Abstract
Tumors are prime examples of cell growth in unfavorable environments that elicit cellular stress. The high metabolic demand and insufficient vascularization of tumors cause a deficiency of oxygen and nutrients. Oncogenic mutations map to signaling events via mammalian target of rapamycin (mTOR), metabolic pathways, and mitochondrial function. These alterations have been linked with cellular stresses, in particular endoplasmic reticulum (ER) stress, hypoxia, and oxidative stress. Yet tumors survive these challenges and acquire highly energy-demanding traits, such as overgrowth and invasiveness. In this review we focus on stresses that occur in cancer cells and discuss them in the context of mTOR signaling. Of note, many tumor traits require mTOR complex 1 (mTORC1) activity, but mTORC1 hyperactivation eventually sensitizes cells to apoptosis. Thus, mTORC1 activity needs to be balanced in cancer cells. We provide an overview of the mechanisms contributing to mTOR regulation by stress and suggest a model wherein stress granules function as guardians of mTORC1 signaling, allowing cancer cells to escape stress-induced cell death.
Keywords: apoptosis, balance, cancer, cell death, ER stress, hypoxia, hyperactivation, mammalian target of rapamycin, mTORC1, mTORC2, oxidative stress, RNA granules, stress granules, survival
Abbreviations
- 4E-BP1
4E-binding protein 1
- 5′TOP
5′ terminal oligopyrimidine
- AMPK
AMP-activated protein kinase
- ATF4
activating transcription factor 4
- ATF6
activating transcription factor 6
- ATG
autophagy regulated protein
- ATM
ataxia telangiectasia mutated
- CAD
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase
- CaMKKbeta
calmodulin-dependent protein kinase kinase β
- CMA
chaperon-mediated autophagy
- DYRK3
dual specificity tyrosine-phosphorylation-regulated kinase 3
- eIF2α
eukaryotic translation initiation factor 2α
- eIF4B
eukaryotic translation initiation factor 4B
- eIF4E
eukaryotic translation initiation factor 4E
- ER
endoplasmic reticulum
- FIP200
FAK family kinase-interacting protein of 200 kDa
- FLCN
folliculin
- FMRP
fragile X mental retardation protein
- FoxO1/3A
forkhead box O1/3A
- G3BP
Ras-GTPase activating protein SH-3 domain binding protein
- GAP
GTPase-activating protein
- GCN2
general control nonderepressible 2
- GLUT4
glucose transporter 4
- Grb10
growth factor receptor-bound protein 10
- GSK
glycogen synthase kinase
- HIF
hypoxia inducible factor
- hnRNP-A1
heterogeneous nuclear ribonucleoprotein A1
- HRI
hemin-regulated inhibitor
- HSF1
heat shock factor protein 1
- Hsp70
70 kDa heat shock protein
- IR
insulin receptor
- Ire1
inositol-requiring protein 1
- IRES
internal ribosomal entry sites
- IRS
insulin receptor substrate
- JNK
c-Jun NH(2)-terminal kinase
- LARP1
La-related protein 1
- LDH
lactate dehydrogenase
- MAPK
mitogen activated protein kinase
- mSin1
mammalian stress-activated protein kinase interacting protein 1
- mtDNA
mitochondrial DNA
- mTOR
mammalian target of rapamycin
- mTORC1
mTOR complex 1
- mTORC2
mTOR complex 2
- NFL
negative feedback loop
- NOX
NADPH oxidase
- Nrf2
nuclear factor erythroid 2-like 2
- PABP1
polyadenylate-binding protein 1
- PDK1
3-phosphoinositide-dependent kinase-1
- PERK
protein kinase RNA-like ER kinase
- PI3K
phosphatidylinositol 3-kinases
- PIP2
phosphatidylinositol-3,4-biphosphate
- PIP3
phosphatidylinositol-3,4,5-triphosphate
- PKR
double-stranded RNA activated protein kinase
- PPP
pentose phosphate pathway
- PTEN
phosphatase and tensin homolog
- R5P
ribose-5-phosphate
- raptor
regulatory associated protein of mTOR
- RACK1
signaling scaffold protein receptor of activated protein kinase C 1
- REDD1
regulated in development and DNA damage responses 1
- rheb
ras-homolog-enriched-in-brain
- rictor
rapamycin-insensitive companion of mTOR
- ROS
reactive oxygen species
- S6K
S6 kinase
- SREBP
sterol regulatory element-binding protein
- TCA cycle
tricarboxylic acid cycle
- TIA-1
T cell intracellular antigen 1
- TIAR
TIA-1-related protein
- TNFα
tumor necrosis factor alpha
- TRAF2
TNF receptor-associated factor 2
- TRB3
tribbles homolog 3
- TSC1
hamartin (tuberous sclerosis 1 protein)
- TSC2
tuberin (tuberous sclerosis 2 protein)
- ULK1
unc-51-like kinase
- uORF
upstream open reading frame
- UPR
unfolded protein response
- USP10
ubiquitin-specific protease 10
- VEGF
vascular endothelial growth factor
Why do Cancer Cells Profit From mTOR Activation?
The mTOR signaling network (Fig. 1) is hyperactivated in many tumors (reviewed by Yecies et al.1). mTOR kinase is present in 2 multiprotein complexes, mTORC1 and mTORC2.2 mTORC1 contains the essential specific scaffold protein regulatory associated protein of mTOR (raptor) and functions as a master regulator of cell growth and metabolism by favoring anabolic processes in the presence of nutrients and energy.3,4 mTORC2 contains the specific proteins rapamycin-insensitive companion of mTOR (rictor) and mammalian stress-activated protein kinase interacting protein 1 (mSin1) (reviewed by Shimobayashi et al.2). mTORC2 senses nutrients and growth factors and modulates lipid and glucose metabolism5 and cytoskeleton reorganization (reviewed by Oh et al.6). The cancer drug rapamycin directly binds and inhibits mTORC1, but can also have indirect long-term effects on mTORC2.7,8
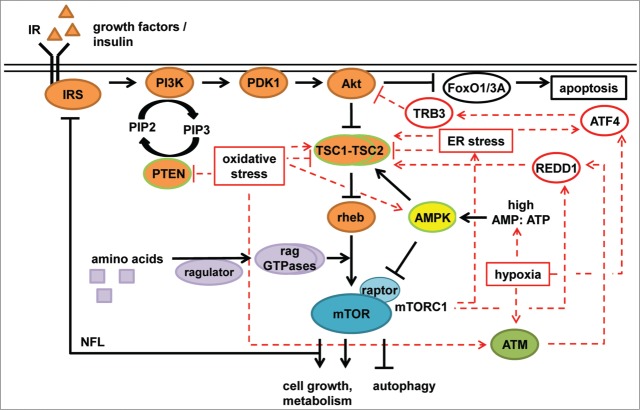
Figure 1.
mTORC1 and stress. mTORC1 is regulated by amino acids, growth factors (i.e., insulin), and energy status (AMP:ATP). Amino acids are sensed by the ragulator complex and the rag GTPases, mediating re-localization of mTORC1 to lysosomes where it encounters rheb. Insulin activates the IR, which then activates the IRS. Active IRS induces PI3K, which converts PIP2 to PIP3. PIP3 accumulation results in the recruitment of PDK1 and Akt to the plasma membrane where Akt is activated by PDK1. Akt phosphorylates and inhibits the TSC1–TSC2 complex, which inhibits rheb. Akt also inhibits the FoxO1/3A transcription factors, which positively regulate apoptosis. AMPK is activated by a high AMP:ATP ratio and inhibits mTORC1 by activating TSC1–TSC2 as well as by direct phosphorylation of the mTORC1 component raptor. Activation of mTORC1 inhibits IRS and Grb10 (not shown), resulting in negative feedback regulation of the PI3K–Akt branch. mTORC1 hyperactivation can lead to ER stress, which can activate or inhibit the TSC1–TSC2 complex. In addition, ER stress induces ATF4 translation, which can induce expression of the negative Akt regulator TRB3. Hypoxia also induces ATF4 translation, and activates AMPK. Induction of HIFs by hypoxia (via ATM) induces expression of REDD1, which activates the TSC1–TSC2 complex, inhibiting mTORC1. This results in a negative feedback loop, as mTORC1 controls REDD1 stability. Oxidative stress inhibits the tumor suppressors PTEN, and inhibits or activates TSC1–TSC2. Furthermore, oxidative stress can activate ATM and AMPK, both of which inhibit mTORC1. Tumor suppressors are framed in green. Stress inputs are shown in red.
Amino acids activate mTORC1 via the rag GTPases,9,10 which function in conjunction with the guanine nucleotide exchange factor (GEF) ragulator complex11 and the GTPase activating protein (GAP) folliculin (FLCN)12 to modulate the translocation of mTORC1 to the lysosomal membrane in a glutaminolysis-dependent manner13 (reviewed by Bar-Peled et al.14). At the lysosome, mTORC1 encounters the small GTPase ras-homolog-enriched-in-brain (rheb), which activates mTORC1 in response to growth factors (e.g., insulin).15 Amino acid deprivation leads to recruitment of the hamartin (TSC1)–tuberin (TSC2) heterocomplex (TSC1–TSC2) to the lysosomal membrane in a rag GTPase-dependent manner.16 The tumor suppressor TSC1–TSC2 functions as a GAP for the GTPase rheb and thereby inhibits mTORC1.17
Acting through insulin receptor substrate (IRS), the insulin receptor (IR) activates class I phosphatidylinositol 3-kinases (PI3K), whose subunits are often mutated in tumors. PI3K phosphorylates phosphatidylinositol-3,4-biphosphate (PIP2) to generate phosphatidylinositol-3,4,5-triphosphate (PIP3). Binding of PIP3 to the oncogenic kinase Akt (also termed protein kinase B, PKB) and 3-phosphoinositide-dependent kinase-1 (PDK1) enables their translocation to the plasma membrane, where PDK1 phosphorylates and activates Akt. Akt acts as an inhibitor of the TSC1–TSC2 complex by phosphorylating TSC2; phosphorylation of TSC2 by Akt leads to dissociation of the TSC1–TSC2 complex from lysosomes18 and enables mTORC1 activation. The PI3K antagonist phosphatase and tensin homolog (PTEN) is a tumor suppressor that counteracts growth factor-dependent mTORC1 activation by dephosphorylating PIP3 to generate PIP2 (reviewed by Laplante et al.19).
mTORC1 responds to cellular energy status via the heterotrimeric AMP-activated protein kinase (AMPK). AMPK is activated by 2 mechanisms. On the one hand, kinases such as the tumor suppressor kinase LKB1 and calmodulin-dependent protein kinase kinase β (CaMKKβ) phosphorylate AMPK in its activation loop. Furthermore, when the cellular ATP:AMP ratio is low, AMP directly binds to AMPK and allosterically activates it (reviewed by Hardie et al20). AMPK inhibits mTORC1 by phosphorylating raptor21 and by an activating phosphorylation on TSC2.22 Furthermore, the ATP-sensitive Tel2–Tti1–Tti2 (TTT)–RUVBL1/2 complex activates mTORC1 by favoring mTORC1 assembly and its lysosomal localization in a rag GTPase-dependent manner.23
Cancer cell growth depends on ATP-demanding anabolic processes including protein, lipid, and nucleotide biosynthesis. mTORC1 controls ATP supply by inducing mitochondrial biogenesis, the tricarboxylic acid (TCA) cycle, and aerobic respiration.24-26 Furthermore, mTORC1 promotes the delivery of substrates to the TCA cycle by inducing glucose uptake27 and glutamine catabolism.28 A major anabolic function of mTORC1 in cancer is its stimulating role in translation29 (reviewed by Ma et al.30). mTORC1 phosphorylates and inhibits eukaryotic translation initiation factor 4E-binding protein 1 (4E‑BP1), an inhibitor of 5´cap-dependent translation. Phosphorylation of 4E-BP1 decreases its binding to the eIF4F complex component eukaryotic translation initiation factor 4E (eIF4E), which upon release from 4E-BP1 assembles into the eIF4F complex. The eIF4F complex mediates the scanning process by which ribosomes reach the start codon. Furthermore, mTORC1 enhances the cellular protein biosynthesis capacity by activating ribosomal RNA (rRNA) transcription and processing31 (reviewed by Iadevaia et al.32) and the biosynthesis of ribosomal proteins and elongation factors; these proteins are often encoded by transcripts that contain 5´ terminal oligopyrimidine (5´TOP) tracts,33 whose translation depends on 4E-BP1 inactivation.26,34 In addition, the raptor interacting protein La-related protein 1 (LARP1) binds to the mRNA 5´cap in an mTORC1-dependent manner, which seems to particularly affect translation of RNAs containing 5´TOP motifs.35 Furthermore, 5´TOP regulation by mTOR has been reported to also occur in a 4E-BP1- and mTORC1-independent manner,36,37 in particular under hypoxic conditions.37 S6 kinase (S6K), another mTORC1 substrate, phosphorylates S638 and the eIF4F component eukaryotic translation initiation factor 4B (eIF4B),39,40 which may contribute to translational control by mTORC1 but not by translational regulation of 5´TOP mRNAs.41 In addition, S6K promotes mRNA expression of ribosome biogenesis genes, thereby probably increasing overall translation capacity.42 The PI3K–Akt–mTORC1 pathway upregulates the synthesis of lipids via the sterol regulatory element-binding protein (SREBP) transcription factors,5,43-46 which regulate genes involved in lipid and sterol synthesis.47 mTORC1 stimulates nucleotide biosynthesis via direct phosphorylation of the trifunctional enzyme carbamoyl-phosphate synthetase 2-aspartate transcarbamylase-dihydroorotase (CAD), which catalyzes the first 3 steps of de novo pyrimidine synthesis.48,49 In addition, mTORC1 promotes the expression of genes encoding enzymes of the oxidative branch of the pentose phosphate pathway (PPP),45 which generates ribose-5-phosphate (R5P) and NADPH for biosynthesis. R5P and ATP are needed for the synthesis of 5-phosphoribosyl-1-phosphate, which is required for the synthesis of purines and pyrimidines. Hence, cancer cells likely profit from mTORC1 activation, as this promotes building block biosynthesis and thereby contributes to abnormal proliferation. It should, however, be noted that mTORC1 inhibits the oncogene Akt via negative feedback loops (NFLs) dependent on IRS50–52 and growth factor receptor-bound protein 10 (Grb10)53,54. Akt inhibits apoptosis by inhibiting the transcription factor forkhead box O1/3A (FoxO1/3A).55 Furthermore, Mounir et al.56 have shown that Akt directly phosphorylates and inhibits the ER stress sensor protein kinase RNA-like ER kinase (PERK), thereby preventing its hyperactivation and subsequent cell death. Thus, chronic mTORC1 activation via NFLs results in Akt inhibition and thereby facilitates apoptosis (reviewed by Apenzeller-Herzog et al.57). Consequently, cancer cells need to balance mTORC1 activity to keep biosynthetic processes and Akt active at the same time.
