Abstract
Oncosuppressor miRNAs inhibit cancer cell proliferation by targeting key components of the cell cycle machinery. In our recent report we showed that miR-340 is a novel tumor suppressor in non-small cell lung cancer. miR-340 inhibits neoplastic cell proliferation and induces p27KIP1 by targeting multiple translational and post-translational regulators of this cyclin-dependent kinase inhibitor.
Keywords: miR-340, pumilio, p27KIP1, SKP2, tumor suppressor
Abbreviations
- CDK
cyclin-dependent kinase
- CDKN1B, cyclin-dependent kinase inhibitor 1B (p27
Kip1)
- NSCLC
non-small cell lung cancer
- PUM1/2
pumilio RNA-binding family member 1/2
- SKP2, S-phase kinase-associated protein 2
E3 ubitiquitin ligase
Oncosuppressor miRNAs have emerged as powerful post-transcriptional inhibitors of genetic programs controlling cancer cell proliferation, survival, invasion, metastasis, and stemness. Moreover, the in vivo inhibition of tumor growth achieved in mouse cancer models by re-expression of well-characterized tumor suppressor miRNAs, such as miR-34 and let-7, suggests strong therapeutic potential.
Experimentally validated bioinformatics analyses show that cell cycle components are highly enriched among targets of the major tumor suppressor miRNAs. Several G1- and S-phase cyclins (D1, D3, and E2) and cyclin-dependent kinases (CDK4 and CDK6) represent key targets of let-7, miR-15/16, and miR-34 families. Conversely, several oncomiRs target the expression of CDK inhibitors; for example members of the miR-17–92 and miR-106b–25 oncogenic clusters target the p21CIP1 transcript.
The CDK inhibitor p27KIP1 is lost or inactivated in cancer cells by multiple mechanisms, including decreased synthesis, increased proteolysis, and mislocalization. The p27KIP1 and p57KIP2 transcripts are critical targets of the closely related miR-221 and miR-222 oncomiRs, which are overexpressed in multiple solid tumors including non-small cell lung cancer (NSCLC).
Downregulation of miR-340 has been reported in multiple tumors such as breast, colon, neuroblastoma, and osteosarcoma, in which mR-340 expression positively correlates with better prognosis. Experimentally validated miR-340 targets include disparate cellular components such as the tyrosine kinase MET in breast cancer,1 the transcription factors SOX2 in neuroblastoma2 and MITF in melanoma,3 and the cytoskeletal regulator ROCK1 in osteosarcoma.4
We recently characterized miR-340 as a novel tumor suppressor in lung cancer and glioblastoma. miR-340 expression inversely correlates with clinical staging in NSCLC patients, whereas exogenous miR-340 inhibits proliferation and survival in NSCLC-derived cells. miR-340–induced growth arrest correlates with p27KIP1 accumulation in both lung adenocarcinoma and glioblastoma cells. In A549 cells miR-340 controls p27KIP1 at both translational and post-translational levels by directly targeting 3 negative regulators of p27KIP1 (PUM1, PUM2, and SKP2) (Fig. 1).5
Figure 1.
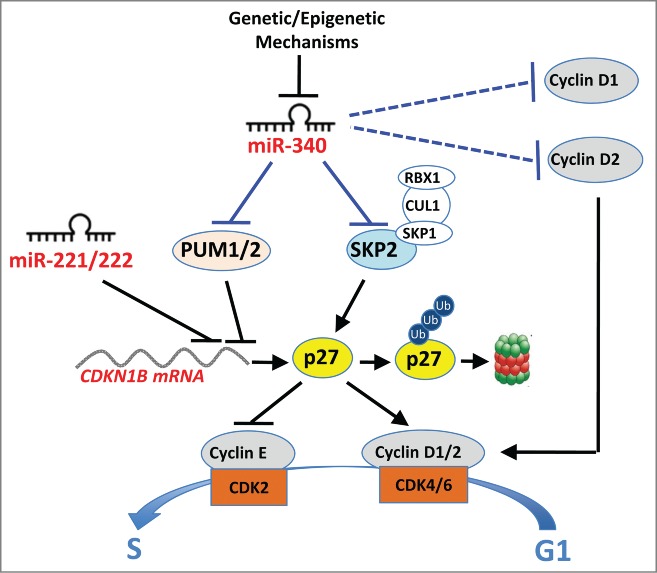
Mechanisms by which miR-340 inhibits the growth of lung cancer cells. miR-340 induces p27 at the translational level by targeting the RNA-binding proteins (PUM1 and PUM2) required for miR-221/222–mediated inhibition of the p27 transcript. miR-340 also induces p27 stabilization by targeting the SKP2 ubiquitin ligase in A549 cells. The blue lines indicate both validated (solid lines: PUM1, PUM2, and SKP2) and preliminarily characterized (dashed lines: cyclins D1 and D2) miR-340 target transcripts in A549 cells.
