Abstract
Our recent study shows that autophagy collaborates with proteasomes to degrade endothelial PAS domain-containing protein 1 (EPAS1, also known as HIF2α) in a manner dependent on Von Hippel-Lindau (VHL) and sequestosome 1 (SQSTM1/p62). The genetic dysregulation of autophagy is a common feature of different subtypes of renal cell carcinoma (RCC).
Keywords: autophagy, HIF2α, p53, p62, proteasome, renal cell carcinoma, VHL
Abbreviations
- ATG
autophagy-related gene
- ATG16L1
autophagy-related 16-like 1
- EPAS1/HIF2α
endothelial PAS domain-containing protein 1
- GABARAP
GABA receptor-associated protein
- GABARAPL2
GABA receptor-associated protein-like 2
- hMSH4
human MutS homolog 4
- MAP1LC3
microtubule-associated protein 1 light chain 3
- RCC
renal cell carcinoma
- SQSTM1/p62
sequestosome 1
- TCGA
The Cancer Genome Atlas
- TP53
tumor protein P53
- ULK2
unc-51 like autophagy activating kinase 2
- UVRAG
UV radiation resistance associated gene
- VBC
VHL-Elogin BC
- VBP1
VHL binding protein 1
- VHL
Von Hippel-Lindau
- WIPI1/2
WD repeat domain phosphoinositide interacting 1/2
Endothelial PAS domain-containing protein 1 (EPAS1, also known as HIF2α) is a transcription factor involved in the induction of genes that drive tumor initiation and metastasis. A complete understanding of the regulatory mechanism of HIF2α homeostasis will provide important clues to the management of tumors that are related to HIF2α dysregulation.
It is well established that HIF2α is degraded by the proteasome, and that the von Hippel-Lindau (VHL)-Elongin BC (VBC) complex mediates the ubiquitination of HIF2α. Our recent study shows that HIF2α is also constitutively degraded by autophagy, another major intracellular protein degradation process.1 This autophagic degradation of HIF2α requires the E3 ligase VHL and sequestosome 1 (SQSTM1, also known as p62). Our study further reveals collaboration between autophagy and the proteasome in HIF2α degradation, and a compensatory role of the proteasome during autophagy inactivation to efficiently remove HIF2α. Considering the presence of this compensatory function of the proteasome we have to be cautious in our interpretation of “negative” results, especially those obtained with cells in which autophagy is chronically inactive. In such cases, it would be necessary to confirm the accumulation of autophagy substrates using protein transient overexpression to overload the proteasome, or alternatively by acute genetic interference that inhibits autophagy before the cells initiate proteasome-mediated compensation.
Our study not only revealed collaboration between the proteasome and autophagy, but also indicated important areas for further enquiry. One such area of investigation is the molecular determinants that direct HIF2α to either autophagy or the proteasome for degradation. In our study, we found that VHL was required for both proteasomal and autophagic degradation of HIF2α, indicating the involvement of ubiquitination in both degradation processes. It is conceivable that different polyubiquitin chain linkages determine which degradation pathway is engaged. We speculate that Lys48-linked polyubiquitination directs HIF2α to proteasomal degradation and, conversely, that Lys63-polyubiqiutinated HIF2α is recognized by the autophagy receptor protein p62 and subjected to autophagic degradation. If this is true, the next issue to address is the molecular mechanism by which the VBC complex switches Lys48- and Lys63-linked ubiquitination of HIF2α. It has recently been reported that overexpression of VHL binding protein 1 (VBP1) favors autophagy-mediated degradation of human MutS homolog 4 (hMSH4), and that VBP1 itself is also degraded.2 Similarly, we detected p62 immunocomplexed with VHL, indicating that p62 is also a VHL binding protein. Based on these findings, it is plausible that these VHL binding proteins first preferentially promote Lys63-linked polyubiquitination and then recruit Lys63-linked polyubiquitinated proteins to the autophagosome for degradation. Some physiological conditions that induce autophagy, such as starvation, inflammation, and ER stress, are known to upregulate p62,3,4 which in turn facilitates autophagy-mediated degradation.
The association between autophagy and cancer has long been proposed, and genetic dysregulation of autophagy has been observed in several cancers.5 Mono-allelic loss of the BECN1 gene encoding the autophagy-related protein beclin1 has been detected in breast, ovarian, and prostate cancers. Deletion of UV radiation resistance associated gene (UVRAG) is associated with colorectal cancer, and somatic mutation of autophagy-related gene 5 (ATG5) is observed in gastrointestinal cancers. Clear cell renal cell carcinoma (RCC) is an angiogenic cancer, and the mechanism of angiogenesis is thought to involve VHL inactivation and consequent HIF2α stabilization. Since HIF2α is subjected to autophagic degradation, we studied the genetic alteration of ATG genes in RCC. Our study revealed that most clear cell RCCs harbor allelic loss and/or mutation of ATG7. Somatic mutations of several other ATG genes were also detected. More importantly, low expression of ATGs involved in the autophagy nucleation step predicts poor prognosis.1 These results indicate an anticancer role of autophagy in clear cell RCC, and suggest that constitutive autophagic degradation of HIF2α acts as a tumor suppression mechanism. Such a broad range of genetic alterations indicates that dysregulation of autophagy is an important hallmark of clear cell RCC.
