Abstract
p53, the revered savior of genomic integrity, receives signals from diverse stress sensors and strategizes to maintain cellular homeostasis. However, the predominance of p53 overshadows the fact that this herculean task is no one-man show; rather, there is a huge army of regulators that reign over p53 at various levels to avoid an unnecessary surge in its levels and sculpt it dynamically to favor one cellular outcome over another. This governance starts right at the time of p53 translation, which is gated by proteins that bind to p53 mRNA and keep a stringent check on p53 protein levels. The same effect is also achieved by ubiquitylases and deubiquitylases that fine-tune p53 turnover and miRNAs that modulate p53 levels, adding precision to this entire scheme. In addition, extensive covalent modifications and differential protein interactions allow p53 to trigger a tailor-made response for a given circumstance. To magnify the marvel, these various tiers of regulation operate simultaneously and in various combinations. In this review, we have tried to provide a glimpse into this bewildering labyrinth. We believe that further studies will result in a better understanding of p53 regulation and that new insights will help unravel many aspects of cancer biology.
Keywords: cancer, genotoxic stress, miRNA, p53, post-translational modifications
Abbreviations
- miRNA
microRNA
- ROS
reactive oxygen species
- UTR
untranslated region
Regulation of any cellular pathway is essential to coordinate the heterogeneity and complexity of functions in multicellular organisms. Among the tumor suppressors, decoding the bewildering number of pathways that p53 is involved in has long been the holy grail of scientists. p53 is a master regulator that integrates signals from diverse nodes and thus it is of no surprise that it is the most commonly mutated gene in a huge array of cancers with varied origins. p53 has many weapons at its disposal to combat stress including cell cycle arrest, senescence, apoptosis, autophagy, and metabolic reprogramming. Paradoxically, some of the outcomes of p53 activation are disparate and contradictory, such as cell cycle arrest, which is prosurvival, versus apoptosis and senescence, which are directed toward eliminating irreversibly damaged cells. This indicates that p53 needs to be educated to sense the extent and type of damage and make an appropriate choice of the kind of response it is going to elicit. Extensive research on the regulation of p53 under diverse kinds of stresses including genotoxic stress, starvation, hypoxia, and oncogene activation clearly indicate that p53 protein is regulated at diverse levels, including synthesis, degradation, covalent modifications, subcellular localization, and differential interaction with other proteins. Moreover, all possible permutations and combinations of these are employed to modulate p53 specificity, tissue heterogeneity, and diversity of function. In light of this, we restrict this review to exclusively discussing the myriad layers of p53 regulation upon genotoxic stress (Fig. 1).
Figure 1.
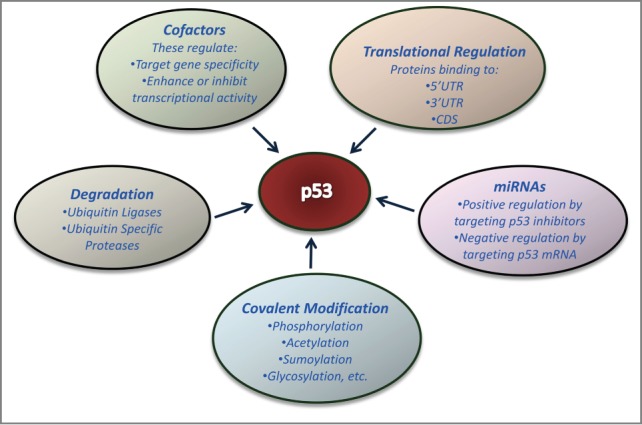
Diverse modes of p53 regulation. The multitude of ways by which p53 can be regulated include regulation at the level of translation and mRNA stability, cellular proteins that bind to and regulate p53 function and stability, and post-translational modifications.
It Begins at the Beginning: Translational Regulation of p53
Transcriptional regulation of p53 is not a major contributor to the modulation of p53 levels. There are reports of various factors that bind to the p53 promoter and can activate its transcription under stress conditions, such as cAMP responsive element binding protein (CREB)1 and Myc.2 p53 is also known to transcriptionally activate itself.3 However, p53 levels are predominantly regulated by a slew of post-transcriptional and post-translational mechanisms. Initially, it was thought that the increase in p53 protein levels upon genotoxic stress is the result of inhibition of its degradation, but translational regulation was later recognized as being critical for maintaining p53 levels. Both the 5’UTR and 3’UTR of p53 mRNA play a significant role in negative regulation of p53 translation.4,5 Moreover, the ribosomal protein RPL26 binds to the 5’UTR of p53 mRNA, and hastens p53 translation under genotoxic stress to elicit a rapid p53 response.6 On the other hand, MDM2 binds to RPL26 and triggers its polyubiquitylation and proteasomal degradation, thereby turning down p53 translation.7 As MDM2 is a p53 transcriptional target, this constitutes an autoregulatory feedback loop that modulates p53 translation. Further exploration of RPL26-dependent translational regulation of p53 showed that 5'UTR–3'UTR base pairing of p53 mRNA is a critical determinant of RPL26 binding and can be manipulated by mutating these sites.8 Providing proof of concept, short oligonucleotides targeting this region inhibit p53 translation and can be used to inhibit expression of mutant p53 protein implicated in tumor progressive mechanisms. This has exciting implications on the clinical front and could be used in targeted therapy to combat many pathologic conditions. The nucleolin protein also binds to the p53 mRNA 5'UTR; however, this interaction inhibits p53 translation under unstressed conditions.6 Pdcd4 performs a similar function to nucleolin to repress p53 translation under normal conditions.9 Furthermore, thymidylate synthase, a folate-dependent enzyme that catalyzes the conversion of dUMP to dTMP, binds to the coding sequence of p53 mRNA and suppresses its translation.10,11 Moreover, the RNA binding proteins HuR and Hzf work hand in glove to bind to the 3’UTR of p53 mRNA in the presence of p19Arf and export it from the nucleus to the cytoplasm in conjunction with its increased translation.12 RNPC1, GAPDH, hnRNP A/B, and hnRNP D also regulate p53 translation by binding to the cytoplasmic polyadenylation elements of p53 mRNA.13,14
In essence, different regulators bind to p53 mRNA 5’UTR, 3’UTR, and coding sequence and collaborate to fine tune p53 abundance in accordance to stress cues. This in turn largely excludes undamaged cells from the ravaging consequences of abundant p53 while ensuring that damaged cells do not escape surveillance.
Auditing p53 Levels: Regulation by Degradation
Ubiquitylases and deubiquitylases are indispensable for protein turnover and are key mediators of signal transduction and thus important players in cancer development.15 Ubiquitin is attached to target proteins through different lysine linkages, namely K6, K11, K27, K29, K33, K48, and K63. K48-linked polyubiquitylation mediates degradation and K63-polyubiquitylation coordinates signaling. In this section we have focused on the ubiquitin-mediated degradation of p53; the role of ubiquitin as a signal modifier and determinant of p53 cellular localization will be discussed later in this review.
