Abstract
We prospectively studied the feasibility and effectiveness of sputum induction in obtaining good quality sputum and its subsequent bacterial yield in children with clinically suspected acute lower-respiratory-tract infection (aLRTI). Good quality sputum was collected in 89/98 (91%) patients. Sputum cultures revealed ≥1 bacterial pathogens in 22 cases (25%). Adverse events were infrequent and mild (6%). Sputum induction is feasible in young children and leads to an increased number of etiological diagnoses of aLRTI.
Keywords: child, sputum induction, acute lower respiratory tract infection, bacterial pathogens, thoracic X-ray
Introduction
Microbiological confirmation of bacterial infection is rarely achieved in children with acute lower-respiratory-tract infections (aLRTIs) because of the inability to obtain material from the infection site (ie, the lung).1 Treatment is, therefore, mostly empirical, which irrevocably leads to overtreatment and sometimes undertreatment. Sputum induction with nebulized hypertonic saline can help adequately produce sputum in children who are otherwise unable to do so. Sputum induction is routinely used to obtain samples for microbial cultures in pediatric illnesses such as cystic fibrosis and tuberculosis.2,3 It may also be useful in establishing a causative diagnosis of aLRTI in otherwise healthy children. Reported success rates for producing good quality sputum vary considerably.4-6 The purpose of this study was to determine the effectiveness of sputum induction in obtaining good quality sputum in children with clinically suspected aLRTI in the everyday, busy general hospital setting.
Materials and Methods
Study Design
A prospective study in 2 large regional hospitals (Jeroen Bosch Hospital and Máxima Medical Centre) in the Netherlands was conducted between October 2009 and October 2011. Children aged 6 months to 18 years presenting with suspected aLRTI were eligible for inclusion. Suspected aLRTI was a clinical diagnosis of the attending physician, with tachypnea and ≥1 of the following symptoms: dyspnea, fever, cough, and/or abdominal pain. Exclusion criteria for participation were a recent severe asthma exacerbation, oxygen saturation ≤92%, anatomical airway abnormalities, and the use of β-blockers or diuretics. The study protocol was reviewed and approved by the local independent ethics committees.
Study Procedures and Specimen Collection
Blood inflammation parameters (ie, leukocyte count and C-reactive protein [CRP] level), blood culture, and chest X-ray were performed before initiation of the study procedure as part of routine diagnostics. Two radiologists retrospectively assessed the presence of pneumonic infiltrates.
Patients were pretreated before sputum induction with inhaled salbutamol to prevent bronchoconstriction. Sputum induction was performed with hypertonic saline (NaCl 5.8%) inhalation using a nebulizing device with oxygen flow. When a patient was unable to expectorate sputum after induction, a sterile suction catheter was used to obtain secretions from the oropharynx. The sputum quality was determined microscopically before culture. Specimens were considered of good quality if <25 squamous epithelial cells and >25 leukocytes were present per low-power field (10x lens objective).7 Only good quality sputum specimens were used for microbiological diagnostics. Sputum samples were cultured for 48 hours using routine microbiological procedures. A concomitant nasopharyngeal lavage sample was also obtained and used for routine bacterial culture. Also, infection with influenza A, influenza B, respiratory syncytial virus (RSV), and human metapneumovirus (hMPV) was routinely investigated by polymerase chain reaction on nasopharyngeal specimens, if considered necessary by the treating physician, taking into account anticipated seasonal circulation.
Statistical Analysis
Categorical data were compared using either χ2 or Fisher exact test. Continuous data were first analyzed for normality using stem-and-leaf plots and quantile-quantile plots, after which either a t-test or Mann-Whitney U test for nonparametric data was performed. To assess the agreement between sputum and nasopharyngeal bacterial cultures, a simple κ coefficient was calculated. All statistical tests were 2-tailed, and P < .05 was considered statistically significant. SPSS (version 22.0; IBM Corp, Armonk, NY) was used for all statistical analyses.