Human PUM1 and PUM2 genes encode 2 evolutionary conserved RNA-binding proteins related to the Pumilio gene products in Drosophila and fem-3 in C. elegans. The functional role of the PUM1/2 binding sites has been characterized for only a few human transcripts. Binding of PUM1 to the p27KIP1 3′-UTR induces a conformational switch that positively controls miR-221/222 accessibility. PUM2 has been suggested to act redundantly with PUM1. Consequently, p27KIP1 expression is affected by PUM1/2 expression levels. Growth factor-induced phosphorylation of PUM1 Ser714 increases its RNA-binding activity, suggesting a role of PUM1 post-translational modification in the control of cell cycle re-entry.6
The PUM1 and PUM2 transcripts share miR-340 target elements in their otherwise divergent 3′-UTRs. Our results show that miRNA-mediated downregulation of PUM1 and PUM2 antagonizes the miR-221/222–mediated inhibition of p27KIP1. Remarkably, transcriptome-wide analyses of PUM1- and PUM2-bound mRNAs show significant enrichment for multiple cell cycle regulators in addition to p27KIP1. Therefore, the miR-340–PUM1/2 axis might control cell cycle progression by targeting multiple transcripts in addition to CDKN1B, which encodes p27KIP1.7 For example, PUM1/2 have also been implicated in the miRNA-mediated control of the cell cycle regulator E2F3 in bladder cancer cells.8
PUM1 belongs to a growing list of RNA-binding proteins including HuR, Dnd1, CRD-BP, and PTB that are implicated in the modulation of miRNA targeting in mammalian cells. Interestingly, the miR-340 target site in the MITF 3′-UTR is controlled by the CRD-BP RNA-binding protein, which interferes with miR-340 binding thus protecting the MITF transcript from miR-340–mediated degradation.3 Intriguingly, in addition to PUM1/2, miR-340 also targets 2 distinct RNA-binding proteins, PBP1/hnRNP1 and hnRNPA2, in colorectal cancer, suggesting a complex interplay between miR-340 and RNA-binding proteins in cancer.9
p27KIP1 levels largely depend on protein stability, which is reduced by SCFSKP2-mediated ubiquitylation. Through investigation of the mechanism of p27KIP1 stabilization in miR-340-overexpressing cells we have identified S-phase kinase-associated protein 2, E3 ubitiquitin ligase (SKP2), the substrate-recognizing component of the SCFSKP2 complex, as a target of miR-340. To our knowledge, this is the first evidence of miRNA-mediated regulation of the human SKP2 oncoprotein. In summary, in NSCLC cells miR-340 induces p27KIP1 accumulation by affecting both synthesis (through PUM1/2) and degradation (through SKP2) of the CDK inhibitor.
Single nucleotide polymorphisms (SNPs) or 3′-UTR shortening events are known to affect the miRNA binding sites of transcripts coding for oncoproteins, such as KRAS. The identification of a SKP2 mRNA species harboring a short 3′-UTR lacking the miR-340 target site suggests that, depending on the splicing pattern, some tumors could express a SKP2 transcript isoform that is resistant to miR-340–mediated repression.