MDM2, an E3 ligase, has the honor of being the first regulator of p53 to be investigated. It keeps p53 levels in check under conditions of no stress through polyubiquitylation and proteasomal degradation.16 MDM2 binds to the N-terminal transactivation domain of p53 and ubiquitylates its 6 C-terminal lysines,17 thus targeting it for proteasomal degradation. As MDM2 is in turn a p53 transcriptional target this constitutes an autoregulatory feedback loop.18 Upon genotoxic stress various DNA damage sensors (e.g., ATM kinases, DNA-PK, Chk2, HIPK2) relieve p53 of association with MDM2 through phosphorylation of p53 N-terminal serine residues, resulting in an increase in p53 levels.19 After DNA damage ATM-mediated phosphorylation of MDM2 elicits its autoubiquitylating activity, decreasing levels of ATM and MDMX, which in turn increases p53 levels.20,21 Although MDM2 is considered the key E3 ligase for p53, continued research has revealed the involvement of a myriad of other E3 ligases in maintaining p53 levels. Of these, Cop-1 and Pirh-2 E3 ligases function similarly to MDM2 in that they are also transcriptionally induced by p53 and constitute an autoregulatory loop. ARF-BP1, Trim24, Trim28, Trim39, E4F1, the cullin family of E3 ligases, Hades, Carp1 and 2, synovilin, and SUMO E3 ligase also degrade p53 under basal conditions.22 Trim24 has been shown to function in a similar manner as MDM2 in regulating p53 levels and is inactivated by ATM-kinases.23
Deubiquitylases remove the chains of ubiquitin that are attached by E3 ligases. To date, USP10, USP42, USP7 (HAUSP), and USP2a have been studied in detail with respect to p53 deubiquitylation. USP7 has a dualistic role in the context of p53 stabilization and degradation. MDM2 and p53 have both been shown to be deubiquitylated and stabilized by USP7, highlighting the complexity of p53 regulation.24 Further studies suggest that following genotoxic stress TRIM21 ubiquitylates GMPS, a nucleotide metabolism enzyme, causing it to be translocated to the cytoplasm where it associates with USP7. This interaction causes a shift in substrate specificity of USP7 from MDM2 to p53.25 Unlike USP7, USP10 is a p53-specific deubiquitylase. Under normal conditions, it is localized in the cytosol where it deubiquitylates monoubiquitylated p53 sequestered in the cytoplasm and sends it back to the nucleus for degradation by MDM2. However, under genotoxic stress conditions ATM-mediated phosphorylation of USP10 promotes its nuclear import where it protects polyubiquitylated p53 from degradation by deubiquitylating it.26 USP42 has also been shown to deubiquitylate p53 in response to genotoxic stress but its regulatory role needs to be further explored.27 On the other hand, USP2a downregulates p53 by deubiquitylating its E3 ligases MDM2 and MDMX.28
Individual interactors of p53 and MDM2 also influence the p53–MDM2 interaction as well as p53 ubiquitylation, thus affecting p53 levels. p14Arf was the first known example of this type of regulation; it binds to MDM2 and inhibits MDM2-mediated p53 degradation.29 Ribosomal protein L11 also binds to and inactivates MDM2 by inducing its nucleolar localization.30 However, Pax3, Twist, Niban, Smurf1/2, and TCTP aid p53 ubiquitylation by enhancing MDM2 activity.22 WIP1 is a phosphatase that dephosphorylates MDM2 thereby increasing its stability and affinity for p53 and resulting in enhanced p53 ubiquitination and degradation.31 p53 also interacts with other transcription factors that modulate its ubiquitylation status such as ATF3, which binds to the C-terminus of p53 and inhibits its ubiquitylation,32 and TFII31, which binds to the p53 N-terminus of p53 and prevents it from binding to MDM2.33 Recently, Bouafia et al. reported that the stress sensor USF1 functions as a stabilizer of p53 by inhibiting the p53–MDM2 interaction after DNA damage. Being present in cells at a high level all times, USF1 binds to p53 as soon as genomic danger is encountered. Studies have shown that USF1 is equally as efficient as Nutlin-3a treatment and superior to other transcription factors such ATF3 and TFII31 in p53 stabilization during genotoxic stress.34
Taken together, these findings indicate that ubiquitylation-dependent pathways ensure minimal levels of p53 in normal cells whereas inhibition of these factors by messengers of DNA damage induces p53 to protect cells from transformation-inducing alterations.
Different Strokes for Different Folks: p53 Regulation Through Covalent Modifications
One of the most important mechanisms for regulating p53 function and stability is post-translational modification. Phosphorylation, acetylation, and ubiquitylation are the prominent modifications of p53 while ubiquitin-like modifiers (e.g., SUMOylation and NEDDylation), glycosylation (O-linked N-acetylgucosamine), prolyl isomerization, and ADP-ribosylation play niche roles in p53 regulation (Fig. 2). The importance of these modifications is brought to the forefront when context-dependent p53 effector functions need to be executed. These modifications also act as a barcode that is read by cellular proteins for association with p53.
Figure 2.
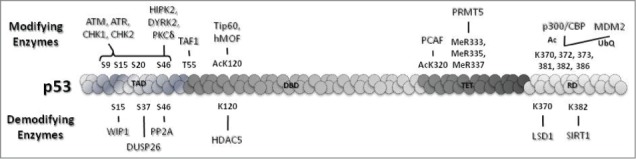
Selected modifiers and demodifiers of p53. These modifications act as determinants of p53 levels and p53-mediated cell fate decisions.
Phosphorylation was the first functionally relevant post-transcriptional modification of p53 to be discovered35 and since then 23 phosphorylation sites on p53 have been uncovered. Several serines and threonines of p53 have been shown to be differentially phosphorylated by kinases, some under genotoxic stress (S6, S9, S15, S20, S46, S215, S366, S376, T388, S392) and others under basal conditions (T55, S376). The extensive redundancy of phosphorylation sites and the respective kinases involved make the implications of phosphorylation even more mystifying, for example S15 is phosphorylated by at least 8 kinases and CHK2 phosphorylates p53 at 7 different sites. ATM, ATR, and their downstream kinases CHK1 and CHK2 play a central role in genome surveillance and mediate p53 S9, S15, S20, S46 phosphorylations.36 S15/S20 phosphorylations play an important role in disrupting the binding between p53 and MDM2 to stabilize p53 and facilitate its transcriptional activity.37 This observation is supported by defective apoptosis and delayed tumor development in mice expressing the p53 S15/20A mutant.38 S6 and S9 phosphorylations have importance in both development and cancer and are thought to be mediated by CK1 family members.39 S46 phosphorylation, which is important for the induction of apoptosis40 is primarily regulated by HIPK2,41 DYRK2,42 and PKCδ43 in response to DNA damage. S392 phosphorylation, which is increased by UV radiation, stabilizes tetramer formation of p53 and hence increases its transcriptional activity.44 Additionally, the immediate stabilization of p53 in response to UV-induced damage has been attributed to phosphorylation of p53 T18 by the serine-threonine kinase VRK1. VRK1 remains associated with p53 even in the absence of damage signals and phosphorylates p53 as soon as cells are exposed to UV,45 thus providing an immediate regulatory response against DNA damage. Phosphorylation events also have an inhibitory effect on p53 activation. ATM-dependent dephosphorylation of S376 activates p5346 and TAF1-mediated phosphorylation of T55 prevents binding of p53 to its target promoters.47 Further studies have shown that TAF1 coordinates with cellular ATP fluctuations caused by DNA damage and facilitates global inhibition of p53 target genes in unstressed cells.48 Consistent with this study, the T55A p53 mutant shows enhanced apoptosis compared to wild-type p53.