mTOR Regulation by Stresses in Cancer Cells
The capacity for uncontrolled cellular growth and proliferation brings about challenges, such as certain stresses, that a tumor cell has to cope with in order to survive. Nutrient and oxygen depletion in conjunction with a hyperactive metabolism, mitochondrial dysfunction, and oncogenic mTOR signaling are common conditions in cancer cells58–62 and often correlate with cellular stresses. We focus here on ER stress, hypoxia, and oxidative stress and their interaction with mTOR and cancer cell metabolism (Fig. 1).
mTORC1 under ER stress
Numerous studies report an accelerated unfolded protein response (UPR) in cancer cells. ER stress results from imbalances between protein synthesis and protein folding capacity that lead to accumulation of unfolded proteins in the ER lumen (reviewed by Clarke et al.63 and Fels et al.64). Several factors can contribute to the phenomenon of ER stress (Fig. 2). When tumors outgrow the vascular system they eventually face a shortage in oxygen and nutrients.64,65 Decreased glucose supply restricts ATP synthesis, which is required for chaperone activity in the ER (reviewed by Braakman et al.66). Thus, decreased ATP levels can result in impaired protein folding and ER stress. Glucose is not only used for ATP synthesis but is also a major source of carbon molecules for the synthesis of cellular building blocks (lipids, nucleotides, and amino acids). Proliferating cells require lipids for membrane formation and ER expansion. A lipid shortage, and hence reduced membrane synthesis, can induce ER stress67–69 and apoptosis.70,71 These observations suggest that glucose limitation is a trigger for ER stress. However, studies on cancer metabolism have reported the Warburg effect, namely aerobic glycolysis and accumulation of lactate.72,73 The Warburg effect is defined by an enhanced glycolytic rate under normoxic conditions. Cells that exhibit the Warburg effect consume glucose relatively rapidly and therefore require a sufficient supply of glucose.74 These 2 seemingly contradictory views on glucose levels in cancer cells may be relevant at different stages of tumor progression. In the initial stages, increased levels of glucose transporters75,76 allow the cell to take up as many nutrients as the environment allows. Enhanced glucose uptake, in conjunction with hyperactivation of the mTOR pathway, is prone to induce ER stress as increased protein synthesis can overwhelm the protein folding capacity of the ER.63,77 In contrast, at advanced tumor stages the outgrowth from the vascular system results in nutrient shortage, which also leads to ER stress as discussed earlier.
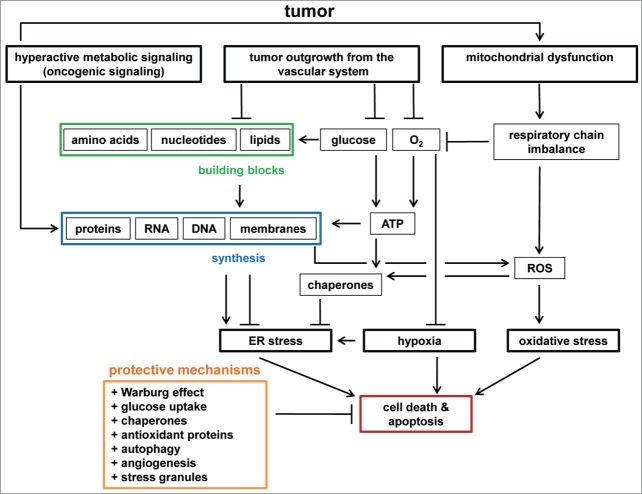
Figure 2.
Stresses in tumors. Hyperactive metabolic signaling (e.g., induced by oncogenes) can result in increased synthesis of proteins, RNA, DNA, and membranes. Lipid synthesis is required for ER homeostasis, whereas hyperactive protein synthesis can induce ER stress. Tumors eventually outgrow the vascular system, leading to a shortage in glucose, oxygen, and building blocks (amino acids, nucleotides, lipids). Glucose is required for ATP synthesis and is a carbon source for building block synthesis. Lack of ATP and building blocks inhibits lipid biosynthesis and chaperone activity. Therefore, ATP depletion enhances ER stress. Oxygen is required for ATP synthesis, and oxygen depletion results in hypoxia. ROS induce oxidative stress and originate from dysfunctions in mitochondria, for example triggered by oncogenic signaling and mtDNA damage, respiratory chain imbalances, and lipid and protein biosynthesis. ER stress, hypoxia, and oxidative stress induce stress responses to restore cellular homeostasis, and eventually trigger apoptosis. Cancer cells have protective mechanisms to prevent the induction of apoptosis by chronic stresses. Examples of such mechanisms are metabolic transformation (the Warburg effect), glucose uptake, chaperone and antioxidant protein synthesis, autophagy, angiogenesis, and stress granule formation.
The ER has its own sensors for the detection of unfolded proteins and to restore ER homeostasis via the UPR (reviewed by Hetz et al.78). The 3 sensors inositol-requiring protein 1 (Ire1), activating transcription factor 6 (ATF6), and PERK are membrane embedded proteins that synergistically re-establish ER homeostasis. For example, they induce chaperone synthesis79,80 to increase protein folding capacity, and inhibit translation81,82 to relieve protein overload. In addition, autophagy (see below) has emerged as the major mechanism for the clearance of misfolded proteins in the ER,83,84 as ER stress suppresses proteasome-mediated degradation.85,86 If cells are unable to restore homeostasis persistent ER stress leads to apoptosis, which needs to be circumvented by cancer cells.
The regulatory interaction between mTORC1 and ER stress can be understood as a bidirectional cross talk (reviewed by Appenzeller-Herzog et al.57) (Fig. 1). Mutations or knock out of the TSC1 and TSC2 genes that lead to mTORC1 hyperactivation sensitize cells to ER stress and apoptosis. This depends on mTORC1 as it can be reversed by raptor inhibition,77,87 further supporting the notion that TSC1–TSC2 and mTORC1 jointly modulate ER stress. Conversely, ER stress may also modulate the activity of mTORC1 via the TSC1–TSC2 complex. In neuronal cells, short-term periods of ER stress result in TSC1–TSC2 inactivation and subsequent mTORC1 activation, whereas prolonged stress activates the TSC1–TSC2 complex.88 Whether this also occurs in cells other than neurons remains to be explored. Akt is another important mediator of ER stress-dependent mTORC1 regulation. ER stress induces translation of activating transcription factor 4 (ATF4); this induces apoptosis by transcriptional activation of stress-related proteins, including tribbles homolog 3 (TRB3),89 which inhibits Akt. In addition, ER stress inhibits mTORC2 and its substrate Akt in a glycogen synthase kinase (GSK) 3-β–dependent manner.90 Furthermore, activation of mTORC1 by ER stress inhibits Akt via the mTORC1-dependent NFLs, followed by activation of the Ire1–c-Jun NH(2)-terminal kinase (JNK) pathway, which in turn induces apoptosis.91 This suggests that cancer cells under chronic ER stress must cope with Akt inactivation by multiple mechanisms.89–91 As active mTORC188 contributes to Akt inhibition and apoptosis susceptibility,77,87,88,91 cancer cells need to prevent mTORC1 hyperactivation to maintain sufficient Akt activity and ensure their survival under ER stress.
mTORC1 under hypoxia
The outgrowth of the tumor from the vascular system entails a shortage not only in glucose supply but also in oxygen (Fig. 2). This phenomenon is termed “hypoxia” and induces a stress response that can be monitored by upregulation of the hypoxia inducible factors (HIFs).58 Oxygen shortage restricts the cellular capacity for ATP production because the respiratory chain requires aerobic conditions. Consequently, pyruvate is not entirely consumed by the TCA cycle but is, at least partially, converted into lactate to maintain the cellular redox balance.58
The hypoxia stress response adapts cells to low levels of oxidative respiration. Thus, hypoxia reduces energy consumption, activates glycolysis, and improves oxygen supply (reviewed by Majmundar et al.92). The HIF transcription factors are key to the hypoxia-induced stress response. HIF1α induces gene products such as the vascular endothelial growth factors (VEGFs),93 which activate growth of the vascular network (angiogenesis)94 to restore oxygen availability. In addition, HIFs induce glycolysis and autophagy (see below). Of note, in cancer cells HIF upregulation often occurs without hypoxic conditions and thereby contributes to the Warburg effect (see below). In this case, HIFs can be induced by oncogenic signaling via mTORC195,96 and promote cell growth, proliferation, and survival. In addition to the HIFs, histone modifications have been reported to contribute to HIF-independent transcriptional regulation under hypoxia,97 but the underlying mechanisms and their potential interaction with mTOR signaling remain to be explored.
Hypoxia inactivates mTORC1 by different mechanisms (Fig. 1). First, hypoxia increases the AMP:ATP ratio, which activates AMPK.98,99 Second, hypoxia activates the DNA damage response protein ataxia telangiectasia mutated (ATM) in the cytosol in a DNA damage-independent manner.100 ATM phosphorylates HIF1α, resulting in induction of regulated in development and DNA damage responses 1 (REDD1).100 REDD1 and mTORC1 are connected via a NFL: REDD1 inhibits mTORC1 via TSC1–TSC2 activation,101–103 whereas mTORC1 is necessary to stabilize the REDD1 protein.104,105 Furthermore, mTORC1 activity is also required for HIF1α expression.95,106 Thus, hypoxic cells require mTORC1 to re-establish homeostasis through the HIF1α- and REDD1-dependent stress response. On the other hand, mTORC1 needs to be restricted, because otherwise the mTORC1-dependent NFLs inhibit Akt, leading to apoptosis sensitization. This is particularly relevant under hypoxia as Akt may be further inhibited by ATF4 induction.107 Thus, hypoxia inhibitory and stimulatory inputs contribute to net mTORC1 activity.
mTORC1 under oxidative stress
A third challenge that is commonly encountered in cancer cells is oxidative stress (Fig. 2). Oxidative stress is induced by the accumulation of reactive oxygen species (ROS). To comply with their high proliferation rate, cancer cells exhibit an accelerated metabolism which entails an increased activity of the respiratory chain and mitochondrial biogenesis.108 This not only increases ATP production but may also increase cellular ROS108 as a result of temporary imbalances between reduction and oxidation at the level of Complexes I and III of the respiratory chain.109 Also, dysfunction of mitochondria in cancer cells110 may contribute to increased ROS levels. Mutations in cancer cells tend to accumulate in mitochondrial DNA (mtDNA)111,112 and are enriched in genes coding for subunits of Complexes I, III, and IV of the electron transport chain,113 which may eventually lead to ROS release. This also occurs during therapeutic intervention, as chemotherapies preferentially induce mutations in mtDNA, correlating with increased ROS formation.114,115 Of note, ROS formation in cancer cells has been often linked with an induction of oncogenic signaling,116 for example of the mitogen activated protein kinase (MAPK) and PTEN/Akt pathways.117–120 For example, H-Ras activates the ROS-producing NADPH oxidase (NOX)121 enzymes and suppresses the antioxidant molecule Sestrin 1.122 Akt increases the activity of several respiratory complexes in a 4E‑BP1-dependent manner, 120 thus increasing the potential for ROS formation, but the underlying mechanism remains elusive. Hence, multiple processes contribute to ROS formation in cancer cells.
How do cancer cells cope with these increased ROS levels? The response to oxidative stress is partially induced by the ROS themselves. ROS can oxidize cysteines, leading to disulfide bond formation in proteins and thereby altering their activity (reviewed by Groitl et al.123). Through this mechanism, ROS activate chaperones to refold damaged proteins. One prominent example is the 2-Cys peroxiredoxin PrxII, whose chaperone activity is induced by cysteine oxidation under oxidative stress.124 In addition, oxidative stress induces the key stress transcription factor nuclear factor erythroid 2-like 2 (Nrf2), which controls the expression of several hundred genes including chaperones, antioxidant enzymes, or proteins involved in the inflammatory and immune response (reviewed by Sosa et al.108). For example, cancer cells show upregulation of the antioxidative proteins glutathione, superoxide dismutase, catalase, and thioredoxin (reviewed by Watson et al.125), at least in part as a result of Nrf2-induced oncogenic signaling (reviewed by DeNicola et al.126).