In addition to the genetic alteration of autophagy genes in clear cell RCC, we further reported that chromophobe RCC is frequently associated with loss of one copy of BECN1, ATG4A/B/C, ATG5, ATG9A, Unc-51 like autophagy activating kinase 2 (ULK2), WD repeat domain phosphoinositide interacting 1 (WIPI1), microtubule-associated protein 1 light chain 3 gamma (MAP1LC3C), GABA receptor-associated protein (GABARAP), and autophagy related 16-Like 1 (ATG16L1). In contrast, papillary RCC is associated with copy number gain of GABARAP, BECN1, ATG9B, WIPI1/2, ULK2, GABARAP-like 2 (GABARAPL2) and microtubule-associated protein 1 light chain 3 beta (MAP1LC3B) (Fig. 1A). These findings indicate that genetic dysregulation of autophagy is a common feature of different subtypes of RCC, and might be used as a molecular marker for RCC classification and prediction. Furthermore, we found that the copy number of tumor protein P53 (TP53, best known as p53) was decreased in chromophobe RCC and increased in papillary RCC, which is in line with the changes observed in the ATG genes (Fig. 1A). Thus, changes in autophagy-related genes are not in the same direction in variant forms of RCC and mechanistic evaluation of autophagic dysregulation in chromophobe and papillary RCC is necessary to fully understand the implications of these findings. The role of autophagy in tumorigenesis is context-dependent, and a recent study revealed that p53 status determines the function of autophagy.6 In the presence of functional p53, loss of autophagy blocks Kras-driven pancreatic tumor progression; in the absence of p53, autophagy inactivation accelerates tumor onset associated with enhanced glucose uptake and anabolic pathways.6 If the working model of autophagy in pancreatic tumor also applies to RCC, we would expect that coordinated changes in p53 and ATGs, either increased or decreased expression, would promote renal tumorigenesis. On the other hand, clear cell RCC is generally associated with allelic loss and/or mutation of VHL, but not p53 (Fig. 1A), and we suspect that VHL rather than p53 might determine the function of autophagy in clear cell RCC. Clear cell RCC exhibits chromosomal instability and both autophagy and VHL have been demonstrated to be important for chromosomal stability.7,8 However, animal models of either Vhl or Atg7 loss fail to phenocopy human clear cell RCC,9,10 and we therefore hypothesize that coordinated loss of VHL and ATG7 is an obligatory early event in clear cell RCC carcinogenesis through the induction of chromosomal instability, and single inactivation is necessary but not sufficient.
Figure 1.
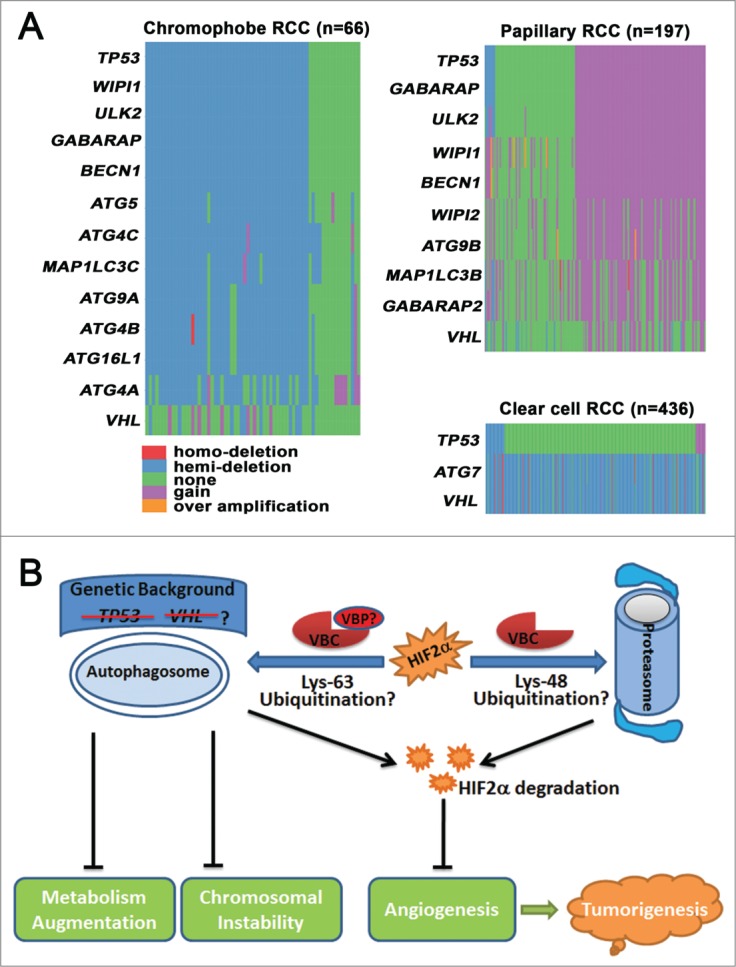
The function and genetic dysregulation of autophagy in renal cell carcinoma (RCC). (A) Copy number changes of autophagy-related genes (ATGs) in different subtypes of RCC. Copy number data for chromophobe RCC, papillary RCC, and clear cell RCC were obtained from The Cancer Genome Atlas (TCGA), and data processing was conducted with statistical computing tools. ATGs that were abnormal in more than 50% of cases are listed. The distribution of genes was drawn in the order of tumor protein P53 (TP53). (B) Autophagy suppresses tumorigenesis by degrading endothelial PAS domain-containing protein 1 (EPAS1, also known as HIF2α), stabilizing chromosomes, and moderating metabolism. Autophagy cooperates with the proteasome to degrade HIF2α and prevent angiogenesis. Lys48-linked polyubiquitination directs HIF2α to the proteasome for degradation, whereas Lys63-polyubiqiutinated HIF2α is subjected to autophagic degradation. VHL binding proteins (VBPs), such as VBP1 and sequestosome 1 (SQSTM1/p62), may preferentially promote Lys63-linked polyubiquitination and recruit Lys63-linked polyubiquitinated HIF2α to the autophagosome for degradation. Autophagy also suppresses renal tumorigenesis by preventing chromosomal instability and by moderating metabolism. The anticancer roles of autophagy may be more important in the absence of tumor suppressor genes, such as Von Hippel-Lindau (VHL) or TP53.