Ablation of these phosphorylations by distinct phosphatases adds another layer of complexity to the regulation of p53. DUSP26, PP1, and PP2A are among the key phosphatases that balance the DNA damage response by regulating the p53–MDM2 interaction and p53 transcriptional target preferences. These phosphatases are mainly required to reinitiate the cell cycle after DNA repair is accomplished. DUSP26 specifically dephosphorylates S20 and S37 and inhibits apoptotic functions of p53.49 On the same note, PP1 dephosphorylates S15 and S37 of p53 thereby downregulating its transcriptional activity and apoptotic functions.50 Ionizing radiation-induced S46 phosphorylation of p53 is reversed by PP2A, resulting in attenuation of p53-mediated apoptosis.51 Recent studies show the requirement for different phosphatases to overcome G1 and G2 arrest upon resolution of DNA damage. PP4 rescues cells from G1 arrest by dephosphorylating KAP1, which then associates with p53 and acts as a transcriptional repressor to inhibit p21 expression. WIP1, on the other hand, dephosphorylates p53 at S15 and relieves CCNB1 repression to rescue cells from G2 arrest of the cell cycle.52
Acetylation of p53 at key DNA binding and C-terminal regulatory domain lysine residues (K120, K164, K320, K370, K372, K373, K381, K382, and K386) is a critical factor in determination of cellular outcome upon genomic insult. Acetylation plays an important role in transcriptional regulation by p53 and influences the recruitment of repressors, activators, and other modulators.53 Histone acetyl transferases (HATs), including p300/CBP, PCAF, hMOF, and Tip60, and the deacetylases SIRT1 and HDACs (HDAC1 and HDAC5) harmonize to bring about cell type-specific and stimulus-specific responses. p300/CBP acetylates all 6 C-terminal lysines that are targeted by MDM2 and promotes p53-mediated transactivation, emphasizing the competitive nature of binding of these proteins at the same site. p53 K320 acetylation by PCAF promotes preferential activation of cell cycle arrest as a DNA damage response and decreases apoptosis.38 p53 acetylation by Tip60/hMOF at K120 promotes cell death as a response to DNA damage.54,55 Mutation of lysine to a non-acetylable arginine (R) residue has become the benchmark method to study the functional consequences of each acetylation separately (e.g., p53K120R), C-terminal acetylation of K370, K372, K373, K381, K382, and K386 all at once (p536KR), core domain containing K120 and K164 together (p532KR), or all key acetylations (K120, K164, K370, K372, K373, K381, K382, and K386) at once (p538KR).56 p538KR is completely inert in vivo, showing the relevance of acetylation for p53 stability and transcriptional activity. p532KR cells show complete loss of p53 transactivation function and stability compared to p536KR cells, which show relatively increased apoptosis after IR radiation exposure. p53K120R cells undergo cell cycle arrest and senescence upon genotoxic stress but p53-mediated apoptosis is completely abrogated.57
To maintain homeostasis, p53 is also regulated by deacetylases that counteract the effect of different acetylations. SIRT1 is known to deacetylate p53 at K382 and hence inhibits DNA damage-mediated apoptosis.58 The intricacies of the relationship between SIRT1 and p53 are highlighted by the presence of hyperacetylated p53 in mice lacking SIRT159 and tumors overexpressing SIRT1 with inactivated p53.60,61 Another deacetylase that plays a critical role in modulating the p53 response to genotoxic stress is HDAC5, a class IIa deacetylase. In the early phase of genotoxic stress, HDAC5 binds to and deacetylates p53 at the K120 residue. This induces cell cycle arrest and clearance of reactive oxygen species (ROS). At the late phase of genotoxic stress, high levels of ROS lead to CamKII-mediated nuclear exit of HDAC5, which facilitates p53 K120 acetylation and induction of apoptosis; in contrast, restraining HDCA5 nuclear export promotes senescence. Thus, in mice subjected to genotoxic stress inhibition of HDCA5 nuclear export extends protection from genotoxic stress whereas abrogation of HDAC5 expression accelerates the onset of p53-mediated apoptosis.62
Methylation is another modification of p53 lysines. There are 3 lysine methyl transferases (KMTs) that monomethylate p53 and 2 KMTs that dimethylate p53. Monomethylation of p53 by the SET family of proteins (SET7, SET8, and SET9) and Smyd2 and dimethylation by G9a and GLP are known to modulate p53 transcriptional activity. K370 methylation by Smyd263 and K382 methylation by SET8 repress p53 transactivation.64 Methylation of K372 by SET7/9 has been shown to positively regulate acetylation of K120 by Tip60 upon DNA damage.65 However, this observation is contradicted by an another study showing a lack of significance of SET7/9 methylation in p53 acetylation and transcriptional functions.66 Dimethylation of K370 by a currently unidentified methylase is known to promote association of p53 to 53BP1 upon DNA damage. The demethylase LSD1 inhibits this association by removing methyl moieties from K370.67 G9a and GLP specifically methylate K373 and interfere with apoptotic functions of p53.68 Arginine methylations in the tetramerization domain of p53 (R333, R335, R337) have also been shown to be catalyzed by PRMT5.69 Arginine methylation regulates p53 target gene specificity thereby promoting apoptosis over cell cycle arrest.