Early evidence for the regulation of mTORC1 complex by ROS came from UV irradiation experiments. UV radiation activates mTORC1 during the first 7 hours, with a subsequent decrease over time,127–129 and mTORC1 activation can be prevented by hydrogen peroxide scavengers.129 Additionally, chemical treatments with hydrogen peroxide or sodium arsenite130 affect mTORC1 in a dosage- and time-dependent manner. Generally speaking, short treatments and low concentrations seem to induce mTORC1, whereas prolonged treatments and high concentrations diminish or abolish mTORC1 activity.131–134 It should be noted, however, that the dosage- and time-dependent effects of ROS on mTORC1 are highly context- and cell type-dependent. The tumor suppressor PTEN135–137 is redox sensitive and directly inactivated by cysteine oxidation; in addition, TSC1–TSC2 has been suggested to be directly oxidized by ROS138 (Fig. 1). Thus, in cancer cells ROS possibly contribute to chronic TSC1–TSC2 and PTEN inactivation and mTORC1-dependent metabolic induction. In contrast, Zhang et al.132 reported recently that mTORC1 can also be inactivated by ROS, and that this depends on peroxisomal localization of TSC2. Furthermore, ROS activates cytoplasmic ATM139,140 and AMPK, which both inhibit mTORC1 (reviewed by Hardie et al.99). Thus, ROS have activating and inhibitory effects on mTORC1, whose net regulation (i.e., activation or inhibition) depends on the cellular context, persistence, and strength of the ROS stress.
Regulation of mTORC2 by stresses
Relatively little is known about the response of mTORC2 to stress, therefore in this review we focus mostly on mTORC1. It should be noted, however, that increasing evidence additionally suggests mTORC2 as an important component of stress signaling. There are activating and inhibiting inputs on the mTORC2 network during different stresses. Examples are the inhibition of mTORC2 by ER stress90 and oxidative stress,141,142 and the activation of mTORC2 during hypoxia.143 ER stress results in GSK3β-dependent phosphorylation of rictor, which decreases the affinity of mTORC2 for its substrates,90 whereas oxidative stress leads to mTORC2 disruption and inactivation.141,142 The mechanism by which mTORC2 is activated during hypoxia is not understood. mTORC2 activation during hypoxia is needed for the hypoxia stress response as mTORC2 induces transcription of HIF1α and HIF2α,106 and positively modulates hypoxia-induced proliferation.143
Interconnection of ER stress, hypoxia, and oxidative stress
Oxidative stress, hypoxia, and ER stress are closely intertwined and cannot be viewed separately. For example, lack of oxygen inhibits ATP production by the respiratory chain,73 which at least in the short term mitigates chaperone-mediated protein folding and thus induces ER stress. In addition, oxygen is the preferred terminal electron acceptor for disulphide bond formation (oxidative protein folding) within the ER.144,145 Thus, hypoxia is able to induce ER stress.146,147 Conversely, severe ER stress induces oxidative protein folding148 that leads to ROS formation, which in a vicious cycle can lead to protein damage and reinforce the ER stress.149 Furthermore, glucose starvation150,151 and hypoxia152,153 can induce ROS formation in tumor cells, but the underlying mechanisms are poorly understood. In conclusion, cancer cell traits are prone to induce stress at different levels; as oxidative stress, hypoxia, and ER stress can induce each other they often occur in conjunction, and cancer cells thus have to cope with chronic stress conditions that are prone to induce apoptosis.154–159 However, cancer cells acquire properties that enable them to escape programmed cell death131,160,161 (see below).
Regulation of Glucose and Protein Homeostasis by mTORC1 During Stress
Hyperactive biosynthesis in proliferating cells creates a high demand for ATP and building blocks, but oxidative phosphorylation is also a source of cellular ROS, as discussed earlier. How do cancer cells cope with this challenge? During glycolysis one glucose molecule is converted into 2 ATP molecules and pyruvate. Under normoxic conditions, pyruvate is introduced into the TCA cycle, which theoretically generates 36 ATP molecules via aerobic respiration. However, under hypoxic conditions pyruvate is converted by lactate dehydrogenase (LDH) to lactate in the cytosol, without further generation of ATP. Cancer cells “ferment” glucose into lactate even under normoxic conditions (aerobic glycolysis).72 Although the ATP yield is low, aerobic conversion of glucose to lactate is fast, generates less ROS, and delivers carbon backbones for building block synthesis (reviewed by Hsu et al.162). This metabolic transformation, which was discovered by Otto Warburg nearly 100 years ago, is named the “Warburg effect."72 Another shift of glucose metabolism in cancer cells is induction of the PPP (reviewed by Sosa et al.108). Diverting carbon from glycolysis into the PPP supplies increases levels of (1) R5P for nucleotide synthesis, which is needed for DNA replication and transcription (reviewed by DeBerardinis et al.163); and (2) NADPH, which supplies electrons for biosynthesis and eliminates ROS, thereby providing protection from oxidative stress. Diversion of glucose into the PPP and thus into lactate is modulated by several mTOR network components that positively regulate glucose uptake and glycolysis: Akt promotes glucose uptake, for example, by stimulating translocation of glucose transporter 4 (GLUT4)164,165 to the plasma membrane. Furthermore, AMPK inactivation is tumorigenic as AMPK inhibits the Warburg effect in a HIF1α-dependent manner.166 This may in fact be mediated by mTORC1, which is activated upon AMPK inhibition. mTORC1 increases HIF1α levels,95,96 which in turn can activate the expression of almost all glycolytic enzymes.167
mTORC1 and stresses also impinge on autophagy, a cell autonomous process that maintains protein homeostasis (Fig. 3). During autophagy, proteins and cell organelles are targeted to the lysosomes for degradation. In cancer cells, autophagy has an ambiguous function. On the one hand, autophagy has been suggested to prevent tumorigenesis, but on the other hand autophagy seems to promote stress survival in established tumors (reviewed by Yang et al.168). There are 3 different types of autophagy (reviewed in Boya et al.169 and Marino et al.170): macroautophagy, microautophagy, and chaperon-mediated autophagy (CMA). Macroautophagy, hereafter called autophagy, is divided into tightly regulated steps. First, a phagophore emulates and elongates to surround a cytoplasmic fraction. The resulting autophagosome docks and fuses with hydrolase-containing lysosomes, enabling digestion of proteins and organelles. The resulting autolysosome consists of the inner membrane of the previous autophagosome and enables digestion of the proteins and organelles within the surrounded cytoplasmic fraction. The building blocks that are released by this process can be reused by the cell. Autophagy initiation (emulation and elongation of the phagophore) is positively controlled by the unc-51–like kinase 1 (ULK1) complex, comprising the proteins ULK1, autophagy regulated proteins 13 and 110 (ATG13, ATG110), and FAK family kinase-interacting protein of 200 kDa (FIP200).171,172 mTORC1 and AMPK phosphorylate ULK1 on different sites and thereby respectively inhibit or activate autophagy.173 mTORC1 phosphorylates ULK1173 and ATG13,172 reducing ULK1 complex stability and ULK1 kinase activity.174,175 In contrast, AMPK binds to the mTORC1-bound ULK1 complex and phosphorylates raptor176 and ULK1173 to activate autophagy. Another modulator of autophagy initiation is the Bcl‑2/Beclin 1 complex, which inhibits phagophore maturation.177 ER stress, hypoxia, and oxidative stress affect autophagy via mTORC1, AMPK, and Bcl‑2/Beclin 1. The ER stress-induced UPR results in Ire1 and JNK activation. JNK phosphorylates Bcl-2,178,179 disrupting its binding to Beclin 1 and inducing autophagy. ER stress also induces autophagy when inhibiting the PI3K–Akt pathway180 and mTORC1.181 Both ER stress and hypoxia induce ATF4, which directly upregulates ULK1 transcription and ULK1 complex activity.182,183 In addition, ATF4 induces TRB3 expression89,184 resulting in inhibition of Akt, which may potentially induce autophagy via mTORC1 inhibition. Furthermore, hypoxia induces autophagy by activating AMPK185 and BNIP3/BNIP3L,186–188 negative modulators of the Bcl-2/Beclin 1 complex. Little is known about autophagy regulation by oxidative stress. Oxidative stress induces AMPK, correlating with induction of autophagy.189 In addition, oxidative stress also activates CMA,190 a process in which proteins are unfolded and directly trans-localized through the lysosomal membrane.
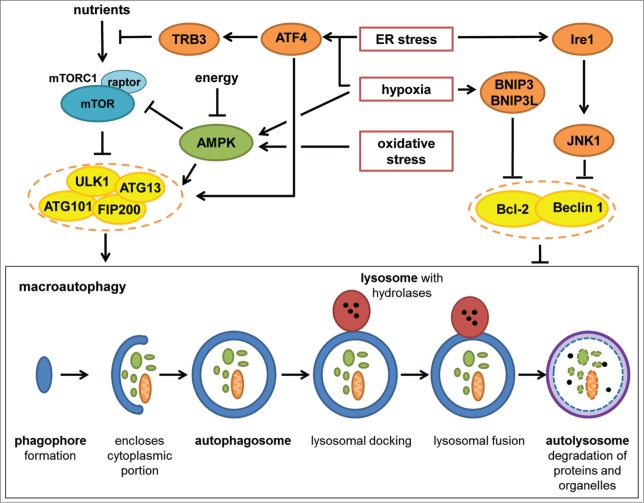
Figure 3.
Autophagy regulation by stress. The ULK1 complex (ULK1, ATG13, ATG101, and FIP200) and the Bcl-2–Beclin 1 complex are major autophagy regulators. Autophagy can be divided into 3 different steps: (1) phagophore formation and enlargement (autophagosome); (2) lysosomal docking and fusion with the autophagosome (autolysosome); (3) degradation of proteins and organelles in the autolysosome. The ULK1 complex is needed for autophagy initiation, whereas assemble of the Bcl-2–Beclin 1 complex prevents Beclin 1 from triggering autophagy. The ULK1 complex is inhibited by mTORC1 and activated by AMPK. AMPK also directly inhibits mTORC1. ER stress induces ATF4, which controls transcription of stress factors such as TRB3, which is a negative effector upstream of mTORC1 (Akt inhibition). In addition, ATF4 has a positive effect on the ULK1 complex. ER stress activates Ire1 kinase, which induces JNK1, leading to disassembly of the Bcl-2–Beclin 1 complex. Hypoxia also induces ATF4 expression and activates AMPK. In addition, hypoxia induces autophagy by BNIP3/BNIP3L-dependent disassembly of the Bcl-2–Beclin 1 complex. Oxidative stress induces autophagy in an AMPK-dependent manner.
In cancer cells, autophagy is necessary to maintain the building block supply, especially under starvation conditions. In addition, autophagy is able to counteract stresses like ER stress and oxidative stress by degrading damaged proteins and cell organelles. In keeping with this, inactivation of the negative AMPK regulator FLCN leads to stress resistance via autophagy induction.191 Furthermore, autophagy inhibition correlates with induction of apoptosis during cancer-related hypoxia and thus seems to have an important function in tumor cell survival under endogenous stress.192 In addition, autophagy induction often correlates with cancer resistance to chemotherapeutics.193,194 In contrast, prolonged autophagy induction has been suggested to result in cell death (reviewed by Loos et al.195 and Marino et al.170). Given that mTORC1 is a potent inhibitor of autophagy, it seems paradoxical that both mTORC1 and autophagy are required for cancer cell survival. This suggests that cancer cells need to maintain a delicate balance between mTORC1 activity and autophagy in order to benefit from both.
Balancing mTORC1 Under Stress: Stress Granules as Guardians of Cancer Cells?
mTORC1 activity contributes to many aspects of cancer cell survival. However, chronic mTORC1 hyperactivation eventually inhibits autophagy and induces cell death, and therefore needs to be counterbalanced. Several inputs into the mTOR network, mainly those impinging on TSC1–TSC2, Akt, and AMPK, restrict mTORC1 activity under stress and thereby not only limit cellular growth, but also potentially enable autophagy and suppress cell death. Stress granules (SGs) represent an additional buffer system in stressed cells. SGs form under a variety of stresses including hypoxia, ER, oxidative, heat, nutrient, osmotic, and cold stress.196–198 Protein synthesis is inhibited during stress, and polysome disassembly can be induced by many different stress sensors. The most prominent examples are eukaryotic translation initiation factor 2α (eIF2α) kinases (reviewed by Donnelly et al.199), which phosphorylate eIF2α at serine 51. eIF2α is a subunit of eIF2, which together with t-RNAfMet and GTP forms a ternary complex that is required for formation of the 48S translation preinitiation complex. Phosphorylation of eIF2α prevents ternary complex formation, leading to polysome disassembly and producing a non-canonical 48S* complex that is unable to recruit the 60S ribosomal subunit. In mammals, 4 eIF2α kinases have been described: hemin-regulated inhibitor (HRI), double-stranded RNA activated protein kinase (PKR), general control nonderepressible 2 (GCN2), and PERK. These kinases allow the cell to respond to a broad spectrum of stresses including oxidative stress,200 ER stress,201 and amino acid starvation.202 Polysome disassembly changes the fate of many proteins involved in mRNA processing, leading to accumulation of mRNAs that disassemble from polysomes. The morphological consequence of this process is the formation of cytoplasmic SGs, which are protein–RNA assemblies.203 SGs have an antiapoptotic function under stress,131,204 and their formation after chemotherapy or radiotherapy in cancer correlates with therapy resistance.205,206 Thus, SGs could help the tumor to balance stress signaling and prevent apoptosis under stresses elicited by the tumor environment or therapeutic interventions.