In summary, our study reveals the involvement of autophagy in HIF2α homeostasis and the dysregulation of autophagy in RCC. Further study will be required to investigate the molecular mechanism that determines whether proteins slated for destruction are shuttled to the proteasome or to the autophagosome. It will also be important to characterize the functional interaction between autophagy and tumor suppressor genes, such as VHL and p53, during RCC development. Generation of kidney-specific autophagy-deficient animal models associated with VHL or p53 inactivation will be necessary to address these issues.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Renee Kaye Cure Fur Cancer Grant and the MD Anderson Cancer Center Kidney Cancer Research Program.
References
- 1. Liu XD, Yao J, Tripathi DN, Ding Z, Xu Y, Sun M, Zhang J, Bai S, German P, Hoang A, et al. . Autophagy mediates HIF2alpha degradation and suppresses renal tumorigenesis. Oncogene 2014; PMID: 24998849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu Y, Her C. VBP1 facilitates proteasome and autophagy-mediated degradation of MutS homologue hMSH4. FASEB J 2013; 27:4799-810; PMID:23964080; http://dx.doi.org/ 10.1096/fj.13-235127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu XD, Ko S, Xu Y, Fattah EA, Xiang Q, Jagannath C, Ishii T, Komatsu M, Eissa NT. Transient aggregation of ubiquitinated proteins is a cytosolic unfolded protein response to inflammation and endoplasmic reticulum stress. J Biol Chem 2012; 287:19687-98; PMID:22518844; http://dx.doi.org/ 10.1074/jbc.M112.350934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sahani MH, Itakura E, Mizushima N. Expression of the autophagy substrate SQSTM1p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy 2014; 10:431-41; PMID:24394643; http://dx.doi.org/ 10.4161/auto.27344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jiang P, Mizushima N. Autophagy and human diseases. Cell Res 2014; 24:69-79; PMID:24323045; http://dx.doi.org/ 10.1038/cr.2013.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R, et al. . p53 status determines the role of autophagy in pancreatic tumour development. Nature 2013; 504:296-300; PMID:24305049; http://dx.doi.org/ 10.1038/nature12865 [DOI] [PubMed] [Google Scholar]
- 7. Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev 2007; 21:1367-81; PMID:17510285; http://dx.doi.org/ 10.1101/gad.1545107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thoma CR, Toso A, Gutbrodt KL, Reggi SP, Frew IJ, Schraml P, Hergovich A, Moch H, Meraldi P, Krek W. VHL loss causes spindle misorientation and chromosome instability. Nat Cell Biol 2009; 11:994-1001; PMID:19620968; http://dx.doi.org/ 10.1038/ncb1912 [DOI] [PubMed] [Google Scholar]
- 9. Jiang M, Wei Q, Dong G, Komatsu M, Su Y, Dong Z. Autophagy in proximal tubules protects against acute kidney injury. Kidney Int 2012; 82:1271-83; PMID:22854643; http://dx.doi.org/ 10.1038/ki.2012.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frew IJ, Thoma CR, Georgiev S, Minola A, Hitz M, Montani M, Moch H, Krek W. pVHL and PTEN tumour suppressor proteins cooperatively suppress kidney cyst formation. EMBO J 2008; 27:1747-57; PMID:18497742; http://dx.doi.org/ 10.1038/emboj.2008.96 [DOI] [PMC free article] [PubMed] [Google Scholar]