Results
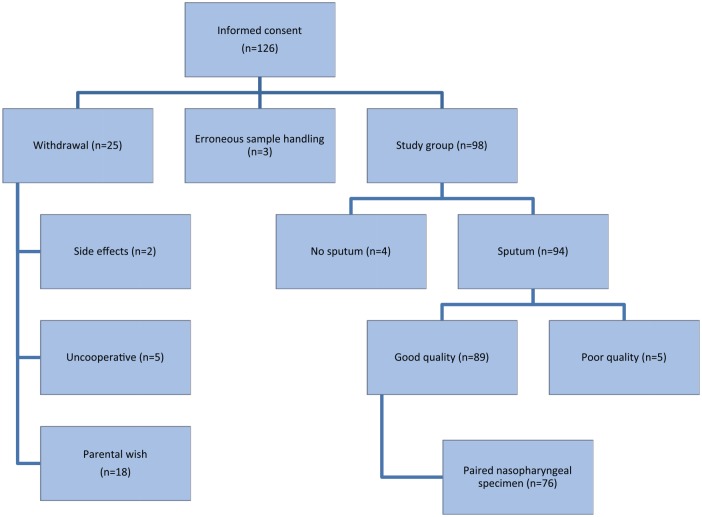
Informed consent was obtained for 126 patients with suspected aLRTI, but 28 patients (22%) did not complete the study procedure and were excluded in the analysis of the microbiological sputum data (Figure 1). In 3 patients, erroneous sputum sample handling resulted in exclusion. The final study group consisted of 98 patients, with a median age of 2.6 years (range = 0.5-16.8 years). The study and drop-out groups were not significantly different in terms of age (P = .694), sex (P = .365), leukocyte count (P = .725), CRP level (P = .444), or radiological diagnosis (P = .289). None of the blood cultures (n = 112) showed associated bacteremia.
Figure 1.
Flow diagram of the study group and sputum samples.
In 89/98 (91%) patients, a good quality sputum sample was obtained. Only 4 patients were unable to produce sputum after completing the induction procedure; 5 samples were of poor quality and were, therefore, not processed (Figure 1). Mild adverse reactions were reported in 6 patients: cough (n = 3), vomiting (n = 1), wheezing (n = 1), transiently decreased oxygen saturation (n = 1), and mild epistaxis (n = 1). No treatment was needed for the adverse reactions; no serious adverse events were reported. However, children often experienced the sputum collection procedure as unpleasant.
Bacterial pathogens were isolated in 22/89 sputum cultures (25%); 13 samples grew only 1 pathogen: Haemophillus influenzae (n = 8), Streptococcus pneumoniae (n = 1), Staphylococcus aureus (n = 2), Moraxella catarrhalis (n = 1), Sphingomonas paucimobilis (n = 1). In 9 samples, 2 bacterial pathogens were isolated: H influenzae + M catarrhalis (n = 3), H influenzae + Streptococcus pneumoniae (n = 5), and Streptococcus pyogenes + Pseudomonas aeruginosa (n = 1). The detection of bacteria in sputum samples was similar in patients with and without radiologically confirmed pneumonia (24% vs 27%, P = .483).
When comparing the detection of bacteria in sputum cultures with their concomitant nasopharyngeal bacterial cultures (n = 76), different pathogens were frequently found. In 15 sputum samples, a bacterial pathogen was also found in the concomitant nasopharyngeal culture; 19 nasopharyngeal cultures yielded bacterial pathogens that were not found in concomitant sputum cultures. In 6 sputum samples, the bacterial pathogen was only detected in sputum and not in the concomitant nasopharyngeal aspirate: Moraxella catarrhalis (n = 3), Haemophilus influenzae (n = 4), and Sphingomonas paucimobilis (n = 1). No significant associations between the sputum and nasopharyngeal bacterial yield were found, with the exception of Haemophilus influenzae (κ = 0.538).
Viral (co)infections (RSV, hMPV, influenza A, and influenza B) were found in 35/75 (47%) nasopharyngeal specimens. The viral presence did not differ between the bacterial sputum culture–positive and –negative groups (P = .484) or between the children with and without radiologically confirmed pneumonia (P = .87).
Discussion
This study shows that good quality sputum samples could be obtained by an induced sputum collection procedure in our everyday, busy general hospital setting in 91% of children with clinically suspected aLRTI. Our success rate is significantly higher than previously reported, using similar definitions of sputum quality.4,5 Bacterial pathogens were found in 25% of induced sputum samples. Not unexpectedly, Haemophilus influenzae, Streptococcus pneumoniae, and Moraxella catarrhalis were found most frequently. In 29% of positive sputum cultures, the bacterial pathogen was found only in sputum and not in the concomitant nasopharyngeal aspirate culture, suggesting its origin from the lower respiratory tract. These findings support our hypothesis that sputum induction is useful to support etiological diagnoses in every day practice because its yield is significantly higher when compared with blood culture or acute phase serology.8
The relatively low percentage of bacterial pathogens detected from induced sputum samples is probably a result of the fact that many aLRTIs were in fact viral infections, as is common in young children. Induced sputum not only identifies bacterial pathogens but can also help exclude bacterial infections and, thus, reduce overtreatment with antibiotics. This can help minimize the development of resistance at a population level.
When comparing our results with those of a similar study performed by Mermond et al,5 there is a difference in bacterial sputum yield (25% vs 50%). However, their good quality sputum yield was significantly lower than ours (91% vs 25%), which was attributed by the authors to methodological problems. This resulted in a sputum culture group of only 26 patients. Another study by Lahti et al4 found evidence for a bacterial infection from induced sputum in 79% of cases but only included patients with radiologically confirmed pneumonia. In our cohort, we found no difference between the presence of common viral and/or bacterial pathogens and radiological diagnosis. Because the use of radiology for the diagnosis of aLRTIs has been a major point of discussion,9 study inclusion based on clinical suspicion of aLRTI probably gives a more realistic reflection of everyday practice.