The first phases in SG aggregation or nucleation depend on SG nucleating proteins, which bind to the disrupted 48S*-mRNA complex. Overexpression of nucleators is often sufficient to induce SGs in vitro.207,208 Thus, overexpression of nucleators in vivo has the potential to promote SG formation in cancer cells. Examples of nucleators are Ras-GTPase activating protein SH-3 domain binding protein 1 and 2 (G3BP),207,209 T cell intracellular antigen (TIA-1) and TIA-1–related protein (TIAR),210,211 polyadenylate-binding protein 1 (PABP1),208 and fragile X mental retardation protein (FMRP).212 Protein levels of SG nucleation factors are induced in several tumor entities.213–215 For example, French et al.213 analyzed 22 breast cancer samples, all of which showed elevated G3BP1. After the nucleation and aggregation phases, further proteins that have intrinsic mRNA binding capacity or that bind to SG proteins by piggy back recruitment, are assembled into SGs.216 Upon stress relief, SGs dissolve and SG proteins relocate to their previous compartments.197,208,217 SGs are thought of as sites of RNA storage and triage during stress.218 In addition, there is increasing evidence that SGs interfere with stress signaling pathways (reviewed by Kedersha et al.216). Proteins involved in apoptosis can be recruited to SGs, which thereby promote survival. For example, SG recruitment of signaling scaffold protein receptor of activated protein kinase C 1 (RACK1) prevents induction of apoptosis by the genotoxic stress-activated p38 and JNK–MAPK pathways204, and ubiquitin-specific protease 10 (USP10) has been reported to exert an antioxidant apoptosis-preventing activity that depends on recruitment of USP10 to SGs.219 Recruitment of TNF receptor-associated factor 2 (TRAF2) to SGs inhibits proinflammatory tumor necrosis factor α (TNFα)–NF-κB signaling.220
SG assembly in both yeast and human cells can inhibit TORC1/mTORC1 signaling (Fig. 4) by sequestering mTOR complex components or the mTORC1 upstream modulator dual specificity tyrosine-phosphorylation-regulated kinase 3 (DYRK3).131,208,217 In cancer cells, DYRK3 integrates mTORC1 activity with SG formation via a dual mechanism.217 During prolonged stress, DYRK3 is sequestered into SGs where it prevents SG dissolution and mTORC1 release. After stress release, DYRK3 phosphorylates and inhibits the mTORC1-inhibitor PRAS40,221–227 thus contributing to mTORC1 reactivation. Furthermore, the adaptor protein astrin disassembles mTORC1 by sequestering raptor into SGs.131 Through this recruitment SGs restrict mTORC1 assembly and prevent its hyperactivation and mTORC1-dependent oxidative stress-induced apoptosis. Thus, inhibition of astrin induces mTORC1-triggered apoptosis in cancer cells.131 Like other SG proteins, astrin is frequently overexpressed in tumors, and has been correlated with an unfavorable prognosis in human breast cancers and non-small cell lung (NSCL) cancers.228, 229 This suggests that high astrin levels render cancer cells apoptosis resistant by counteracting mTORC1 hyperactivation. Also in yeast, SG induction by heat shock or PABP1 overexpression leads to TOR inhibition by sequestration into SGs, and TORC1 reactivation after stress correlates with its release from SGs.208 Thus, SG formation has a conserved inhibitory effect on TORC1/mTORC1 in eukaryotic cells. However, mTORC1 activity is also needed for SG formation in mammalian cells;206 for example, formation of 5´cap–eIF4F complexes requires phosphorylation of 4EBP1 by mTORC1.230 Thus, SGs and mTORC1 are connected via a NFL in which mTORC1 positively regulates SGs, whereas SGs inhibit mTORC1 (Fig. 4).
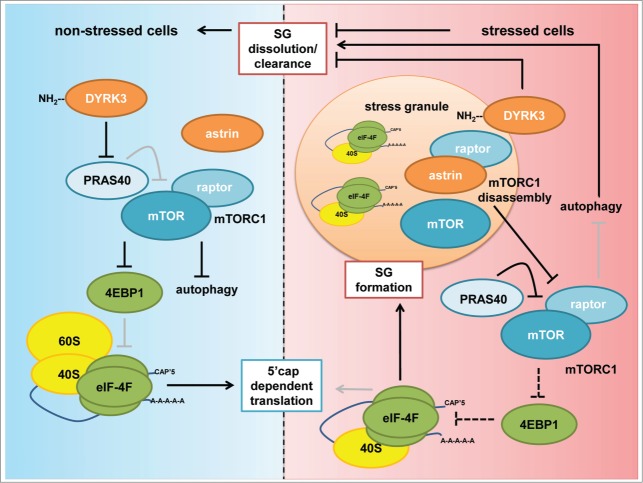
Figure 4.
Stress granules and mTORC1. Under non-stressed conditions DYRK3 phosphorylates and inactivates the mTORC1 inhibitor PRAS40. Active mTORC1 inhibits 4E‑BP1, allowing for eIF4F–5´cap–mRNA complex formation, ribosomal binding, and translation initiation. Stressed conditions induce translational arrest, polysome disassembly, and SG formation. mTORC1 is disassembled, and the mTORC1 components mTOR and raptor are recruited to SGs. Kinase-inactive DYRK3 localizes through its N-terminus to SGs, where it promotes SG stability and prevents mTOR release. Astrin binds to raptor and recruits it to SGs, thereby mediating SG-dependent mTORC1 disassembly. mTORC1 inactivation results in induction of autophagy, which is required for SG clearance after stress release and for SG formation. However, inhibition of 4E-BP1 by mTORC1 is required for SG formation, as 5´cap–eIF4F complexes and binding of the 40S ribosomal subunit are required for SG formation. Thus, SGs restrict mTORC1 activity, but some mTORC1 activity is needed for SG assembly (indicated by dashed arrows). Black arrows represent active connections, gray arrows represent inactive connections in stressed versus non-stressed cells.
mTORC1 and SGs have both been linked to the regulation of translation and autophagy and it is interesting to consider how they may interact to control protein synthesis and autophagy under stress. During stress, 5´cap-dependent translation is reduced, and this is linked to mTORC1 inhibition. For example, the SG components TIA-1 and TIAR inhibit translation of 5´TOP mRNAs by promoting their assembly into SGs when mTORC1 is inhibited.231 However, in a background of mTORC1 inhibition and reduced overall translation levels, stress response proteins still need to be expressed232 although active translation requires mTORC1 activity. Thus, there is a seemingly contradictory requirement for mTORC1 activation/inhibition during stress. SGs have emerged as excellent candidates for balancing mTORC1 activity and the dependent translational events. Both mTORC1 and SGs control translation of stress related factors,29,34,233–235 and SGs have been suggested as sites of stress-specific translation initiation.236 Translation under stress depends on upstream open reading frames (uORFs) and internal ribosomal entry sites (IRES).218,237–239 mTORC1 induces both IRES-mediated240,241 and uORF-dependent translation via eIF4GI,242 a member of the eIF4F complex. For example, the stress-related proteins heat shock factor protein 1 (HSF1), heterogeneous nuclear ribonucleoprotein A1 (hnRNP-A1), and 70 kDa heat shock protein (Hsp70) require mTORC1 for their expression under oxidative stress.131 hnRNP-A1 is required for IRES-mediated translation under stress in tumor cells,243,244 whereas HSF1 mediates transcriptional events under stress, including Hsp70 expression.233 Additionally, ATF4 protein expression under stress is regulated by mTORC1.131 The ATF4 mRNA contains 2 uORFs, leading to increased ATF4 translation in response to stress-related eIF2α phosphorylation.239 ATF4 induces autophagy under ER stress and hypoxia (see above). Of note, autophagy is required for SG clearance in yeast and mammalian cells,245,246 and inhibition of autophagy results in mistargeting of proteins to SGs.246 Thus, it seems that, while mTORC1 must be active to enable expression of stress factors, mTORC1 activity needs to be restricted to enable autophagy. mTORC1 and autophagy-mediated SG turnover may therefore represent a mechanism of feedback regulation that balances mTORC1 activity under stress.
Therapeutic Implications: mTORC1 in Stress as a Target in Cancer?
mTORC1 signaling is mostly perceived as a prosurvival and antiapoptotic process. However, there is ample evidence that dysregulated hyperactive signaling via mTORC1, for example in response to TSC1–TSC2 inactivation, is prone to elicit cell death. How do cancer cells survive the inactivation of major negative regulators (i.e., tumor suppressors) of mTORC1 signaling in conjunction with a hyperactive metabolism and high stress levels? Persistent stresses eventually trigger apoptosis in healthy cells. However, short-term stresses and their consequences need to be buffered to prevent the induction of cell death by transient imbalances in cellular signaling, metabolism, and redox homeostasis. Therefore, signaling, transcription, translation, and metabolic networks are stabilized by multiple feedback loops and buffer systems. SGs represent one such buffer system. It is likely that cancer cells hijack this system by overexpressing SG components. This may render the tumor cells resistant to hyperactive signaling induced by oncogenic mutations, hyperactive metabolism, and stresses, as well as therapeutic interventions such as chemotherapy (genotoxic stress) or irradiation. Signaling and metabolic networks that are hyperactive in cancer, such as mTORC1 signaling or glycolysis, often represent vital cellular functions that cannot be therapeutically targeted without major side effects on healthy tissues. SGs, in contrast, are likely to be more essential for cancer cells than for healthy tissues to overcome a stressed cellular environment. Thus, SG modulation represents a promising orthogonal approach to complement existing therapies involving targeted drugs or chemotherapeutics.
Acknowledgments
We thank Antje Thien for critical reading of this manuscript.
A patent entitled “Modulators of the interaction of astrin and raptor, and uses thereof in cancer therapy” has been filed, on which KT is a co-inventor: publication number WO2014108532 A1, priority date January 11, 2013.
Funding
KT and BMB are recipients of Rosalind Franklin Fellowships, University of Groningen, NL. This work was supported in part by the Royal Society, UK (SNG and KT, IE131392), the Excellence Initiative of the German Federal and State Governments (EXC 294 to KT, FRIAS LifeNet to KT, GSC-4, Spemann Graduate School to MTP), and the Top Institute Food and Nutrition, NL (Tifn, to KvE).