In addition to modulating degradation, ubiquitylation also determines the endocytosis, transcriptional regulation, and subcellular localization of p53.70 Monoubiquitylated p53 shuttles from the nucleus to cytoplasm, which lowers p53 transactivation activity. MDM2 levels in the cell also act as a determinant for subcellular localization of p53. Low levels of MDM2 monoubiquitylate p53 and result in its sequestration in the cytoplasm.71 MSL2 specifically monoubiquitylates K351 and K357 of p53 independent of MDM2, and mediates its nuclear export.72 Similarly, other E3 ligases such as CUL9/PARC and WWP1 ligase monoubiquitylate p53 and facilitate its nuclear export.73,74 Paradoxically, monoubiquitylation by E4F1 facilitates association of p53 with chromatin, eliciting its cell cycle arrest functions.75
Modification by SUMOylation and NEDDylation involves the conjugation of small ubiquitin-like proteins. While SUMOylation of K386 of p53 by the PIAS family and TOPORS enhances transcriptional activity,76 NEDDylation of p53 at different C-terminal residues by MDM2 (K370, K372 and K373) and FBXO11 (K320 and K321) represses its activity.77,78 Prolyl isomerization by Pin179 and glycosylation80 also direct p53 transactivation after DNA damage. In response to DNA damage, p53 also becomes poly(ADPribosyl)ated (PARylated) within its core domain (E255, D256, and E268), which inhibits its nuclear export and hence contributes to increased transactivation.81
It Takes Two To Tango: the p53 Interactors
The p53 interactome includes many proteins that bind to p53 to regulate its transactivation function. Some of these proteins play a crucial role in target selection whereas others mediate assembly of the transcription complex. An important regulator of p53 apoptotic functions is the ASPP family. ASPP family members ASPP1, ASPP2, and iASPP bind to the DNA-binding domain of p53 to modulate selectivity of binding to p53 target promoters. ASPP1 and ASPP2 guide p53 toward induction of the apoptotic response whereas iASPP competes for the same site to inhibit p53 binding to apoptotic targets and promote cell cycle arrest.82,83 The p53 target Hzf also binds to the DNA-binding domain of p53 and enhances p53 selectivity for cell cycle arrest genes in response to genotoxic stress.84 Brn3 family members Brn3a and Brn3b, APAK, YB1, Muc1, hCAS/CSE1L, p18/Hamlet, c-Abl, and the p52 subunit of NF-kB are other guiding partners of p53 that add heterogeneity to its functional outcomes.53 Brn3a, YB1, and Muc1 direct p53 toward cell survival mechanisms by either inducing cell cycle arrest or senescence. In contrast, Brn3b, hCAS/CSE1L, p18/Hamlet, and c-Abl channel p53 toward apoptosis induction. APAK is unique among these proteins as it binds to p53 and facilitates repression of its transcriptional activity by recruiting HDAC1 under normal conditions.85 NF-Y occupies a special place among transcriptional regulators of p53 in that it has a dual function. On one hand, NF-Y binds CCAAT sequences and subsequent binding of p53 to the NF-YA subunit of the NF-Y complex induces transcription of apoptotic genes lacking a p53 response element. On the other hand, NF-Y recruits HDAC corepressors to repress genes involved in the rescue of G2/M arrest.86 CBP/p300, pCAF, JMY, MAML1, TAFII-32/70, GPS2, and ADA3 coactivate p53 transcriptional activity but do not partake in target selection.53 These proteins play an important role in histone modification and chromatin remodeling, and also facilitate the recruitment of components of the transcription initiation machinery.
Interplay between post-translational modification and cofactor binding also determines cell fate decisions. S46 phosphorylation of p53 upon stress induces binding of Pin1 to its N-terminal domain, displacing iASPP from the core domain of p53 and thereby triggering apoptosis.87 K120 acetylation inhibits binding of MDM2 as a corepressor and induces binding of p53 to apoptotic targets.56 Association of p53 with p68 subunit of Drosha microRNA processor is also enhanced when p53 is acetylated on K120 and is important for nuclear primary miRNA processing of miRNA-203. Subsequently, miRNA-203 degrades antiapoptotic Bcl-w, leading to increased apoptosis.88 Moreover, DNA damage induces K382 acetylation and S392 phosphorylation of p53, which augments the interaction of p53 with MDC1, a mediator of the DNA damage response.89
Custodial Custody: miRNAs Regulating p53
MicroRNAs (miRNAs) act as insurance in the mesh of regulatory networks that maintain discipline at the molecular level. These endogenously expressed 20–25 nucleotide long RNAs can base pair to the 3’UTRs of target mRNA (mRNA), inhibiting their translation and/or affecting their stability. Interestingly, miRNAs both target and are targeted by p53. p53 takes a multivalent approach to the induction of miRNAs; it not only transactivates the miRNA coding genes but also has the potential to steer the processing machinery in favor of a particular miRNA family (e.g., mir-16–1, mir-143, and mir-145). It accomplishes this by enhancing the interaction of DEAD BOX RNA helicase p68 with the DROSHA complex, thereby facilitating the processing of microRNAs.90 To further strengthen the network, p53 regulates miRNA target selection. p53 induces the RNA binding protein p38, which binds to the 3'UTRs of target mRNAs including p53 targets such as p21 and DR5 and obscures the miRNA binding site, thereby competing with miRNAs for binding and inhibiting their activity.91
miRNAs also play a key role in maintaining p53 levels (Fig. 3). miRNAs that positively regulate p53 include miR-29, which targets CDC42 (a Rho GTPase) and p85α (the regulatory subunit of phosphatidyl inositol 3 kinase) and increases p53 levels through a mdm-2–dependent mechanism.92 miR-34 targets Sirt1 and potentiates p53 by inhibiting its deacetylation.93 miR-542–3p contributes to p53 stability by weakening the interaction between p53 and MDM2.94 miR-506 inhibits expression of the NF-κB p65 subunit, leading to ROS accumulation and subsequent p53 activation.95
Figure 3.
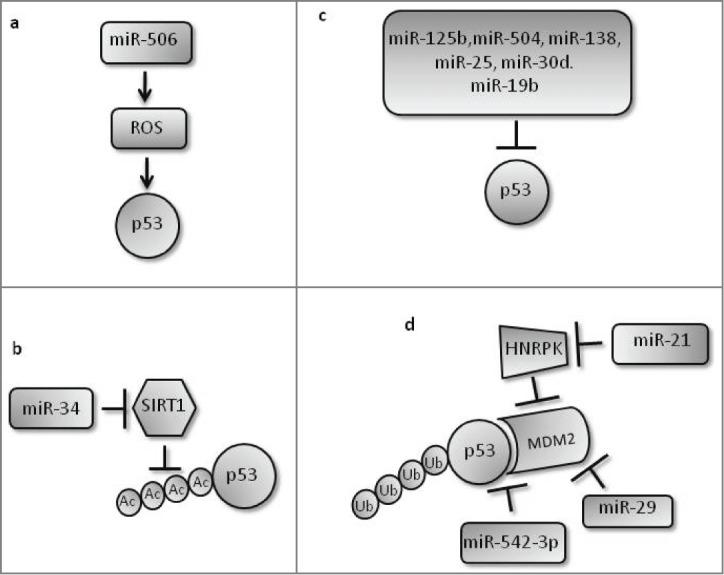
Various mechanisms by which miRNAs positively and negatively regulate p53 to maintain cellular homeostasis. (a) miR-506 leads to DNA damage via ROS generation and hence activates p53 through the DNA damage pathway. (b) miR-34 positively regulates p53 by inhibiting its deacetylase SIRT1. (c) Various miRNAs negatively regulate p53 through direct base pairing with its 3’UTR. (d) Several miRNAs target MDM2-mediated p53 ubiquitylation: miR-542–3p and miR-29 positively regulate p53 by targeting p53-MDM2 interaction and MDM2, respectively; miR-21 negatively regulates p53 by inhibiting HNRPK, an inhibitor of MDM2, leading to increased MDM2 activity and p53 ubiquitylation.
miRNAs that negatively regulate p53 and further enhance the precision of the system include miR-21 that targets HNRPK, which is known to stabilize p53 by interfering with MDM2 activity. Downregulation of HNRPK thus results in increased MDM2-mediated ubiquitylation and degradation of p53.96 miRNAs such as miR-125b, a brain enriched miRNA, can also directly target the 3'UTR of p53 and lead to its mRNA degradation. Upon genotoxic stress, miR-125b is downregulated to allow accumulation of p53.97 The p53 3’UTR has 2 response elements for miR-504, and overexpression of miR-504 leads to reduced p53 protein levels and tumor suppressive functions.98 miR-138 also targets the 3’UTR of p53 mRNA, significantly reducing its expression.99 More recently, miR-25, miR-30d,100 and miR-19b101 have also been recognized as negative regulators of p53 that bind directly to its 3’UTR. miRNAs thus play a significant role as agonists and antagonists of p53, allowing buffering of the p53 response and preventing extreme responses in its execution.
