Abstract
Telomeres are nucleoprotein complexes that protect the natural ends of chromosomes from fusion and degradation and prevent them eliciting a checkpoint response. This protective function, which is called telomere capping, is largely mediated by telomere-binding proteins that suppress checkpoint activation and DNA repair activities. Telomere dysfunction through progressive shortening or removal of capping proteins leads to a checkpoint-mediated block of cell proliferation, which acts as a cancer-suppressor mechanism. However, genetic alterations that inactivate the checkpoint can lead to further telomere erosion and increased genomic instability that, coupled with the activation of mechanisms to restabilize telomeres, can drive the oncogenic process.
Keywords: cancer, checkpoint, double-strand breaks, genomic stability, telomere
Introduction
The natural ends of chromosomes must be distinguished from intrachromosomal DNA double-strand breaks (DSBs), which activate a DNA damage response (DDR) including checkpoint-mediated cell cycle arrest and DNA repair/recombination pathways. Protection of chromosome ends, referred to as capping, is achieved by packaging into protective structures called telomeres.1,2 A number of proteins bind the telomeric DNA and protect it from fusion, degradation, and recognition as a DSB that would otherwise lead to chromosome instability and cell death.
In most eukaryotes, telomeric DNA is highly repetitive and consists of short TG-rich nucleotide repeats in the strand running 5′ to 3′ toward the chromosome end. In addition to double-stranded telomeric DNA, the 3’ end (G-strand) extends beyond its complementary strand (C-strand) to form a single-stranded overhang called the G-tail.2
Most telomeric DNA is replicated by standard semiconservative DNA replication. As DNA polymerases replicate DNA only in the 5′ to 3′ direction and need a primer to initiate DNA synthesis, removal of the terminal RNA primer at the 5′ ends of newly replicated strands leaves a gap that cannot be filled in by the canonical DNA replication machinery. As a result, telomeric DNA sequences become shorter with each round of DNA replication. In most eukaryotes, this loss of telomeric DNA is counteracted by a ribonucleoprotein enzyme called telomerase, which uses its RNA component as a template to add telomere repeats at the telomeric 3′ overhang in a reverse transcriptase reaction.3 In mammalian cells, the minimal catalytic core of telomerase consists of the telomerase reverse transcriptase (TERT) and the telomerase RNA (TERC). In Saccharomyces cerevisiae, the telomerase consists of the reverse transcriptase Est2, the template RNA TLC1, and two accessory proteins Est1 and Est3.2
Telomeres can function both as tumor suppressors by limiting the number of cell divisions and as tumor promoters by inducing genome instability. Although telomerase is continuously expressed in unicellular eukaryotes, its expression is downregulated in most human somatic tissues.4 The inability of the replication machinery to fully replicate DNA ends, coupled with low/absent telomerase activity, results in progressive telomere shortening that causes cells to stop dividing in a process called replicative senescence.5,6 This phenomenon was first described by Hayflick and Moorhead, who reported that cultured human fibroblasts undergo only a limited number of cell divisions and then stop dividing.7 Similarly, elimination of telomerase in S. cerevisiae leads to progressive telomere shortening and loss of viability within approximately 60–80 population doublings, indicating that replicative senescence also occurs in this unicellular eukaryote.8 In both organisms, loss of telomeric DNA repeats results in telomere uncapping that activates a DNA damage checkpoint response, which in turn leads to cell cycle arrest and senescence.9-13 In the absence of other genetic changes, these cells can remain in a quiescent state that essentially functions as an anticancer mechanism for long-lived species like humans. However, genetic alterations that cause a failure to activate the checkpoint response may allow additional cell divisions, during which the dysfunctional telomeres continue to erode until they eventually become too short to protect the chromosome termini from unscheduled DNA repair events. The cells then enter a period called “crisis”, during which the chromosomes ends undergo end-to-end fusion events and enter a breakage-bridge fusion cycle that leads to genomic instability. Although cells entering this state can be eliminated by apoptosis, most cancer cells exhibit upregulation of telomerase activity, indicating that rare surviving cells can avert senescence and crisis by restabilizing telomeres.14,15 In this review, we discuss the mechanisms by which telomeres are capped in S. cerevisiae and mammals and the consequences of telomere uncapping for genome stability and carcinogenesis.