References
- 1. Yecies JL, Manning BD. mTOR links oncogenic signaling to tumor cell metabolism. J Mol Med 2011; 89:221-8; PMID:21301797; http://dx.doi.org/ 10.1007/s00109-011-0726-6 [DOI] [PubMed] [Google Scholar]
- 2. Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol 2014; 15:155-62; PMID:24556838; http://dx.doi.org/ 10.1038/nrm3757 [DOI] [PubMed] [Google Scholar]
- 3. Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002; 110:163-75; PMID:12150925; http://dx.doi.org/ 10.1016/S0092-8674(02)00808-5 [DOI] [PubMed] [Google Scholar]
- 4. Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 2002; 110:177-89; PMID:12150926; http://dx.doi.org/ 10.1016/S0092-8674(02)00833-4 [DOI] [PubMed] [Google Scholar]
- 5. Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, Trapani F, Terracciano L, Heim MH, Ruegg MA, Hall MN. Hepatic mTORC2 Activates Glycolysis and Lipogenesis through Akt, Glucokinase, and SREBP1c. Cell Metab 2012; 15:725-38; PMID:22521878; http://dx.doi.org/ 10.1016/j.cmet.2012.03.015 [DOI] [PubMed] [Google Scholar]
- 6. Oh WJ, Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle 2011; 10:2305-16; PMID:21670596; http://dx.doi.org/ 10.4161/cc.10.14.16586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 2012; 335:1638-43; PMID:22461615; http://dx.doi.org/ 10.1126/science.1215135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. MolCell 2006; 22:159-68 [DOI] [PubMed] [Google Scholar]
- 9. Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 2008; 10:935-45; PMID:18604198; http://dx.doi.org/ 10.1038/ncb1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008; 320:1496-501; PMID:18497260; http://dx.doi.org/ 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 2012; 150:1196-208; PMID:22980980; http://dx.doi.org/ 10.1016/j.cell.2012.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell 2013; 52:495-505; PMID:24095279; http://dx.doi.org/ 10.1016/j.molcel.2013.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duran RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, Hall MN. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell 2012; 47:349-58; PMID:22749528; http://dx.doi.org/ 10.1016/j.molcel.2012.05.043 [DOI] [PubMed] [Google Scholar]
- 14. Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell biol 2014; 24:400-6; PMID:24698685; http://dx.doi.org/ 10.1016/j.tcb.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. CurrBiol 2005; 15:702-13; http://dx.doi.org/ 10.1016/j.cub.2005.02.053 [DOI] [PubMed] [Google Scholar]
- 16. Demetriades C, Doumpas N, Teleman AA. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 2014; 156:786-99; PMID:24529380; http://dx.doi.org/ 10.1016/j.cell.2014.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 2003; 17:1829-34; PMID:12869586; http://dx.doi.org/ 10.1101/gad.1110003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 2014; 156:771-85; PMID:24529379; http://dx.doi.org/ 10.1016/j.cell.2013.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell 2012; 149:274-93; PMID:22500797; http://dx.doi.org/ 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hardie DG, Ashford ML. AMPK: regulating energy balance at the cellular and whole body levels. Physiology 2014; 29:99-107; PMID:24583766; http://dx.doi.org/ 10.1152/physiol.00050.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 2008; 30:214-26; PMID:18439900; http://dx.doi.org/ 10.1016/j.molcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003; 115:577-90; PMID:14651849; http://dx.doi.org/ 10.1016/S0092-8674(03)00929-2 [DOI] [PubMed] [Google Scholar]
- 23. Kim SG, Hoffman GR, Poulogiannis G, Buel GR, Jang YJ, Lee KW, Kim BY, Erikson RL, Cantley LC, Choo AY, et al. Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol Cell 2013; 49:172-85; PMID:23142078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schieke SM, Phillips D, McCoy JP, Jr., Aponte AM, Shen RF, Balaban RS, Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem 2006; 281:27643-52; PMID:16847060; http://dx.doi.org/ 10.1074/jbc.M603536200 [DOI] [PubMed] [Google Scholar]
- 25. Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 2007; 450:736-40; PMID:18046414; http://dx.doi.org/ 10.1038/nature06322 [DOI] [PubMed] [Google Scholar]
- 26. Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 2013; 18:698-711; PMID:24206664; http://dx.doi.org/ 10.1016/j.cmet.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 27. Buller CL, Loberg RD, Fan MH, Zhu Q, Park JL, Vesely E, Inoki K, Guan KL, Brosius FC, 3rd. A GSK-3/TSC2/mTOR pathway regulates glucose uptake and GLUT1 glucose transporter expression. Am J Physiol Cell Physiol 2008; 295:C836-43; PMID:18650261; http://dx.doi.org/ 10.1152/ajpcell.00554.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 2013; 153:840-54; PMID:23663782; http://dx.doi.org/ 10.1016/j.cell.2013.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012; 485:55-61; PMID:22367541; http://dx.doi.org/ 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 2009; 10:307-18; PMID:19339977; http://dx.doi.org/ 10.1038/nrm2672 [DOI] [PubMed] [Google Scholar]
- 31. Iadevaia V, Zhang Z, Jan E, Proud CG. mTOR signaling regulates the processing of pre-rRNA in human cells. Nucleic Acids Res 2012; 40:2527-39; PMID:22121221; http://dx.doi.org/ 10.1093/nar/gkr1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iadevaia V, Huo Y, Zhang Z, Foster LJ, Proud CG. Roles of the mammalian target of rapamycin, mTOR, in controlling ribosome biogenesis and protein synthesis. Biochem Soc Trans 2012; 40:168-72; PMID:22260684; http://dx.doi.org/ 10.1042/BST20110682 [DOI] [PubMed] [Google Scholar]
- 33. Levy S, Avni D, Hariharan N, Perry RP, Meyuhas O. Oligopyrimidine tract at the 5' end of mammalian ribosomal protein mRNAs is required for their translational control. Proc Natl Acad Sci U S A 1991; 88:3319-23; PMID:2014251; http://dx.doi.org/ 10.1073/pnas.88.8.3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012; 485:109-13; PMID:22552098; http://dx.doi.org/ 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5'TOP mRNA translation. Genes Dev 2014; 28:357-71; PMID:24532714; http://dx.doi.org/ 10.1101/gad.231407.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patursky-Polischuk I, Stolovich-Rain M, Hausner-Hanochi M, Kasir J, Cybulski N, Avruch J, Ruegg MA, Hall MN, Meyuhas O. The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol Cell Biol 2009; 29:640-9; PMID:19047368; http://dx.doi.org/ 10.1128/MCB.00980-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miloslavski R, Cohen E, Avraham A, Iluz Y, Hayouka Z, Kasir J, Mudhasani R, Jones SN, Cybulski N, Ruegg MA, et al. Oxygen sufficiency controls TOP mRNA translation via the TSC-Rheb-mTOR pathway in a 4E-BP-independent manner. J Mol Cell Biol 2014; 6:255-66; PMID:24627160; http://dx.doi.org/ 10.1093/jmcb/mju008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 1992; 69:1227-36; PMID:1377606; http://dx.doi.org/ 10.1016/0092-8674(92)90643-Q [DOI] [PubMed] [Google Scholar]
- 39. Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 2004; 23:1761-9; PMID:15071500; http://dx.doi.org/ 10.1038/sj.emboj.7600193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kroczynska B, Kaur S, Katsoulidis E, Majchrzak-Kita B, Sassano A, Kozma SC, Fish EN, Platanias LC. Interferon-dependent engagement of eukaryotic initiation factor 4B via S6 kinase (S6K)- and ribosomal protein S6K-mediated signals. Mol Cell Biol 2009; 29:2865-75; PMID:19289497; http://dx.doi.org/ 10.1128/MCB.01537-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D, Avruch J, Meyuhas O. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. MolCell Biol 2001; 21:8671-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chauvin C, Koka V, Nouschi A, Mieulet V, Hoareau-Aveilla C, Dreazen A, Cagnard N, Carpentier W, Kiss T, Meyuhas O, et al. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 2014; 33:474-83; PMID:23318442; http://dx.doi.org/ 10.1038/onc.2012.606 [DOI] [PubMed] [Google Scholar]
- 43. Porstmann T, Griffiths B, Chung YL, Delpuech O, Griffiths JR, Downward J, Schulze A. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene 2005; 24:6465-81; PMID:16007182 [DOI] [PubMed] [Google Scholar]
- 44. Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab 2008; 8:224-36; PMID:18762023; http://dx.doi.org/ 10.1016/j.cmet.2008.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 2010; 39:171-83; PMID:20670887; http://dx.doi.org/ 10.1016/j.molcel.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yecies JL, Zhang HH, Menon S, Liu S, Yecies D, Lipovsky AI, Gorgun C, Kwiatkowski DJ, Hotamisligil GS, Lee CH, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab 2011; 14:21-32; PMID:21723501; http://dx.doi.org/ 10.1016/j.cmet.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab 2012; 23:65-72; PMID:22154484; http://dx.doi.org/ 10.1016/j.tem.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 2013; 339:1323-8; PMID:23429703; http://dx.doi.org/ 10.1126/science.1228792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 2013; 339:1320-3; PMID:23429704; http://dx.doi.org/ 10.1126/science.1228771 [DOI] [PubMed] [Google Scholar]
- 50. Myers MG, Jr., Grammer TC, Wang LM, Sun XJ, Pierce JH, Blenis J, White MF. Insulin receptor substrate-1 mediates phosphatidylinositol 3'-kinase and p70S6k signaling during insulin, insulin-like growth factor-1, and interleukin-4 stimulation. J Biol Chem 1994; 269:28783-9; PMID:7961833 [PubMed] [Google Scholar]
- 51. Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. CurrBiol 2004; 14:1650-6; http://dx.doi.org/ 10.1016/j.cub.2004.08.026 [DOI] [PubMed] [Google Scholar]
- 52. Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol 2004; 166:213-23; PMID:15249583; http://dx.doi.org/ 10.1083/jcb.200403069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 2011; 332:1317-22; PMID:21659604; http://dx.doi.org/ 10.1126/science.1199498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 2011; 332:1322-6; PMID:21659605; http://dx.doi.org/ 10.1126/science.1199484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999; 96:857-68; PMID:10102273; http://dx.doi.org/ 10.1016/S0092-8674(00)80595-4 [DOI] [PubMed] [Google Scholar]
- 56. Mounir Z, Krishnamoorthy JL, Wang S, Papadopoulou B, Campbell S, Muller WJ, Hatzoglou M, Koromilas AE. Akt determines cell fate through inhibition of the PERK-eIF2alpha phosphorylation pathway. Sci signal 2011; 4:ra62; PMID:21954288; http://dx.doi.org/ 10.1126/scisignal.2001630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Appenzeller-Herzog C, Hall MN. Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Biol 2012; 22:274-82; PMID:22444729; http://dx.doi.org/ 10.1016/j.tcb.2012.02.006 [DOI] [PubMed] [Google Scholar]
- 58. Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer 2011; 11:393-410; PMID:21606941; http://dx.doi.org/ 10.1038/nrc3064 [DOI] [PubMed] [Google Scholar]
- 59. Cornu M, Albert V, Hall MN. mTOR in aging, metabolism, and cancer. Curr Opin genet Dev 2013; 23:53-62; PMID:23317514; http://dx.doi.org/ 10.1016/j.gde.2012.12.005 [DOI] [PubMed] [Google Scholar]
- 60. Modica-Napolitano JS, Singh KK. Mitochondrial dysfunction in cancer. Mitochondrion 2004; 4:755-62; PMID:16120430; http://dx.doi.org/ 10.1016/j.mito.2004.07.027 [DOI] [PubMed] [Google Scholar]
- 61. Kumimoto H, Yamane Y, Nishimoto Y, Fukami H, Shinoda M, Hatooka S, Ishizaki K. Frequent somatic mutations of mitochondrial DNA in esophageal squamous cell carcinoma. Int J Cancer J Int du Cancer 2004; 108:228-31; PMID:14639607; http://dx.doi.org/ 10.1002/ijc.11564 [DOI] [PubMed] [Google Scholar]
- 62. Liang J, Mills GB. AMPK: a contextual oncogene or tumor suppressor? Cancer Res 2013; 73:2929-35; PMID:23644529; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Clarke HJ, Chambers JE, Liniker E, Marciniak SJ. Endoplasmic reticulum stress in malignancy. Cancer Cell 2014; 25:563-73; PMID:24823636; http://dx.doi.org/ 10.1016/j.ccr.2014.03.015 [DOI] [PubMed] [Google Scholar]
- 64. Fels DR, Koumenis C. The PERK/eIF2alpha/ATF4 module of the UPR in hypoxia resistance and tumor growth. Cancer Biol Ther 2006; 5:723-8; PMID:16861899; http://dx.doi.org/ 10.4161/cbt.5.7.2967 [DOI] [PubMed] [Google Scholar]
- 65. Brahimi-Horn MC, Chiche J, Pouyssegur J. Hypoxia and cancer. J Mol Med 2007; 85:1301-7; PMID:18026916; http://dx.doi.org/ 10.1007/s00109-007-0281-3 [DOI] [PubMed] [Google Scholar]
- 66. Braakman I, Hebert DN. Protein folding in the endoplasmic reticulum. Cold Spring Harbor perspectives Biol 2013; 5:a013201; PMID:23637286; http://dx.doi.org/ 10.1101/cshperspect.a013201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. van der Sanden MH, Houweling M, van Golde LM, Vaandrager AB. Inhibition of phosphatidylcholine synthesis induces expression of the endoplasmic reticulum stress and apoptosis-related protein CCAAT/enhancer-binding protein-homologous protein (CHOP/GADD153). Biochem J 2003; 369:643-50; PMID:12370080; http://dx.doi.org/ 10.1042/BJ20020285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Little JL, Wheeler FB, Fels DR, Koumenis C, Kridel SJ. Inhibition of fatty acid synthase induces endoplasmic reticulum stress in tumor cells. Cancer Res 2007; 67:1262-9; PMID:17283163; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-1794 [DOI] [PubMed] [Google Scholar]