Conclusion and Future Direction
Among the diverse stresses known to activate p53, genotoxic stress has achieved benchmark status in research endeavors to understand p53 regulation. Genotoxic drugs have been effectively used as a weapon to kill tumor cells by eliciting heightened p53 levels. p53, a savior for stressed cells and messenger of death for irreparable cells, is regulated in every probable dimension to make life and death decisions. An enhanced understanding of the kinetics of these regulatory mechanisms using systems biology approaches is an important approach that might bring clarity to p53 regulation. Moreover, p53 regulatory mechanisms provide the opportunity to exploit its function in cancer cells by regulating the selectivity of p53 toward its target promoters. The emergence of p53 post-translational modifications and new interaction partners as important mediators of its functions lends credence to the search for small molecules that specifically target the enzymatic activities of regulators of post-translational modifications and the binding affinities of interaction partners. Such agents could be used in combination with genotoxic drugs for better therapeutic outcomes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the members of the Molecular Oncology Laboratory for helpful discussions. We apologize to researchers whose work could not be referenced due to space limitations.
Funding
The authors would like to acknowledge the financial support from NII Core Fund.
References
- 1. Okoshi R, Ando K, Suenaga Y, Sang M, Kubo N, Kizaki H, Nakagawara A, Ozaki T. Transcriptional regulation of tumor suppressor p53 by cAMP-responsive element-binding protein/AMP-activated protein kinase complex in response to glucose deprivation. Genes Cells 2009; 14:1429-40; PMID:19930465; http://dx.doi.org/ 10.1111/j.1365-2443.2009.01359.x [DOI] [PubMed] [Google Scholar]
- 2. Reisman D, Elkind NB, Roy B, Beamon J, Rotter V. c-Myc trans-activates the p53 promoter through a required downstream CACGTG motif. Cell Growth Differ 1993; 4:57-65; PMID:8494784 [PubMed] [Google Scholar]
- 3. Deffie A, Wu H, Reinke V, Lozano G. The tumor suppressor p53 regulates its own transcription. Mol Cell Biol 1993; 13:3415-23; PMID:7684498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fu L, Benchimol S. Participation of the human p53 3'UTR in translational repression and activation following gamma-irradiation. EMBO J 1997; 16:4117-25; PMID:9233820; http://dx.doi.org/ 10.1093/emboj/16.13.4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mosner J, Mummenbrauer T, Bauer C, Sczakiel G, Grosse F, Deppert W. Negative feedback regulation of wild-type p53 biosynthesis. EMBO J 1995; 14:4442-9; PMID:7556087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 2005; 123:49-63; PMID:16213212; http://dx.doi.org/ 10.1016/j.cell.2005.07.034 [DOI] [PubMed] [Google Scholar]
- 7. Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell 2008; 32:180-9; PMID:18951086; http://dx.doi.org/ 10.1016/j.molcel.2008.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen J, Kastan MB. 5'-3'-UTR interactions regulate p53 mRNA translation and provide a target for modulating p53 induction after DNA damage. Genes Dev 2010; 24:2146-56; PMID:20837656; http://dx.doi.org/ 10.1101/gad.1968910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wedeken L, Singh P, Klempnauer KH. Tumor suppressor protein Pdcd4 inhibits translation of p53 mRNA. J Biol Chem 2011; 286:42855-62; PMID:22033922; http://dx.doi.org/ 10.1074/jbc.M111.269456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chu E, Copur SM, Ju J, Chen TM, Khleif S, Voeller DM, Mizunuma N, Patel M, Maley GF, Maley F, et al. Thymidylate synthase protein and p53 mRNA form an in vivo ribonucleoprotein complex. Mol Cell Biol 1999; 19:1582-94; PMID:9891091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ju J, Pedersen-Lane J, Maley F, Chu E. Regulation of p53 expression by thymidylate synthase. Proc Natl Acad Sci U S A 1999; 96:3769-74; PMID:10097112; http://dx.doi.org/ 10.1073/pnas.96.7.3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakamura H, Kawagishi H, Watanabe A, Sugimoto K, Maruyama M, Sugimoto M. Cooperative role of the RNA-binding proteins Hzf and HuR in p53 activation. Mol Cell Biol 2011; 31:1997-2009; PMID:21402775; http://dx.doi.org/ 10.1128/MCB.01424-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J, Cho SJ, Shu L, Yan W, Guerrero T, Kent M, Skorupski K, Chen H, Chen X. Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev 2011; 25:1528-43; PMID:21764855; http://dx.doi.org/ 10.1101/gad.2069311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rosenstierne MW, Vinther J, Mittler G, Larsen L, Mann M, Norrild B. Conserved CPEs in the p53 3' untranslated region influence mRNA stability and protein synthesis. Anticancer Res 2008; 28:2553-9; PMID:19035278 [PubMed] [Google Scholar]
- 15. Satija YK, Bhardwaj A, Das S. A portrayal of E3 ubiquitin ligases and deubiquitylases in cancer. Int J Cancer 2013; 133:2759-68; PMID:23436247 [DOI] [PubMed] [Google Scholar]
- 16. Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 1997; 420:25-7; PMID:9450543; http://dx.doi.org/ 10.1016/S0014-5793(97)01480-4 [DOI] [PubMed] [Google Scholar]
- 17. Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol 2000; 20:8458-67; PMID:11046142; http://dx.doi.org/ 10.1128/MCB.20.22.8458-8467.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev 1993; 7:1126-32; PMID:8319905; http://dx.doi.org/ 10.1101/gad.7.7a.1126 [DOI] [PubMed] [Google Scholar]
- 19. Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res 2003; 1:1001-8; PMID:14707283 [PubMed] [Google Scholar]
- 20. Pereg Y, Shkedy D, de Graaf P, Meulmeester E, Edelson-Averbukh M, Salek M, Biton S, Teunisse AF, Lehmann WD, Jochemsen AG, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci U S A 2005; 102:5056-61; PMID:15788536; http://dx.doi.org/ 10.1073/pnas.0408595102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J 2004; 23:1547-56; PMID:15029243; http://dx.doi.org/ 10.1038/sj.emboj.7600145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hock AK, Vousden KH. The role of ubiquitin modification in the regulation of p53. Biochim Biophys Acta 2014; 1843:137-49; PMID:23742843; http://dx.doi.org/ 10.1016/j.bbamcr.2013.05.022 [DOI] [PubMed] [Google Scholar]