Similar mechanisms might affect the p27KIP1 and/or PUM1/2 3′-UTRs. Interestingly, we have identified cell lines in which p27KIP1 is unaffected by miR-340. Since miR-340 retains its antiproliferative activity in these cell lines, we investigated other putative miR-340 targets. Among various oncogenically relevant target transcripts, our preliminary experiments identified both cyclin D1 and cyclin D2, whose expression shows a significant inverse correlation with that of the miR-340 host gene (RNF130). Therefore, miR-340 could influence G1/S transition by affecting the accumulation of cyclins D1/D2 and the activity of cyclin D/CDK4/6 complexes, together with the induction of p27KIP1 (via PUM1/2 and SKP2) and inhibition of the cyclin E/CDK2 complex. In addition, having observed that miR-340 is responsive to serum induction we postulate that miR-340 might participate in the control of cell cycle progression in response to extracellular mitogenic signals.
In addition to further studies aimed at the transcriptome-wide identification of target mRNAs and oncogenic networks modulated by miR-340, future investigations will address the applications of miR-340. Importantly, systemic delivery of pre-miR-340 has recently been shown to inhibit the growth of xenografts of human colorectal cancer cells in mice.10 Therefore, multiple lines of evidence point to miR-340 as a novel, highly promising bullet for miRNA-based anticancer therapeutics.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by funding from AIRC (Associazione Italiana per la Ricerca sul Cancro) Grant-10489 and AICR (Association for International Cancer Research, UK) Grant-08–182 to Pasquale Verde.
References
- 1. Wu Z-S, Wu Q, Wang C-Q, Wang X-N, Huang J, Zhao J-J, Mao S-S, Zhang G-H, Xu X-C, Zhang N. miR-340 inhibition of breast cancer cell migration and invasion through targeting of oncoprotein c-met. Cancer 2011; 117:2842-52; PMID:21692045; http://dx.doi.org/ 10.1002/cncr.25860 [DOI] [PubMed] [Google Scholar]
- 2. Das S, Bryan K, Buckley PG, Piskareva O, Bray IM, Foley N, Ryan J, Lynch J, Creevey L, Fay J, et al. . Modulation of neuroblastoma disease pathogenesis by an extensive network of epigenetically regulated microRNAs. Oncogene 2012; 32:2927-36; PMID:22797059; http://dx.doi.org/ 10.1038/onc.2012.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goswami S, Tarapore RS, Teslaa JJ, Grinblat Y, Setaluri V, Spiegelman VS. MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor mRNA is inhibited by the coding region determinant-binding protein. J Biol Chem 2010; 285:20532-40; PMID:20439467; http://dx.doi.org/ 10.1074/jbc.M110.109298 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4. Zhou X, Wei M, Wang W. MicroRNA-340 suppresses osteosarcoma tumor growth and metastasis by directly targeting ROCK1. Biochem Biophysi Res Commun 2013; 437:653-8; PMID:23872151; http://dx.doi.org/ 10.1016/j.bbrc.2013.07.033 [DOI] [PubMed] [Google Scholar]
- 5. Fernandez S, Risolino M, Mandia N, Talotta F, Soini Y, Incoronato M, Condorelli G, Banfi S, Verde P. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene 2014; 25; PMID:25151966; http://dx.doi.org/ 10.1038/onc.2014.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JAF, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol 2010; 12:1014-20; PMID:20818387; http://dx.doi.org/ 10.1038/ncb2105 [DOI] [PubMed] [Google Scholar]
- 7. Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS ONE 2008; 3:e3164; PMID:18776931; http://dx.doi.org/ 10.1371/journal.pone.0003164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miles WO, Tschöp K, Herr A, Ji J-Y, Dyson NJ. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev 2012; 26:356-68; PMID:22345517; http://dx.doi.org/ 10.1101/gad.182568.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu Y. miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the warburg effect. Oncol Rep 2012; 28:1346–1352; PMID: 22895557; http://dx.doi.org/ 10.3892/or.2012.1958 [DOI] [PubMed] [Google Scholar]
- 10. Takeyama H, Yamamoto H, Yamashita S, Wu X, Takahashi H, Nishimura J, Haraguchi N, Miyake Y, Suzuki R, Murata K, et al. . Decreased miR-340 expression in bone marrow is associated with liver metastasis of colorectal cancer. Mol Cancer Ther 2014; 13:976–985; PMID:24448820; http://dx.doi.org/ 10.1158/1535-7163.MCT-13-0571 [DOI] [PubMed] [Google Scholar]