- 69. Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol 2009; 187:525-36; PMID:19948500; http://dx.doi.org/ 10.1083/jcb.200907074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pizer ES, Jackisch C, Wood FD, Pasternack GR, Davidson NE, Kuhajda FP. Inhibition of fatty acid synthesis induces programmed cell death in human breast cancer cells. Cancer Res 1996; 56:2745-7; PMID:8665507 [PubMed] [Google Scholar]
- 71. Mashima T, Seimiya H, Tsuruo T. De novo fatty-acid synthesis and related pathways as molecular targets for cancer therapy. British J Cancer 2009; 100:1369-72; PMID:19352381; http://dx.doi.org/ 10.1038/sj.bjc.6605007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Warburg O. On the origin of cancer cells. Science 1956; 123:309-14; PMID:13298683; http://dx.doi.org/ 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- 73. Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov 2012; 2:881-98; PMID:23009760; http://dx.doi.org/ 10.1158/2159-8290.CD-12-0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 2011; 11:325-37; PMID:21508971; http://dx.doi.org/ 10.1038/nrc3038 [DOI] [PubMed] [Google Scholar]
- 75. Szablewski L. Expression of glucose transporters in cancers. Biochim Biophys Acta 2013; 1835:164-9; PMID:23266512 [DOI] [PubMed] [Google Scholar]
- 76. Young CD, Lewis AS, Rudolph MC, Ruehle MD, Jackman MR, Yun UJ, Ilkun O, Pereira R, Abel ED, Anderson SM. Modulation of glucose transporter 1 (GLUT1) expression levels alters mouse mammary tumor cell growth in vitro and in vivo. PloS One 2011; 6:e23205; PMID:21826239; http://dx.doi.org/ 10.1371/journal.pone.0023205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ozcan U, Ozcan L, Yilmaz E, Duvel K, Sahin M, Manning BD, Hotamisligil GS. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Mol Cell 2008; 29:541-51; PMID:18342602; http://dx.doi.org/ 10.1016/j.molcel.2007.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 2012; 13:89-102; PMID:22251901 [DOI] [PubMed] [Google Scholar]
- 79. Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem 1998; 273:33741-9; PMID:9837962; http://dx.doi.org/ 10.1074/jbc.273.50.33741 [DOI] [PubMed] [Google Scholar]
- 80. Yamamoto K, Sato T, Matsui T, Sato M, Okada T, Yoshida H, Harada A, Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell 2007; 13:365-76; PMID:17765680; http://dx.doi.org/ 10.1016/j.devcel.2007.07.018 [DOI] [PubMed] [Google Scholar]
- 81. Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999; 397:271-4; PMID:9930704; http://dx.doi.org/ 10.1038/16729 [DOI] [PubMed] [Google Scholar]
- 82. Prostko CR, Brostrom MA, Brostrom CO. Reversible phosphorylation of eukaryotic initiation factor 2 alpha in response to endoplasmic reticular signaling. Mol Cell Biochem 1993; 127-128: 255-65; PMID:7935356 [DOI] [PubMed] [Google Scholar]
- 83. Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 2006; 26:9220-31; PMID:17030611; http://dx.doi.org/ 10.1128/MCB.01453-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol 2007; 171:513-24; PMID:17620365; http://dx.doi.org/ 10.2353/ajpath.2007.070188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nijholt DA, de Graaf TR, van Haastert ES, Oliveira AO, Berkers CR, Zwart R, Ovaa H, Baas F, Hoozemans JJ, Scheper W. Endoplasmic reticulum stress activates autophagy but not the proteasome in neuronal cells: implications for Alzheimer disease. Cell Death Differ 2011; 18:1071-81; PMID:21252911; http://dx.doi.org/ 10.1038/cdd.2010.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Menendez-Benito V, Verhoef LG, Masucci MG, Dantuma NP. Endoplasmic reticulum stress compromises the ubiquitin-proteasome system. Hum Mol Genet 2005; 14:2787-99; PMID:16103128; http://dx.doi.org/ 10.1093/hmg/ddi312 [DOI] [PubMed] [Google Scholar]
- 87. Kang YJ, Lu MK, Guan KL. The TSC1 and TSC2 tumor suppressors are required for proper ER stress response and protect cells from ER stress-induced apoptosis. Cell Death Differ 2011; 18:133-44; PMID:20616807; http://dx.doi.org/ 10.1038/cdd.2010.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Di Nardo A, Kramvis I, Cho N, Sadowski A, Meikle L, Kwiatkowski DJ, Sahin M. Tuberous sclerosis complex activity is required to control neuronal stress responses in an mTOR-dependent manner. J Neurosci 2009; 29:5926-37; PMID:19420259; http://dx.doi.org/ 10.1523/JNEUROSCI.0778-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J 2005; 24:1243-55; PMID:15775988; http://dx.doi.org/ 10.1038/sj.emboj.7600596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chen CH, Shaikenov T, Peterson TR, Aimbetov R, Bissenbaev AK, Lee SW, Wu J, Lin HK, Sarbassov dos D. ER stress inhibits mTORC2 and Akt signaling through GSK-3beta-mediated phosphorylation of rictor. Sci Signal 2011; 4:ra10; PMID:21343617; http://dx.doi.org/ 10.1126/scisignal.2001731 [DOI] [PubMed] [Google Scholar]
- 91. Kato H, Nakajima S, Saito Y, Takahashi S, Katoh R, Kitamura M. mTORC1 serves ER stress-triggered apoptosis via selective activation of the IRE1-JNK pathway. Cell Death Differ 2012; 19:310-20; PMID:21779001; http://dx.doi.org/ 10.1038/cdd.2011.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mole Cell 2010; 40:294-309; PMID:20965423; http://dx.doi.org/ 10.1016/j.molcel.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol 1996; 16:4604-13; PMID:8756616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Choi KS, Bae MK, Jeong JW, Moon HE, Kim KW. Hypoxia-induced angiogenesis during carcinogenesis. J Biochem Mol Biol 2003; 36:120-7; PMID:12542982; http://dx.doi.org/ 10.5483/BMBRep.2003.36.1.120 [DOI] [PubMed] [Google Scholar]
- 95. Dodd KM, Yang J, Shen MH, Sampson JR, Tee AR. mTORC1 drives HIF-1alpha and VEGF-A signalling via multiple mechanisms involving 4E-BP1, S6K1 and STAT3. Oncogene 2014; http://dx.doi.org/ 10.1038/onc.2014.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sakamoto T, Weng JS, Hara T, Yoshino S, Kozuka-Hata H, Oyama M, Seiki M. Hypoxia-inducible factor 1 regulation through cross talk between mTOR and MT1-MMP. Mol Cell Biol 2014; 34:30-42; PMID:24164895; http://dx.doi.org/ 10.1128/MCB.01169-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Johnson AB, Denko N, Barton MC. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutation Res 2008; 640:174-9; PMID:18294659; http://dx.doi.org/ 10.1016/j.mrfmmm.2008.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gowans GJ, Hardie DG. AMPK: a cellular energy sensor primarily regulated by AMP. Biochem Soc Trans 2014; 42:71-5; PMID:24450630; http://dx.doi.org/ 10.1042/BST20130244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 2012; 13:251-62; PMID:22436748; http://dx.doi.org/ 10.1038/nrm3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1alpha. Mol Cell 2010; 40:509-20; PMID:21095582; http://dx.doi.org/ 10.1016/j.molcel.2010.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG, Jr. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 2004; 18:2893-904; PMID:15545625; http://dx.doi.org/ 10.1101/gad.1256804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol 2005; 25:5834-45; PMID:15988001; http://dx.doi.org/ 10.1128/MCB.25.14.5834-5845.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev 2008; 22:239-51; PMID:18198340; http://dx.doi.org/ 10.1101/gad.1617608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tan CY, Hagen T. mTORC1 dependent regulation of REDD1 protein stability. PloS One 2013; 8:e63970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kimball SR, Do AN, Kutzler L, Cavener DR, Jefferson LS. Rapid Turnover of the mTOR Complex 1 (mTORC1) Repressor REDD1 and Activation of mTORC1 Signaling following Inhibition of Protein Synthesis. JBiolChem 2008; 2-8; 283:3465-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Toschi A, Lee E, Gadir N, Ohh M, Foster DA. Differential dependence of hypoxia-inducible factors 1 alpha and 2 alpha on mTORC1 and mTORC2. J Biol Chem 2008; 283:34495-9; PMID:18945681; http://dx.doi.org/ 10.1074/jbc.C800170200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Tagliavacca L, Caretti A, Bianciardi P, Samaja M. In vivo up-regulation of the unfolded protein response after hypoxia. Biochim Biophys Acta 2012; 1820:900-6; PMID:22450154; http://dx.doi.org/ 10.1016/j.bbagen.2012.02.016 [DOI] [PubMed] [Google Scholar]
- 108. Sosa V, Moline T, Somoza R, Paciucci R, Kondoh H, ME LL. Oxidative stress and cancer: an overview. Ageing Res Rev 2013; 12:376-90; PMID:23123177; http://dx.doi.org/ 10.1016/j.arr.2012.10.004 [DOI] [PubMed] [Google Scholar]
- 109. Desler C, Marcker ML, Singh KK, Rasmussen LJ. The importance of mitochondrial DNA in aging and cancer. J Aging Res 2011; 2011:407536; PMID:21584235; http://dx.doi.org/ 10.4061/2011/407536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Woo DK, Green PD, Santos JH, D'Souza AD, Walther Z, Martin WD, Christian BE, Chandel NS, Shadel GS. Mitochondrial genome instability and ROS enhance intestinal tumorigenesis in APC(Min/+) mice. Am J Pathol 2012; 180:24-31; PMID:22056359; http://dx.doi.org/ 10.1016/j.ajpath.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A 1997; 94:514-9; PMID:9012815; http://dx.doi.org/ 10.1073/pnas.94.2.514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Jr., Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 2010; 464:610-4; PMID:20200521; http://dx.doi.org/ 10.1038/nature08802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Larman TC, DePalma SR, Hadjipanayis AG, Protopopov A, Zhang J, Gabriel SB, Chin L, Seidman CE, Kucherlapati R, Seidman JG. Spectrum of somatic mitochondrial mutations in five cancers. Proc Natl Acad Sci U S A 2012; 109:14087-91; PMID:22891333; http://dx.doi.org/ 10.1073/pnas.1211502109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Carew JS, Zhou Y, Albitar M, Carew JD, Keating MJ, Huang P. Mitochondrial DNA mutations in primary leukemia cells after chemotherapy: clinical significance and therapeutic implications. Leukemia 2003; 17:1437-47; PMID:12886229; http://dx.doi.org/ 10.1038/sj.leu.2403043 [DOI] [PubMed] [Google Scholar]
- 115. Chiara F, Gambalunga A, Sciacovelli M, Nicolli A, Ronconi L, Fregona D, Bernardi P, Rasola A, Trevisan A. Chemotherapeutic induction of mitochondrial oxidative stress activates GSK-3alpha/β and Bax, leading to permeability transition pore opening and tumor cell death. Cell Death Dis 2012; 3:e444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 2009; 8:579-91; PMID:19478820; http://dx.doi.org/ 10.1038/nrd2803 [DOI] [PubMed] [Google Scholar]
- 117. Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell 2002; 9:1031-44; PMID:12049739; http://dx.doi.org/ 10.1016/S1097-2765(02)00520-8 [DOI] [PubMed] [Google Scholar]
- 118. Weyemi U, Lagente-Chevallier O, Boufraqech M, Prenois F, Courtin F, Caillou B, Talbot M, Dardalhon M, Al Ghuzlan A, Bidart JM, et al. ROS-generating NADPH oxidase NOX4 is a critical mediator in oncogenic H-Ras-induced DNA damage and subsequent senescence. Oncogene 2012; 31:1117-29; PMID:21841825; http://dx.doi.org/ 10.1038/onc.2011.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kodama R, Kato M, Furuta S, Ueno S, Zhang Y, Matsuno K, Yabe-Nishimura C, Tanaka E, Kamata T. ROS-generating oxidases Nox1 and Nox4 contribute to oncogenic Ras-induced premature senescence. Genes Cell 2013; 18:32-41; http://dx.doi.org/ 10.1111/gtc.12015 [DOI] [PubMed] [Google Scholar]
- 120. Goo CK, Lim HY, Ho QS, Too HP, Clement MV, Wong KP. PTEN/Akt signaling controls mitochondrial respiratory capacity through 4E-BP1. PloS One 2012; 7:e45806; PMID:23049865; http://dx.doi.org/ 10.1371/journal.pone.0045806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Irani K, Xia Y, Zweier JL, Sollott SJ, Der CJ, Fearon ER, Sundaresan M, Finkel T, Goldschmidt-Clermont PJ. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science 1997; 275:1649-52; PMID:9054359; http://dx.doi.org/ 10.1126/science.275.5306.1649 [DOI] [PubMed] [Google Scholar]
- 122. Kopnin PB, Agapova LS, Kopnin BP, Chumakov PM. Repression of sestrin family genes contributes to oncogenic Ras-induced reactive oxygen species up-regulation and genetic instability. Cancer Res 2007; 67:4671-8; PMID:17510393; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Groitl B, Jakob U. Thiol-based redox switches. Biochim Biophys Acta 2014; 1844:1335-43; PMID:24657586; http://dx.doi.org/ 10.1016/j.bbapap.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Moon JC, Hah YS, Kim WY, Jung BG, Jang HH, Lee JR, Kim SY, Lee YM, Jeon MG, Kim CW, et al. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J Biol Chem 2005; 280:28775-84; PMID:15941719; http://dx.doi.org/ 10.1074/jbc.M505362200 [DOI] [PubMed] [Google Scholar]
- 125. Watson J. Oxidants, antioxidants and the current incurability of metastatic cancers. Open Biol 2013; 3:120144; PMID:23303309; http://dx.doi.org/ 10.1098/rsob.120144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011; 475:106-9; PMID:21734707; http://dx.doi.org/ 10.1038/nature10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Brenneisen P, Wenk J, Wlaschek M, Krieg T, Scharffetter-Kochanek K. Activation of p70 ribosomal protein S6 kinase is an essential step in the DNA damage-dependent signaling pathway responsible for the ultraviolet B-mediated increase in interstitial collagenase (MMP-1) and stromelysin-1 (MMP-3) protein levels in human dermal fibroblasts. J Biol Chem 2000; 275:4336-44; PMID:10660603; http://dx.doi.org/ 10.1074/jbc.275.6.4336 [DOI] [PubMed] [Google Scholar]