- 23. Jain AK, Allton K, Duncan AD, Barton MC. TRIM24 is a p53-Induced E3-Ubiquitin Ligase that undergoes ATM-Mediated Phosphorylation and Autodegradation during DNA Damage. Mol Cell Biol 2014; PMID:24820418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 2004; 13:879-86; PMID:15053880; http://dx.doi.org/ 10.1016/S1097-2765(04)00157-1 [DOI] [PubMed] [Google Scholar]
- 25. Reddy BA, van der Knaap JA, Bot AG, Mohd-Sarip A, Dekkers DH, Timmermans MA, Martens JW, Demmers JA, Verrijzer CP. Nucleotide biosynthetic enzyme GMP synthase is a TRIM21-controlled relay of p53 stabilization. Mol Cell 2014; 53:458-70; PMID:24462112; http://dx.doi.org/ 10.1016/j.molcel.2013.12.017 [DOI] [PubMed] [Google Scholar]
- 26. Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 2010; 140:384-96; PMID:20096447; http://dx.doi.org/ 10.1016/j.cell.2009.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hock AK, Vigneron AM, Carter S, Ludwig RL, Vousden KH. Regulation of p53 stability and function by the deubiquitinating enzyme USP42. EMBO J 2011; 30:4921-30; PMID:22085928; http://dx.doi.org/ 10.1038/emboj.2011.419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J 2007; 26:976-86; PMID:17290220; http://dx.doi.org/ 10.1038/sj.emboj.7601567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 1998; 92:713-23; PMID:9529248; http://dx.doi.org/ 10.1016/S0092-8674(00)81400-2 [DOI] [PubMed] [Google Scholar]
- 30. Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 2003; 3:577-87; PMID:12842086; http://dx.doi.org/ 10.1016/S1535-6108(03)00134-X [DOI] [PubMed] [Google Scholar]
- 31. Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell 2007; 12:342-54; PMID:17936559; http://dx.doi.org/ 10.1016/j.ccr.2007.08.033 [DOI] [PubMed] [Google Scholar]
- 32. Yan C, Lu D, Hai T, Boyd DD. Activating transcription factor 3, a stress sensor, activates p53 by blocking its ubiquitination. EMBO J 2005; 24:2425-35; PMID:15933712; http://dx.doi.org/ 10.1038/sj.emboj.7600712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Buschmann T, Lin Y, Aithmitti N, Fuchs SY, Lu H, Resnick-Silverman L, Manfredi JJ, Ronai Z, Wu X. Stabilization and activation of p53 by the coactivator protein TAFII31. J Biol Chem 2001; 276:13852-7; PMID:11278372; http://dx.doi.org/ 10.1074/jbc.M103786200 [DOI] [PubMed] [Google Scholar]
- 34. Bouafia A, Corre S, Gilot D, Mouchet N, Prince S, Galibert MD. p53 requires the stress sensor USF1 to direct appropriate cell fate decision. PLoS Genet 2014; 10:e1004309; PMID:24831529; http://dx.doi.org/ 10.1371/journal.pgen.1004309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Samad A, Anderson CW, Carroll RB. Mapping of phosphomonoester and apparent phosphodiester bonds of the oncogene product p53 from simian virus 40-transformed 3T3 cells. Proc Natl Acad Sci U S A 1986; 83:897-901; PMID:3006031; http://dx.doi.org/ 10.1073/pnas.83.4.897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 2003; 3:155-68; PMID:12612651; http://dx.doi.org/ 10.1038/nrc1011 [DOI] [PubMed] [Google Scholar]
- 37. Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer 2006; 6:909-23; PMID:17128209; http://dx.doi.org/ 10.1038/nrc2012 [DOI] [PubMed] [Google Scholar]
- 38. Chao C, Wu Z, Mazur SJ, Borges H, Rossi M, Lin T, Wang JY, Anderson CW, Appella E, Xu Y. Acetylation of mouse p53 at lysine 317 negatively regulates p53 apoptotic activities after DNA damage. Mol Cell Biol 2006; 26:6859-69; PMID:16943427; http://dx.doi.org/ 10.1128/MCB.00062-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Higashimoto Y, Saito S, Tong XH, Hong A, Sakaguchi K, Appella E, Anderson CW. Human p53 is phosphorylated on serines 6 and 9 in response to DNA damage-inducing agents. J Biol Chem 2000; 275:23199-203; PMID:10930428; http://dx.doi.org/ 10.1074/jbc.M002674200 [DOI] [PubMed] [Google Scholar]
- 40. Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, et al. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 2000; 102:849-62; PMID:11030628; http://dx.doi.org/ 10.1016/S0092-8674(00)00073-8 [DOI] [PubMed] [Google Scholar]
- 41. D'Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 2002; 4:11-9; PMID:11780126; http://dx.doi.org/ 10.1038/ncb714 [DOI] [PubMed] [Google Scholar]
- 42. Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol Cell 2007; 25:725-38; PMID:17349958; http://dx.doi.org/ 10.1016/j.molcel.2007.02.007 [DOI] [PubMed] [Google Scholar]
- 43. Yoshida K, Liu H, Miki Y. Protein kinase C delta regulates Ser46 phosphorylation of p53 tumor suppressor in the apoptotic response to DNA damage. J Biol Chem 2006; 281:5734-40; PMID:16377624; http://dx.doi.org/ 10.1074/jbc.M512074200 [DOI] [PubMed] [Google Scholar]
- 44. Matsumoto M, Furihata M, Ohtsuki Y. Posttranslational phosphorylation of mutant p53 protein in tumor development. Med Mol Morphol 2006; 39:79-87; PMID:16821145; http://dx.doi.org/ 10.1007/s00795-006-0320-0 [DOI] [PubMed] [Google Scholar]
- 45. Lopez-Sanchez I, Valbuena A, Vazquez-Cedeira M, Khadake J, Sanz-Garcia M, Carrillo-Jimenez A, Lazo PA. VRK1 interacts with p53 forming a basal complex that is activated by UV-induced DNA damage. FEBS Lett 2014; 588:692-700; PMID:24492002; http://dx.doi.org/ 10.1016/j.febslet.2014.01.040 [DOI] [PubMed] [Google Scholar]
- 46. Waterman MJ, Stavridi ES, Waterman JL, Halazonetis TD. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat Genet 1998; 19:175-8; PMID:9620776; http://dx.doi.org/ 10.1038/542 [DOI] [PubMed] [Google Scholar]