Even though we found no significant difference in baseline characteristics between the groups, the drop-out percentage after inclusion (22%) is a limitation of our study, but is similar to that in the study by Lahti et al,4 a study that is currently seen as a justification of sputum induction in aLRTIs.
Another limitation of our study is that we cannot rule out the possibility that, occasionally, the induced sputum did not reflect the etiology of the aLRTI because we did not compare the induced sputum cultures with material retrieved directly from the lungs by bronchoalveolar lavage or thoracocentesis, which would obviously have been unethical to perform in these non–critically ill children. The majority of bacteria found in sputum cultures were also present in the concomitant nasopharyngeal cultures. This could either mean that the sputum sample was contaminated while passing the upper-respiratory tract during expectoration or that an invasive infection by a colonizing bacterial pathogen had taken place, which is a known phenomenon in respiratory tract infections.10 It is likely that many bacteria cultured from induced sputum during an acute respiratory tract infection play a causative role.
In conclusion, we found that sputum induction in children with suspected aLRTIs in a general hospital setting provides good quality sputum in most cases, with infrequent and only mild adverse events. It can be a useful tool for the general pediatrician to promote pathogen-based treatment of aLRTIs in everyday clinical practice.
Author Contributions
IYB contributed to the conception and design; contributed to acquisition, analysis, and interpretation of data; drafted the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy.
MM contributed to the acquisition and interpretation of data; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy.
RvG contributed to acquisition of data; critically revised the analysis and first drafts of the manuscript; could not give final approval (deceased). The senior author (de Vries) accounts for his approval.
MHvL contributed to acquisition and interpretation of data; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy.
MH contributed to analysis of data; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy.
AW contributed to the conception and design; contributed to interpretation; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy.
PCW contributed to the conception and design; contributed to interpretation; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy.
EdV contributed to the conception and design; contributed to acquisition, analysis, and interpretation of data; codrafted the manuscript; critically revised the manuscript; gave final approval; and agrees to be accountable for all aspects of work ensuring integrity and accuracy.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The Jeroen Bosch Hospital funded this study.
References
- 1. Hammitt LL, Murdoch DR, Scott JAG, et al. Specimen collection for the diagnosis of pediatric pneumonia. Clin Infect Dis. 2012;54(suppl 2):S132-S139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Al-Saleh S, Dell SD, Grasemann H, et al. Sputum induction in routine clinical care of children with cystic fibrosis. J Pediatr. 2010;157:1006-1011.e1. [DOI] [PubMed] [Google Scholar]
- 3. Planting NS, Visser GL, Nicol MP, Workman L, Isaacs W, Zar HJ. Safety and efficacy of induced sputum in young children hospitalised with suspected pulmonary tuberculosis. Int J Tuberc Lung Dis. 2014;18:8-12. [DOI] [PubMed] [Google Scholar]
- 4. Lahti E, Peltola V, Waris M, et al. Induced sputum in the diagnosis of childhood community-acquired pneumonia. Thorax. 2009;64:252-257. [DOI] [PubMed] [Google Scholar]
- 5. Mermond S, Zurawski V, D’Ortenzio E, et al. Lower respiratory infections among hospitalized children in New Caledonia: a pilot study for the Pneumonia Etiology Research for Child Health Project. Clin Infect Dis. 2012;54(suppl 2):S180-S189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zar HJ, Tannenbaum E, Hanslo D, Hussey G. Sputum induction as a diagnostic tool for community-acquired pneumonia in infants and young children from a high HIV prevalence area. Pediatr Pulmonol. 2003;36:58-62. [DOI] [PubMed] [Google Scholar]
- 7. Geckler RW, Gremillion DH, McAllister CK, Ellenbogen C. Microscopic and bacteriological comparison of paired sputa and transtracheal aspirates. J Clin Microbiol. 1977;6:396-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cevey-Macherel M, Galetto-Lacour A, Gervaix A, et al. Etiology of community-acquired pneumonia in hospitalized children based on WHO clinical guidelines. Eur J Pediatr. 2009;168:1429-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lynch T, Bialy L, Kellner JD, et al. A systematic review on the diagnosis of pediatric bacterial pneumonia: when gold is bronze. PLoS One. 2010;5:e11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ghaffar F, Friedland IR, McCracken GH. Dynamics of nasopharyngeal colonization by Streptococcus pneumoniae. Pediatr Infect Dis J. 1999;18:638-646. [DOI] [PubMed] [Google Scholar]