- 128. Parrott LA, Templeton DJ. Osmotic stress inhibits p70/85 S6 kinase through activation of a protein phosphatase. J Biol Chem 1999; 274:24731-6; PMID:10455142; http://dx.doi.org/ 10.1074/jbc.274.35.24731 [DOI] [PubMed] [Google Scholar]
- 129. Huang C, Li J, Ke Q, Leonard SS, Jiang BH, Zhong XS, Costa M, Castranova V, Shi X. Ultraviolet-induced phosphorylation of p70(S6K) at Thr(389) and Thr(421)/Ser(424) involves hydrogen peroxide and mammalian target of rapamycin but not Akt and atypical protein kinase C. Cancer Res 2002; 62:5689-97; PMID:12384526 [PubMed] [Google Scholar]
- 130. Wang X, Proud CG. p70 S6 kinase is activated by sodium arsenite in adult rat cardiomyocytes: roles for phosphatidylinositol 3-kinase and p38 MAP kinase. Biochem Biophys Res Commun 1997; 238:207-12; PMID:9299480; http://dx.doi.org/ 10.1006/bbrc.1997.7273 [DOI] [PubMed] [Google Scholar]
- 131. Thedieck K, Holzwarth B, Prentzell MT, Boehlke C, Klasener K, Ruf S, Sonntag AG, Maerz L, Grellscheid SN, Kremmer E, et al. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell 2013; 154:859-74; PMID:23953116; http://dx.doi.org/ 10.1016/j.cell.2013.07.031 [DOI] [PubMed] [Google Scholar]
- 132. Zhang J, Kim J, Alexander A, Cai S, Tripathi DN, Dere R, Tee AR, Tait-Mulder J, Di Nardo A, Han JM, et al. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nature cell biology 2013; 15:1186-96; PMID:23955302; http://dx.doi.org/ 10.1038/ncb2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Bae GU, Seo DW, Kwon HK, Lee HY, Hong S, Lee ZW, Ha KS, Lee HW, Han JW. Hydrogen peroxide activates p70(S6k) signaling pathway. J Biol Chem 1999; 274:32596-602; PMID:10551813; http://dx.doi.org/ 10.1074/jbc.274.46.32596 [DOI] [PubMed] [Google Scholar]
- 134. Zheng M, Wang YH, Wu XN, Wu SQ, Lu BJ, Dong MQ, Zhang H, Sun P, Lin SC, Guan KL, et al. Inactivation of Rheb by PRAK-mediated phosphorylation is essential for energy-depletion-induced suppression of mTORC1. Nat Cell Biol 2011; 13:263-72; PMID:21336308; http://dx.doi.org/ 10.1038/ncb2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Leslie NR, Bennett D, Lindsay YE, Stewart H, Gray A, Downes CP. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J 2003; 22:5501-10; PMID:14532122; http://dx.doi.org/ 10.1093/emboj/cdg513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Chetram MA, Don-Salu-Hewage AS, Hinton CV. ROS enhances CXCR4-mediated functions through inactivation of PTEN in prostate cancer cells. Biochem Biophys Res Commun 2011; 410:195-200; PMID:21627959; http://dx.doi.org/ 10.1016/j.bbrc.2011.05.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Denu JM, Tanner KG. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry 1998; 37:5633-42; PMID:9548949; http://dx.doi.org/ 10.1021/bi973035t [DOI] [PubMed] [Google Scholar]
- 138. Yoshida S, Hong S, Suzuki T, Nada S, Mannan AM, Wang J, Okada M, Guan KL, Inoki K. Redox regulates mammalian target of rapamycin complex 1 (mTORC1) activity by modulating the TSC1/TSC2-Rheb GTPase pathway. J Biol Chem 2011; 286:32651-60; PMID:21784859; http://dx.doi.org/ 10.1074/jbc.M111.238014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science 2010; 330:517-21; PMID:20966255; http://dx.doi.org/ 10.1126/science.1192912 [DOI] [PubMed] [Google Scholar]
- 140. Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, Inoki K, Guan KL, Shen J, Person MD, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci U S A 2010; 107:4153-8; PMID:20160076; http://dx.doi.org/ 10.1073/pnas.0913860107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Wang RH, Kim HS, Xiao C, Xu X, Gavrilova O, Deng CX. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. J Clin Invest 2011; 121:4477-90; PMID:21965330; http://dx.doi.org/ 10.1172/JCI46243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Muders MH, Zhang H, Wang E, Tindall DJ, Datta K. Vascular endothelial growth factor-C protects prostate cancer cells from oxidative stress by the activation of mammalian target of rapamycin complex-2 and AKT-1. Cancer Res 2009; 69:6042-8; PMID:19638584; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-0552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Li W, Petrimpol M, Molle KD, Hall MN, Battegay EJ, Humar R. Hypoxia-induced endothelial proliferation requires both mTORC1 and mTORC2. Circulation Res 2007; 100:79-87; PMID:17110594; http://dx.doi.org/ 10.1161/01.RES.0000253094.03023.3f [DOI] [PubMed] [Google Scholar]
- 144. Tu BP, Weissman JS. The FAD- and O(2)-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol Cell 2002; 10:983-94; PMID:12453408; http://dx.doi.org/ 10.1016/S1097-2765(02)00696-2 [DOI] [PubMed] [Google Scholar]
- 145. Koritzinsky M, Levitin F, van den Beucken T, Rumantir RA, Harding NJ, Chu KC, Boutros PC, Braakman I, Wouters BG. Two phases of disulfide bond formation have differing requirements for oxygen. J Cell Biol 2013; 203:615-27; PMID:24247433; http://dx.doi.org/ 10.1083/jcb.201307185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest 2010; 120:127-41; PMID:20038797; http://dx.doi.org/ 10.1172/JCI40027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Rouschop KM, Dubois LJ, Keulers TG, van den Beucken T, Lambin P, Bussink J, van der Kogel AJ, Koritzinsky M, Wouters BG. PERK/eIF2alpha signaling protects therapy resistant hypoxic cells through induction of glutathione synthesis and protection against ROS. Proc Natl Acad Sci U S A 2013; 110:4622-7; PMID:23471998; http://dx.doi.org/ 10.1073/pnas.1210633110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 2004; 18:3066-77; PMID:15601821; http://dx.doi.org/ 10.1101/gad.1250704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Malhotra JD, Kaufman RJ. Endoplasmic reticulum stress and oxidative stress: a vicious cycle or a double-edged sword? Antioxid Redox Signal 2007; 9:2277-93; PMID:17979528; http://dx.doi.org/ 10.1089/ars.2007.1782 [DOI] [PubMed] [Google Scholar]
- 150. Blackburn RV, Spitz DR, Liu X, Galoforo SS, Sim JE, Ridnour LA, Chen JC, Davis BH, Corry PM, Lee YJ. Metabolic oxidative stress activates signal transduction and gene expression during glucose deprivation in human tumor cells. Free Radical Biol Med 1999; 26:419-30; PMID:9895234; http://dx.doi.org/ 10.1016/S0891-5849(98)00217-2 [DOI] [PubMed] [Google Scholar]
- 151. Spitz DR, Sim JE, Ridnour LA, Galoforo SS, Lee YJ. Glucose deprivation-induced oxidative stress in human tumor cells. A fundamental defect in metabolism? Ann New York Acad Sci 2000; 899:349-62; PMID:10863552; http://dx.doi.org/ 10.1111/j.1749-6632.2000.tb06199.x [DOI] [PubMed] [Google Scholar]
- 152. Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, Schumacker PT. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J Biol Chem 2000; 275:25130-8; PMID:10833514; http://dx.doi.org/ 10.1074/jbc.M001914200 [DOI] [PubMed] [Google Scholar]
- 153. Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci U S A 1998; 95:11715-20; PMID:9751731; http://dx.doi.org/ 10.1073/pnas.95.20.11715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Lu M, Lawrence DA, Marsters S, Acosta-Alvear D, Kimmig P, Mendez AS, Paton AW, Paton JC, Walter P, Ashkenazi A. Cell death. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science 2014; 345:98-101; PMID:24994655; http://dx.doi.org/ 10.1126/science.1254312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Hiramatsu N, Messah C, Han J, LaVail MM, Kaufman RJ, Lin JH. Translational and posttranslational regulation of XIAP by eIF2alpha and ATF4 promotes ER stress-induced cell death during the unfolded protein response. Mol Biol Cell 2014; 25:1411-20; PMID:24623724; http://dx.doi.org/ 10.1091/mbc.E13-11-0664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Win S, Than TA, Fernandez-Checa JC, Kaplowitz N. JNK interaction with Sab mediates ER stress induced inhibition of mitochondrial respiration and cell death. Cell Death Dis 2014; 5:e989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 1998; 394:485-90; PMID:9697772; http://dx.doi.org/ 10.1038/28867 [DOI] [PubMed] [Google Scholar]
- 158. Kim JY, Ahn HJ, Ryu JH, Suk K, Park JH. BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia-inducible factor 1alpha. J Exp Med 2004; 199:113-24; PMID:14699081; http://dx.doi.org/ 10.1084/jem.20030613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Li G, Scull C, Ozcan L, Tabas I. NADPH oxidase links endoplasmic reticulum stress, oxidative stress, and PKR activation to induce apoptosis. J Cell Biol 2010; 191:1113-25; PMID:21135141; http://dx.doi.org/ 10.1083/jcb.201006121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Singhapol C, Pal D, Czapiewski R, Porika M, Nelson G, Saretzki GC. Mitochondrial telomerase protects cancer cells from nuclear DNA damage and apoptosis. PloS One 2013; 8:e52989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Delbridge AR, Valente LJ, Strasser A. The role of the apoptotic machinery in tumor suppression. Cold Spring Harbor Perspect Biol 2012; 4; PMID:23125015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell 2008; 134:703-7; PMID:18775299; http://dx.doi.org/ 10.1016/j.cell.2008.08.021 [DOI] [PubMed] [Google Scholar]
- 163. Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev 2008; 18:54-61; PMID:18387799; http://dx.doi.org/ 10.1016/j.gde.2008.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Kohn AD, Summers SA, Birnbaum MJ, Roth RA. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. J Biol Chem 1996; 271:31372-8; PMID:8940145; http://dx.doi.org/ 10.1074/jbc.271.49.31372 [DOI] [PubMed] [Google Scholar]
- 165. Garrido P, Moran J, Alonso A, Gonzalez S, Gonzalez C. 17beta-estradiol activates glucose uptake via GLUT4 translocation and PI3K/Akt signaling pathway in MCF-7 cells. Endocrinology 2013; 154:1979-89; PMID:23546602; http://dx.doi.org/ 10.1210/en.2012-1558 [DOI] [PubMed] [Google Scholar]
- 166. Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong Z, Dupuy F, Chambers C, Fuerth BJ, Viollet B, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell Metab 2013; 17:113-24; PMID:23274086; http://dx.doi.org/ 10.1016/j.cmet.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev 2010; 20:51-6; PMID:19942427; http://dx.doi.org/ 10.1016/j.gde.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther 2011; 10:1533-41; PMID:21878654; http://dx.doi.org/ 10.1158/1535-7163.MCT-11-0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol 2013; 15:713-20; PMID:23817233; http://dx.doi.org/ 10.1038/ncb2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 2014; 15:81-94; PMID:24401948; http://dx.doi.org/ 10.1038/nrm3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 2009; 5:649-62; PMID:19287211; http://dx.doi.org/ 10.4161/auto.5.5.8249 [DOI] [PubMed] [Google Scholar]
- 172. Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 2009; 284:12297-305; PMID:19258318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13:132-41; PMID:21258367; http://dx.doi.org/ 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009; 20:1992-2003; PMID:19225151; http://dx.doi.org/ 10.1091/mbc.E08-12-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 2009; 20:1981-91; PMID:19211835; http://dx.doi.org/ 10.1091/mbc.E08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Lee JW, Park S, Takahashi Y, Wang HG. The association of AMPK with ULK1 regulates autophagy. PloS One 2010; 5:e15394; PMID:21072212; http://dx.doi.org/ 10.1371/journal.pone.0015394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122:927-39; PMID:16179260; http://dx.doi.org/ 10.1016/j.cell.2005.07.002 [DOI] [PubMed] [Google Scholar]
- 178. Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 2008; 30:678-88; PMID:18570871; http://dx.doi.org/ 10.1016/j.molcel.2008.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J Biol Chem 2009; 284:2719-28; PMID:19029119; http://dx.doi.org/ 10.1074/jbc.M805920200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ 2007; 14:230-9; PMID:16794605; http://dx.doi.org/ 10.1038/sj.cdd.4401984 [DOI] [PubMed] [Google Scholar]
- 181. Qin L, Wang Z, Tao L, Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy 2010; 6:239-47; PMID:20104019; http://dx.doi.org/ 10.4161/auto.6.2.11062 [DOI] [PubMed] [Google Scholar]
- 182. Rzymski T, Milani M, Pike L, Buffa F, Mellor HR, Winchester L, Pires I, Hammond E, Ragoussis I, Harris AL. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 2010; 29:4424-35; PMID:20514020; http://dx.doi.org/ 10.1038/onc.2010.191 [DOI] [PubMed] [Google Scholar]
- 183. Pike LR, Singleton DC, Buffa F, Abramczyk O, Phadwal K, Li JL, Simon AK, Murray JT, Harris AL. Transcriptional up-regulation of ULK1 by ATF4 contributes to cancer cell survival. Biochem J 2013; 449:389-400; PMID:23078367; http://dx.doi.org/ 10.1042/BJ20120972 [DOI] [PubMed] [Google Scholar]
- 184. Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, Vazquez P, Blazquez C, Torres S, Garcia S, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest 2009; 119:1359-72; PMID:19425170; http://dx.doi.org/ 10.1172/JCI37948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Papandreou I, Lim AL, Laderoute K, Denko NC. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L. Cell Death Differ 2008; 15:1572-81; PMID:18551130; http://dx.doi.org/ 10.1038/cdd.2008.84 [DOI] [PubMed] [Google Scholar]
