038/nmeth.3185. [DOI] [PubMed] [Google Scholar]
- 3.Peterka DS, Takahashi H, Yuste R. Neuron. 2011;69:9–21. doi: 10.1016/j.neuron.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knöpfel T. Nat. Rev. Neurosci. 2012;13:687–700. doi: 10.1038/nrn3293. [DOI] [PubMed] [Google Scholar]
- 5.Tsutsui H, Karasawa S, Okamura Y, Miyawaki A. Nat. Methods. 2008;5:683–685. doi: 10.1038/nmeth.1235. [DOI] [PubMed] [Google Scholar]
- 6.Jin L, et al. Neuron. 2012;75:779–785. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.St-Pierre F, et al. Nat. Neurosci. 2014;17:884–889. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lundby A, Mutoh H, Dimitrov D, Akemann W, Knöpfel T. PLOS ONE. 2008;3:e2514. doi: 10.1371/journal.pone.0002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamel EJ, Grewe BF, Parker JG, Schnitzer MJ. Neuron. 2015;86:140–159. doi: 10.1016/j.neuron.2015.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hochbaum DR, et al. Nat. Methods. 2014;11:825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsunoda SP, et al. Biophys. J. 2006;91:1471–1479. doi: 10.1529/biophysj.106.086421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaner NC, et al. Nat. Methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong Y, Wagner MJ, Zhong Li J, Schnitzer MJ. Nat. Commun. 2014;5:3674. doi: 10.1038/ncomms4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou P, et al. Nat. Commun. 2014;5:4625. doi: 10.1038/ncomms5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilt BA, Fitzgerald JE, Schnitzer MJ. Biophys. J. 2013;104:51–62. doi: 10.1016/j.bpj.2012.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Materials and methods are available as supplementary materials on Science Online.
- 17.Osakada F, et al. Neuron. 2011;71:617–631. doi: 10.1016/j.neuron.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Burkhalter A. J. Comp. Neurol. 2007;502:339–357. doi: 10.1002/cne.21286. [DOI] [PubMed] [Google Scholar]
- 19.Niell CM, Stryker MP. J. Neurosci. 2008;28:7520–7536. doi: 10.1523/JNEUROSCI.0623-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou YH, et al. Nat. Neurosci. 2010;13:439–449. doi: 10.1038/nn.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenett A, et al. Cell Reports. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson RI, Turner GC, Laurent G. Science. 2004;303:366–370. doi: 10.1126/science.1090782. [DOI] [PubMed] [Google Scholar]
- 23.Olsen SR, Bhandawat V, Wilson RI. Neuron. 2007;54:89–103. doi: 10.1016/j.neuron.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lisman JE. Trends Neurosci. 1997;20:38–43. doi: 10.1016/S0166-2236(96)10070-9. [DOI] [PubMed] [Google Scholar]
- 25.Mathy A, et al. Neuron. 2009;62:388–399. doi: 10.1016/j.neuron.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuo Y, et al. Curr. Biol. 2015;25:357–363. doi: 10.1016/j.cub.2014.11.065. [DOI] [PubMed] [Google Scholar]
- 27.Diba K, Amarasingham A, Mizuseki K, Buzsáki G. J. Neurosci. 2014;34:14984–14994. doi: 10.1523/JNEUROSCI.1091-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahnloser RH, Kozhevnikov AA, Fee MS. Nature. 2002;419:65–70. doi: 10.1038/nature00974. [DOI] [PubMed] [Google Scholar]
- 29.Borst A, Haag J, Reiff DF. Annu. Rev. Neurosci. 2010;33:49–70. doi: 10.1146/annurev-neuro-060909-153155. [DOI] [PubMed] [Google Scholar]
- 30.Smith SL, Smith IT, Branco T, Häusser M. Nature. 2013;503:115–120. doi: 10.1038/nature12600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheffield ME, Dombeck DA. Nature. 2015;517:200–204. doi: 10.1038/nature13871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson BO, et al. Front. Neurosci. 2010;4:29. doi: 10.3389/fnins.2010.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prevedel R, et al. Nat. Methods. 2014;11:727–730. doi: 10.1038/nmeth.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.