The Cellular Response to a DSB
The presence of telomeres distinguishes natural chromosome ends from internal chromosome breaks, which are potent stimulators of the DDR. The DDR comprises pathways that repair DNA lesions and surveillance mechanisms called DNA damage checkpoints that inhibit cell cycle progression until DSBs are repaired.16 Repair of DNA DSBs can occur by either non-homologous end joining (NHEJ), which results in ligation of the broken ends, or homologous recombination (HR), which uses a homologous DNA sequence to restore the genetic information lost at the break site. The choice of pathway for DSB repair is regulated during the cell cycle, with HR occurring preferentially during S/G2. This cell cycle specificity is mediated by cyclin-dependent kinases (Cdks), which promote the nucleolytic degradation (resection) of the 5′ DSB ends to yield 3′-ended single-stranded DNA (ssDNA) tails that initiate HR and concomitantly inhibit NHEJ.17
When a DSB occurs, the MRX/MRN complex (Mre11-Rad50-Xrs2 in yeast; Mre11-Rad50-Nbs1 in mammals) and the Ku70/Ku80 heterodimer (hereafter referred to as Ku) are recruited to the break site. Ku allows NHEJ by promoting the binding of NHEJ factors18 and by protecting the DSB ends from degradation during the G1 phase of the cell cycle.19 The MRX/MRN complex, in combination with the Sae2/CtIP protein, is important for initiating resection of the DSB ends to generate 3′-ended ssDNA overhangs that direct repair toward HR.20 Subsequent extensive nucleolytic degradation of the DSB ends is performed by two pathways, one of which is dependent on the 5′-3′ exonuclease Exo1, while the other depends on the nuclease/helicase Dna2 acting in conjunction with the RecQ helicase Sgs1/BLM.21,22
DSBs also elicit a checkpoint response; key players in this response include the protein kinases Mec1 and Tel1 in S. cerevisiae, or the respective mammalian orthologs ATR and ATM.16 Tel1 or ATM is recruited to unprocessed or minimally processed DSBs via the MRX/MRN complex.23 Upon generation of ssDNA, the replication protein A (RPA) complex binds and promotes recruitment of Mec1/ATR.24 Mec1/ATR kinase activity is additionally boosted by the heterotrimeric 9-1-1 complex (Rad17, Mec3, and Ddc1 in S. cerevisiae), which is loaded onto RPA-coated ssDNA by the Rad24-RFC complex.16 Once Tel1/ATM and/or Mec1/ATR are activated by damaged DNA, their checkpoint signals are propagated through the protein kinases Rad53 and Chk1 (Chk2 and Chk1 in mammals, respectively), whose activation requires conserved mediator proteins, including the BRCT domain containing protein Rad9 and its metazoan ortholog 53BP1.16 In turn, both S. cerevisiae Rad9 and human 53BP1 limit the generation of ssDNA at the DSB ends by acting as barriers toward end-processing enzymes.25-27
Once recruited to DSBs, Tel1/ATM facilitates the activation of Mec1/ATR,28-30 possibly by facilitating generation of the ssDNA that transforms the DSB ends from Tel1/ATM substrates to Mec1/ATR substrates.30,31 In turn, S. cerevisiae Mec1/ATR inhibits the generation of ssDNA at DSBs, thus preventing excessive nucleolytic degradation of the DSB ends.32,33 As the single-stranded 3′ overhangs increase in length, Mec1/ATR activation is coupled with ssDNA-dependent loss of Tel1/ATM activation, and this resection-mediated switch from Tel1- to Mec1-dependent signaling activity ensures proper termination of the checkpoint response.33
Capping of Chromosome Ends
The ends of eukaryotic chromosomes are not subjected to DNA repair events and do not activate the DDR, despite being physical DNA ends. Several studies on S. cerevisiae and mammals have revealed that protein complexes with specificity for double-stranded and single-stranded telomeric DNA prevent the natural chromosome ends from being recognized as intrachromosomal DSBs (Fig. 1).1,34 In S. cerevisiae, the telomeric ssDNA is bound by Cdc13, which can be found with Stn1 and Ten1 in a heterotrimeric complex called CST. CST protects telomeric DNA from degradation; exposure of cells harboring cdc13, stn1, or ten1 conditional alleles to restrictive conditions causes telomere degradation by progressive resection of the 5′-ended strand and a checkpoint-dependent cell cycle arrest.2 The 5′-3′ exonuclease Exo1 appears to be the major nuclease that degrades telomeres in cdc13 mutants.35 As Cdc13 binding to the single-stranded telomeric DNA reduces the association of Mec1 with these DNA ends36 and the CST complex bears a structural resemblance to the RPA complex,37 CST binding to the telomere has been proposed to prevent RPA recruitment and subsequent Mec1 activation. A CST-like complex (Ctc1-Stn1-Ten1) has recently been identified in S. pombe, plants, and humans and shown to perform a similar function in telomere capping and telomerase regulation.38

Figure 1. Capping activities at S. cerevisiae and human telomeres. In S. cerevisiae, Rap1 and Rif2 inhibit 5′-3′ resection of the telomeric DNA ends and also repress the NHEJ repair pathway. Rif1 supports the function of the CST complex in preventing excessive resection at telomeric ends and Mec1-dependent checkpoint activation. In humans, TRF2 represses ATM signaling and the NHEJ pathway, whereas POT1 prevents ATR activation by inhibiting the binding of the ssDNA-binding protein RPA. POT1 and RAP1 block HR. ATM, ataxia telangiectasia mutated; ATR, ataxia telangiectasia and Rad3-related protein; CST, Cdc13, Stn1, and Ten1; HR, homologous recombination; NHEJ, non-homologous end joining; RPA, replication protein A.