- 47. Cai X, Liu X. Inhibition of Thr-55 phosphorylation restores p53 nuclear localization and sensitizes cancer cells to DNA damage. Proc Natl Acad Sci U S A 2008; 105:16958-63; PMID:18952844; http://dx.doi.org/ 10.1073/pnas.0804608105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu Y, Lin JC, Piluso LG, Dhahbi JM, Bobadilla S, Spindler SR, Liu X. Phosphorylation of p53 by TAF1 inactivates p53-dependent transcription in the DNA damage response. Mol Cell 2014; 53:63-74; PMID:24289924; http://dx.doi.org/ 10.1016/j.molcel.2013.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shang X, Vasudevan SA, Yu Y, Ge N, Ludwig AD, Wesson CL, Wang K, Burlingame SM, Zhao YJ, Rao PH. Dual-specificity phosphatase 26 is a novel p53 phosphatase and inhibits p53 tumor suppressor functions in human neuroblastoma. Oncogene 2010; 29:4938-46; PMID:20562916; http://dx.doi.org/ 10.1038/onc.2010.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li DW, Liu JP, Schmid PC, Schlosser R, Feng H, Liu WB, Yan Q, Gong L, Sun SM, Deng M, Liu Y. Protein serine/threonine phosphatase-1 dephosphorylates p53 at Ser-15 and Ser-37 to modulate its transcriptional and apoptotic activities. Oncogene 2006; 25:3006-22; PMID:16501611; http://dx.doi.org/ 10.1038/sj.onc.1209334 [DOI] [PubMed] [Google Scholar]
- 51. Mi J, Bolesta E, Brautigan DL, Larner JM. PP2A regulates ionizing radiation-induced apoptosis through Ser46 phosphorylation of p53. Mol Cancer Ther 2009; 8:135-40; PMID:19139122; http://dx.doi.org/ 10.1158/1535-7163.MCT-08-0457 [DOI] [PubMed] [Google Scholar]
- 52. Shaltiel IA, Aprelia M, Saurin AT, Chowdhury D, Kops GJ, Voest EE, Medema RH. Distinct phosphatases antagonize the p53 response in different phases of the cell cycle. Proc Natl Acad Sci U S A 2014; 111:7313-8; PMID:24711418; http://dx.doi.org/ 10.1073/pnas.1322021111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol 2010; 2:a000935; PMID:20679336; http://dx.doi.org/ 10.1101/cshperspect.a000935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, et al. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell 2006; 24:841-51; PMID:17189187; http://dx.doi.org/ 10.1016/j.molcel.2006.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell 2006; 24:827-39; PMID:17189186; http://dx.doi.org/ 10.1016/j.molcel.2006.11.021 [DOI] [PubMed] [Google Scholar]
- 56. Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell 2008; 133:612-26; PMID:18485870; http://dx.doi.org/ 10.1016/j.cell.2008.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 2012; 149:1269-83; PMID:22682249; http://dx.doi.org/ 10.1016/j.cell.2012.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 2001; 107:137-48; PMID:11672522; http://dx.doi.org/ 10.1016/S0092-8674(01)00524-4 [DOI] [PubMed] [Google Scholar]
- 59. Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A 2003; 100:10794-9; PMID:12960381; http://dx.doi.org/ 10.1073/pnas.1934713100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sasca D, Hahnel PS, Szybinski J, Khawaja K, Kriege O, Pante SV, Bullinger L, Strand S, Strand D, Theobald M, et al. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood 2014; 124(1):121-33; PMID:24855208; http://dx.doi.org/ 10.1182/blood-2013-11-538819 [DOI] [PubMed] [Google Scholar]
- 61. Wilking MJ, Singh C, Nihal M, Zhong W, Ahmad N. SIRT1 deacetylase is overexpressed in human melanoma and its small molecule inhibition imparts anti-proliferative response via p53 activation. Arch Biochem Biophys 2014; 563:94-100; PMID:24751483; http://dx.doi.org/ 10.1016/j.abb.2014.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sen N, Kumari R, Singh MI, Das S. HDAC5, a key component in temporal regulation of p53-mediated transactivation in response to genotoxic stress. Mol Cell 2013; 52:406-20; PMID:24120667; http://dx.doi.org/ 10.1016/j.molcel.2013.09.003 [DOI] [PubMed] [Google Scholar]
- 63. Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006; 444:629-32; PMID:17108971; http://dx.doi.org/ 10.1038/nature05287 [DOI] [PubMed] [Google Scholar]
- 64. Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 2007; 27:636-46; PMID:17707234; http://dx.doi.org/ 10.1016/j.molcel.2007.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, Wall D, Li E, Gaudet F. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell 2008; 29:392-400; PMID:18280244; http://dx.doi.org/ 10.1016/j.molcel.2007.12.025 [DOI] [PubMed] [Google Scholar]
- 66. Lehnertz B, Rogalski JC, Schulze FM, Yi L, Lin S, Kast J, Rossi FM. p53-dependent transcription and tumor suppression are not affected in Set7/9-deficient mice. Mol Cell 2011; 43:673-80; PMID:21855805; http://dx.doi.org/ 10.1016/j.molcel.2011.08.006 [DOI] [PubMed] [Google Scholar]
- 67. Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, et al. p53 is regulated by the lysine demethylase LSD1. Nature 2007; 449:105-8; PMID:17805299; http://dx.doi.org/ 10.1038/nature06092 [DOI] [PubMed] [Google Scholar]
- 68. Huang J, Dorsey J, Chuikov S, Perez-Burgos L, Zhang X, Jenuwein T, Reinberg D, Berger SL. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem 2010; 285:9636-41; PMID:20118233; http://dx.doi.org/ 10.1074/jbc.M109.062588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, La Thangue NB. Arginine methylation regulates the p53 response. Nat Cell Biol 2008; 10:1431-9; PMID:19011621; http://dx.doi.org/ 10.1038/ncb1802 [DOI] [PubMed] [Google Scholar]
- 70. Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol 2001; 2:195-201; PMID:11265249; http://dx.doi.org/ 10.1038/35056583 [DOI] [PubMed] [Google Scholar]
- 71. Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 2003; 302:1972-5; PMID:14671306; http://dx.doi.org/ 10.1126/science.1091362 [DOI] [PubMed] [Google Scholar]
- 72. Kruse JP, Gu W. MSL2 promotes Mdm2-independent cytoplasmic localization of p53. J Biol Chem 2009; 284:3250-63; PMID:19033443; http://dx.doi.org/ 10.1074/jbc.M805658200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pei XH, Bai F, Li Z, Smith MD, Whitewolf G, Jin R, Xiong Y. Cytoplasmic CUL9/PARC ubiquitin ligase is a tumor suppressor and promotes p53-dependent apoptosis. Cancer Res 2011; 71:2969-77; PMID:21487039; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Laine A, Ronai Z. Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1. Oncogene 2007; 26:1477-83; PMID:16924229; http://dx.doi.org/ 10.1038/sj.onc.1209924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Le Cam L, Linares LK, Paul C, Julien E, Lacroix M, Hatchi E, Triboulet R, Bossis G, Shmueli A, Rodriguez MS, et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell 2006; 127:775-88; PMID:17110336; http://dx.doi.org/ 10.1016/j.cell.2006.09.031 [DOI] [PubMed] [Google Scholar]
- 76. Melchior F, Hengst L. SUMO-1 and p53. Cell Cycle 2002; 1:245-9; PMID:12429940; http://dx.doi.org/ 10.4161/cc.1.4.131 [DOI] [PubMed] [Google Scholar]