- 186. Tracy K, Dibling BC, Spike BT, Knabb JR, Schumacker P, Macleod KF. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol 2007; 27:6229-42; PMID:17576813; http://dx.doi.org/ 10.1128/MCB.02246-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Azad MB, Chen Y, Henson ES, Cizeau J, McMillan-Ward E, Israels SJ, Gibson SB. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving BNIP3. Autophagy 2008; 4:195-204; PMID:18059169; http://dx.doi.org/ 10.4161/auto.5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 2009; 29:2570-81; PMID:19273585; http://dx.doi.org/ 10.1128/MCB.00166-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Huang Q, Wu YT, Tan HL, Ong CN, Shen HM. A novel function of poly(ADP-ribose) polymerase-1 in modulation of autophagy and necrosis under oxidative stress. Cell Death Differ 2009; 16:264-77; PMID:18974775; http://dx.doi.org/ 10.1038/cdd.2008.151 [DOI] [PubMed] [Google Scholar]
- 190. Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell 2004; 15:4829-40; PMID:15331765; http://dx.doi.org/ 10.1091/mbc.E04-06-0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Possik E, Jalali Z, Nouet Y, Yan M, Gingras MC, Schmeisser K, Panaite L, Dupuy F, Kharitidi D, Chotard L, et al. Folliculin regulates ampk-dependent autophagy and metabolic stress survival. PLoS Genet 2014; 10:e1004273; PMID:24763318; http://dx.doi.org/ 10.1371/journal.pgen.1004273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006; 10:51-64; PMID:16843265; http://dx.doi.org/ 10.1016/j.ccr.2006.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Ajabnoor GM, Crook T, Coley HM. Paclitaxel resistance is associated with switch from apoptotic to autophagic cell death in MCF-7 breast cancer cells. Cell Death Dis 2012; 3:e260; PMID:22278287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Amaral C, Borges M, Melo S, da Silva ET, Correia-da-Silva G, Teixeira N. Apoptosis and autophagy in breast cancer cells following exemestane treatment. PloS One 2012; 7:e42398; PMID:22912703; http://dx.doi.org/ 10.1371/journal.pone.0042398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Loos B, Engelbrecht AM, Lockshin RA, Klionsky DJ, Zakeri Z. The variability of autophagy and cell death susceptibility: Unanswered questions. Autophagy 2013; 9:1270-85; PMID:23846383; http://dx.doi.org/ 10.4161/auto.25560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol 2007; 431:61-81; PMID:17923231; http://dx.doi.org/ 10.1016/S0076-6879(07)31005-7 [DOI] [PubMed] [Google Scholar]
- 197. Hofmann S, Cherkasova V, Bankhead P, Bukau B, Stoecklin G. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol Biol Cell 2012; 23:3786-800; PMID:22875991; http://dx.doi.org/ 10.1091/mbc.E12-04-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp Cell Res 2007; 313:4130-44; PMID:17967451; http://dx.doi.org/ 10.1016/j.yexcr.2007.09.017 [DOI] [PubMed] [Google Scholar]
- 199. Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci 2013; 70:3493-511; PMID:23354059; http://dx.doi.org/ 10.1007/s00018-012-1252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem 2005; 280:16925-33; PMID:15684421; http://dx.doi.org/ 10.1074/jbc.M412882200 [DOI] [PubMed] [Google Scholar]
- 201. Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 2000; 5:897-904; PMID:10882126; http://dx.doi.org/ 10.1016/S1097-2765(00)80330-5 [DOI] [PubMed] [Google Scholar]
- 202. Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol 1995; 15:4497-506; PMID:7623840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Anderson P, Kedersha N. Stressful initiations. J Cell Sci 2002; 115:3227-34; PMID:12140254 [DOI] [PubMed] [Google Scholar]
- 204. Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol 2008; 10:1324-32; PMID:18836437; http://dx.doi.org/ 10.1038/ncb1791 [DOI] [PubMed] [Google Scholar]
- 205. Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell 2004; 5:429-41; PMID:15144951; http://dx.doi.org/ 10.1016/S1535-6108(04)00115-1 [DOI] [PubMed] [Google Scholar]
- 206. Fournier MJ, Coudert L, Mellaoui S, Adjibade P, Gareau C, Cote MF, Sonenberg N, Gaudreault RC, Mazroui R. Inactivation of the mTORC1-eukaryotic translation initiation factor 4E pathway alters stress granule formation. Mol Cell Biol 2013; 33:2285-301; PMID:23547259; http://dx.doi.org/ 10.1128/MCB.01517-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Matsuki H, Takahashi M, Higuchi M, Makokha GN, Oie M, Fujii M. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes cell 2013; 18:135-46; http://dx.doi.org/ 10.1111/gtc.12023 [DOI] [PubMed] [Google Scholar]
- 208. Takahara T, Maeda T. Transient sequestration of TORC1 into stress granules during heat stress. Mol Cell 2012; 47:242-52; PMID:22727621; http://dx.doi.org/ 10.1016/j.molcel.2012.05.019 [DOI] [PubMed] [Google Scholar]
- 209. Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 2003; 160:823-31; PMID:12642610; http://dx.doi.org/ 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 210. Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 α to the assembly of mammalian stress granules. J Cell Biol 1999; 147:1431-42; PMID:10613902; http://dx.doi.org/ 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 2000; 151:1257-68; PMID:11121440; http://dx.doi.org/ 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Didiot MC, Subramanian M, Flatter E, Mandel JL, Moine H. Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol Biol Cell 2009; 20:428-37; PMID:19005212; http://dx.doi.org/ 10.1091/mbc.E08-07-0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. French J, Stirling R, Walsh M, Kennedy HD. The expression of Ras-GTPase activating protein SH3 domain-binding proteins, G3BPs, in human breast cancers. Histochem J 2002; 34:223-31; PMID:12587999; http://dx.doi.org/ 10.1023/A:1021737413055 [DOI] [PubMed] [Google Scholar]
- 214. Guitard E, Parker F, Millon R, Abecassis J, Tocque B. G3BP is overexpressed in human tumors and promotes S phase entry. Cancer Lett 2001; 162:213-21; PMID:11146228; http://dx.doi.org/ 10.1016/S0304-3835(00)00638-8 [DOI] [PubMed] [Google Scholar]
- 215. Luca R, Averna M, Zalfa F, Vecchi M, Bianchi F, La Fata G, Del Nonno F, Nardacci R, Bianchi M, Nuciforo P, et al. The fragile X protein binds mRNAs involved in cancer progression and modulates metastasis formation. EMBO Mol Med 2013; 5:1523-36; PMID:24092663; http://dx.doi.org/ 10.1002/emmm.201302847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci 2013; 38:494-506; PMID:24029419; http://dx.doi.org/ 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 2013; 152:791-805; PMID:23415227; http://dx.doi.org/ 10.1016/j.cell.2013.01.033 [DOI] [PubMed] [Google Scholar]
- 218. Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal 2011; 23:324-34; PMID:20813183; http://dx.doi.org/ 10.1016/j.cellsig.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Takahashi M, Higuchi M, Matsuki H, Yoshita M, Ohsawa T, Oie M, Fujii M. Stress granules inhibit apoptosis by reducing reactive oxygen species production. Mol Cell Biol 2013; 33:815-29; PMID:23230274; http://dx.doi.org/ 10.1128/MCB.00763-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Kim WJ, Back SH, Kim V, Ryu I, Jang SK. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol Cell Biol 2005; 25:2450-62; PMID:15743837; http://dx.doi.org/ 10.1128/MCB.25.6.2450-2462.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Nascimento EB, Snel M, Guigas B, van der Zon GC, Kriek J, Maassen JA, Jazet IM, Diamant M, Ouwens DM. Phosphorylation of PRAS40 on Thr246 by PKB/AKT facilitates efficient phosphorylation of Ser183 by mTORC1. Cell Signal 2010; 22:961-7; PMID:20138985; http://dx.doi.org/ 10.1016/j.cellsig.2010.02.002 [DOI] [PubMed] [Google Scholar]
- 222. Fonseca BD, Smith EM, Lee VH, MacKintosh C, Proud CG. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J Biol Chem 2007; 282:24514-24; PMID:17604271; http://dx.doi.org/ 10.1074/jbc.M704406200 [DOI] [PubMed] [Google Scholar]
- 223. Oshiro N, Takahashi R, Yoshino K, Tanimura K, Nakashima A, Eguchi S, Miyamoto T, Hara K, Takehana K, Avruch J, et al. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem 2007; 282:20329-39; PMID:17517883; http://dx.doi.org/ 10.1074/jbc.M702636200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Wang L, Harris TE, Lawrence JC, Jr. Regulation of proline-rich Akt substrate of 40 kDa (PRAS40) function by mammalian target of rapamycin complex 1 (mTORC1)-mediated phosphorylation. J Biol Chem 2008; 283:15619-27; PMID:18372248; http://dx.doi.org/ 10.1074/jbc.M800723200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Thedieck K, Polak P, Kim ML, Molle KD, Cohen A, Jeno P, Arrieumerlou C, Hall MN. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PloS One 2007; 2:e1217; http://dx.doi.org/ 10.1371/journal.pone.0001217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. MolCell 2007; 25:903-15 [DOI] [PubMed] [Google Scholar]
- 227. Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. NatCell Biol 2007; 9:316-23 [DOI] [PubMed] [Google Scholar]
- 228. Valk K, Vooder T, Kolde R, Reintam MA, Petzold C, Vilo J, Metspalu A. Gene expression profiles of non-small cell lung cancer: survival prediction and new biomarkers. Oncology 2010; 79:283-92; PMID:21412013; http://dx.doi.org/ 10.1159/000322116 [DOI] [PubMed] [Google Scholar]
- 229. Buechler S. Low expression of a few genes indicates good prognosis in estrogen receptor positive breast cancer. BMC Cancer 2009; 9:243; PMID:19619298; http://dx.doi.org/ 10.1186/1471-2407-9-243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Heesom KJ, Denton RM. Dissociation of the eukaryotic initiation factor-4E/4E-BP1 complex involves phosphorylation of 4E-BP1 by an mTOR-associated kinase. FEBS Lett 1999; 457:489-93; PMID:10471835; http://dx.doi.org/ 10.1016/S0014-5793(99)01094-7 [DOI] [PubMed] [Google Scholar]
- 231. Damgaard CK, Lykke-Andersen J. Translational coregulation of 5'TOP mRNAs by TIA-1 and TIAR. Genes Dev 2011; 25:2057-68; PMID:21979918; http://dx.doi.org/ 10.1101/gad.17355911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Yamasaki S, Anderson P. Reprogramming mRNA translation during stress. Curr Opin Cell Biol 2008; 20:222-6; PMID:18356035; http://dx.doi.org/ 10.1016/j.ceb.2008.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Chou SD, Prince T, Gong J, Calderwood SK. mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PloS One 2012; 7:e39679; PMID:22768106; http://dx.doi.org/ 10.1371/journal.pone.0039679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Huo Y, Iadevaia V, Yao Z, Kelly I, Cosulich S, Guichard S, Foster LJ, Proud CG. Stable isotope-labelling analysis of the impact of inhibition of the mammalian target of rapamycin on protein synthesis. Biochem J 2012; 444:141-51; PMID:22428559; http://dx.doi.org/ 10.1042/BJ20112107 [DOI] [PubMed] [Google Scholar]
- 235. Iadevaia V, Wang X, Yao Z, Foster LJ, Proud CG. Evaluation of mTOR-regulated mRNA translation. Methods Mol Biol 2012; 821:171-85; PMID:22125065; http://dx.doi.org/ 10.1007/978-1-61779-430-8_10 [DOI] [PubMed] [Google Scholar]
- 236. Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 2009; 36:932-41; PMID:20064460; http://dx.doi.org/ 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 2005; 6:318-27; PMID:15803138; http://dx.doi.org/ 10.1038/nrm1618 [DOI] [PubMed] [Google Scholar]
- 238. Holcik M, Sonenberg N, Korneluk RG. Internal ribosome initiation of translation and the control of cell death. Trends Genet 2000; 16:469-73; PMID:11050335; http://dx.doi.org/ 10.1016/S0168-9525(00)02106-5 [DOI] [PubMed] [Google Scholar]
- 239. Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 2004; 101:11269-74; PMID:15277680; http://dx.doi.org/ 10.1073/pnas.0400541101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Dai N, Rapley J, Angel M, Yanik MF, Blower MD, Avruch J. mTOR phosphorylates IMP2 to promote IGF2 mRNA translation by internal ribosomal entry. Genes Dev 2011; 25:1159-72; PMID:21576258; http://dx.doi.org/ 10.1101/gad.2042311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Grzmil M, Hemmings BA. Translation regulation as a therapeutic target in cancer. Cancer Res 2012; 72:3891-900; PMID:22850420; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-0026 [DOI] [PubMed] [Google Scholar]
- 242. Ramirez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. J Cell Biol 2008; 181:293-307; PMID:18426977; http://dx.doi.org/ 10.1083/jcb.200710215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Damiano F, Rochira A, Tocci R, Alemanno S, Gnoni A, Siculella L. HnRNP A1 mediates the activation of the IRES-dependent SREBP-1a mRNA translation in response to endoplasmic reticulum stress. Biochem J 2012; http://dx.doi.org/ 10.1042/BJ20120906 [DOI] [PubMed] [Google Scholar]
- 244. Rubsamen D, Blees JS, Schulz K, Doring C, Hansmann ML, Heide H, Weigert A, Schmid T, Brune B. IRES-dependent translation of egr2 is induced under inflammatory conditions. RNA 2012; 18:1910-20; PMID:22915601; http://dx.doi.org/ 10.1261/rna.033019.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 2013; 153:1461-74; PMID:23791177; http://dx.doi.org/ 10.1016/j.cell.2013.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Seguin SJ, Morelli FF, Vinet J, Amore D, De Biasi S, Poletti A, Rubinsztein DC, Carra S. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ 2014; PMID:25034784; http://dx.doi.org/ 10.1038/cdd.2014.103 [DOI] [PMC free article] [PubMed] [Google Scholar]