- 77. Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 2004; 118:83-97; PMID:15242646; http://dx.doi.org/ 10.1016/j.cell.2004.06.016 [DOI] [PubMed] [Google Scholar]
- 78. Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. J Biol Chem 2007; 282:1797-804; PMID:17098746; http://dx.doi.org/ 10.1074/jbc.M609001200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zheng H, You H, Zhou XZ, Murray SA, Uchida T, Wulf G, Gu L, Tang X, Lu KP, Xiao ZX. The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature 2002; 419:849-53; PMID:12397361; http://dx.doi.org/ 10.1038/nature01116 [DOI] [PubMed] [Google Scholar]
- 80. Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, Cho JW. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nature Cell Biology 2006; 8:1074-83; PMID:16964247; http://dx.doi.org/ 10.1038/ncb1470 [DOI] [PubMed] [Google Scholar]
- 81. Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, Fukasawa K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol 2007; 9:1175-83; PMID:17891139; http://dx.doi.org/ 10.1038/ncb1638 [DOI] [PubMed] [Google Scholar]
- 82. Samuels-Lev Y, O'Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S, Campargue I, Naumovski L, Crook T, Lu X. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell 2001; 8:781-94; PMID:11684014; http://dx.doi.org/ 10.1016/S1097-2765(01)00367-7 [DOI] [PubMed] [Google Scholar]
- 83. Bergamaschi D, Samuels Y, Sullivan A, Zvelebil M, Breyssens H, Bisso A, Del Sal G, Syed N, Smith P, Gasco M, et al. iASPP preferentially binds p53 proline-rich region and modulates apoptotic function of codon 72-polymorphic p53. Nat Genet 2006; 38:1133-41; PMID:16964264; http://dx.doi.org/ 10.1038/ng1879 [DOI] [PubMed] [Google Scholar]
- 84. Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW. Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell 2007; 130:624-37; PMID:17719541; http://dx.doi.org/ 10.1016/j.cell.2007.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tian C, Xing G, Xie P, Lu K, Nie J, Wang J, Li L, Gao M, Zhang L, He F. KRAB-type zinc-finger protein Apak specifically regulates p53-dependent apoptosis. Nat Cell Biol 2009; 11:580-91; PMID:19377469; http://dx.doi.org/ 10.1038/ncb1864 [DOI] [PubMed] [Google Scholar]
- 86. Imbriano C, Gnesutta N, Mantovani R. The NF-Y/p53 liaison: well beyond repression. Biochimica et biophysica acta 2012; 1825:131-9; PMID:22138487 [DOI] [PubMed] [Google Scholar]
- 87. Mantovani F, Tocco F, Girardini J, Smith P, Gasco M, Lu X, Crook T, Del Sal G. The prolyl isomerase Pin1 orchestrates p53 acetylation and dissociation from the apoptosis inhibitor iASPP. Nat Struct Mol Biol 2007; 14:912-20; PMID:17906639; http://dx.doi.org/ 10.1038/nsmb1306 [DOI] [PubMed] [Google Scholar]
- 88. Chang J, Davis-Dusenbery BN, Kashima R, Jiang X, Marathe N, Sessa R, Louie J, Gu W, Lagna G, Hata A. Acetylation of p53 stimulates miRNA processing and determines cell survival following genotoxic stress. EMBO J 2013; 32:3192-205; PMID:24219989; http://dx.doi.org/ 10.1038/emboj.2013.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shahar OD, Gabizon R, Feine O, Alhadeff R, Ganoth A, Argaman L, Shimshoni E, Friedler A, Goldberg M. acetylation of lysine 382 and phosphorylation of serine 392 in p53 modulate the interaction between p53 and MDC1 in vitro. PloS One 2013; 8:e78472; PMID:24194938; http://dx.doi.org/ 10.1371/journal.pone.0078472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature 2009; 460:529-33; PMID:19626115; http://dx.doi.org/ 10.1038/nature08199 [DOI] [PubMed] [Google Scholar]
- 91. Leveille N, Elkon R, Davalos V, Manoharan V, Hollingworth D, Oude Vrielink J, le Sage C, Melo CA, Horlings HM, Wesseling J, et al. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat Commun 2011; 2:513; PMID:22027593; http://dx.doi.org/ 10.1038/ncomms1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol 2009; 16:23-9; PMID:19079265; http://dx.doi.org/ 10.1038/nsmb.1533 [DOI] [PubMed] [Google Scholar]
- 93. Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A 2008; 105:13421-6; PMID:18755897; http://dx.doi.org/ 10.1073/pnas.0801613105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wang Y, Huang JW, Castella M, Huntsman DG, Taniguchi T. p53 Is Positively Regulated by miR-542-3p. Cancer Res 2014; 74(12):3218-27; PMID:24762395; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yin M, Ren X, Zhang X, Luo Y, Wang G, Huang K, Feng S, Bao X, Huang K, He X, et al. Selective killing of lung cancer cells by miRNA-506 molecule through inhibiting NF-kappaB p65 to evoke reactive oxygen species generation and p53 activation. Oncogene 2014; 1-13 (epub; preprint); PMID:24469051; http://dx.doi.org/ 10.1038/onc.2013.597 [DOI] [PubMed] [Google Scholar]
- 96. Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res 2008; 68:8164-72; PMID:18829576; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-1305 [DOI] [PubMed] [Google Scholar]
- 97. Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev 2009; 23:862-76; PMID:19293287; http://dx.doi.org/ 10.1101/gad.1767609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song JS, Tang LH, Levine AJ, Feng Z. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol Cell 2010; 38:689-99; PMID:20542001; http://dx.doi.org/ 10.1016/j.molcel.2010.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ye D, Wang G, Liu Y, Huang W, Wu M, Zhu S, Jia W, Deng AM, Liu H, Kang J. MiR-138 promotes induced pluripotent stem cell generation through the regulation of the p53 signaling. Stem Cells 2012; 30:1645-54; PMID:22696098; http://dx.doi.org/ 10.1002/stem.1149 [DOI] [PubMed] [Google Scholar]
- 100. Kumar M, Lu Z, Takwi AA, Chen W, Callander NS, Ramos KS, Young KH, Li Y. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene 2011; 30:843-53; PMID:20935678; http://dx.doi.org/ 10.1038/onc.2010.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Fan Y, Yin S, Hao Y, Yang J, Zhang H, Sun C, Ma M, Chang Q, Xi JJ. miR-19b promotes tumor growth and metastasis via targeting TP53. Rna 2014; 20:765-72; PMID:24742936; http://dx.doi.org/ 10.1261/rna.043026.113 [DOI] [PMC free article] [PubMed] [Google Scholar]