Double-stranded telomeric DNA in S. cerevisiae is bound by the Rap1-Rif1-Rif2 complex. Loss of function of this complex has less catastrophic consequences than CST inactivation; dysfunction of Rap1 or Rif2 leads to increased amounts of telomeric ssDNA and NHEJ-mediated fusion events.39-41 Generation of telomeric ssDNA in cells defective for Rif2 or Rap1 requires the MRX complex,40 suggesting that Rap1 and Rif2 prevent resection at telomeric ends by interfering with the association between MRX and telomeres. On the other hand, inactivation of Rap1 or Rif2 does not leads to checkpoint activation,40,41 suggesting that the exposed telomeric ssDNA is still covered by Cdc13, which limits association of Mec1 with telomeres. Unlike Rif2 and Rap1, Rif1 is not involved in the prevention of telomeric fusions by NHEJ39 and plays a very minor role in protecting telomeres from degradation.40,42 Instead, Rif1 prevents short telomeric ends from activating checkpoint-mediated cell cycle arrest by inhibiting the recruitment of checkpoint proteins to these ends.42,43 Furthermore, it plays a unique role in supporting cell viability43,44 and prevents nucleolytic degradation in mutants defective in the CST complex.44 Interestingly, both CST and Rif1 physically and genetically interact with components of the polα-primase complex,44,45 raising the possibility that Rif1 might promote the ability of CST to fill in the exposed telomeric ssDNA through activation/recruitment of the lagging-strand DNA replication machinery.
Degradation of telomeric DNA is also counteracted by the Ku complex,2 which acts in a different pathway from Rif2.40 In fact, while MRX is primarily responsible for nucleolytic degradation of telomeres in rif2∆ cells,40,41 Exo1 is the nuclease that degrades telomeric DNA in yku70∆ G1 cells.35 Interestingly, Ku protects telomeres in the G1 phase of the cell cycle, when the protective function of CST is dispensable.41
In vertebrates, telomeres are protected from eliciting the DDR and undergoing degradation or fusion events by a specialized group of proteins collectively called shelterin, which includes TRF1, TRF2, RAP1, TIN2, TPP1, and POT1 (Fig. 1).1 Although the shelterin complex represents a functional unit, the individual components have specific protective functions.46 Inactivation of TRF2 in mouse embryo fibroblasts by either gene deletion or overexpression of a dominant-negative variant causes activation of ATM,47,48 as well as accumulation of telomere-induced foci formed by DDR factors such as 53BP1, MRN, ATM, and the histone variant γH2AX.11,49 Moreover, TRF2 also protects telomeres from NHEJ-mediated fusion events.48 In mammals, the telomeric single-stranded overhang can fold back on the double-stranded part of the telomere to form a lariat structure, called a t-loop, which is predicted to prevent the binding of DNA repair/checkpoint proteins to the telomeric DNA. As TRF2 is required for the formation and/or maintenance of t-loops,50 TRF2-dependent remodeling of telomeres into t-loop structures might explain how TRF2 represses NHEJ and ATM signaling at telomeres.
Repression of ATR is performed by POT1 in humans and POT1a in mice.51,52 Based on the finding that POT1 specifically recognizes the telomeric ssDNA overhangs, it has been proposed that POT1 and POT1a block ATR activation by preventing RPA binding to the telomeric ssDNA.53 This switch from RPA to POT1 on telomeric ssDNA is promoted by the heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) protein through a mechanism that is regulated by telomeric repeat containing RNA (TERRA), a non-coding RNA that is transcribed from the telomeric C-rich strand.54 Inhibition of RPA binding by POT1 may also play an important role in preventing HR, which is repressed at telomeres in a redundant manner by POT1 and RAP1.52,55
Even when mouse telomeres are stripped of shelterin and Ku, complete loss of telomeric DNA does not occur unless the DNA damage response protein 53BP1 is also depleted,56 suggesting that multiple pathways act in a highly redundant manner to block telomere degradation.
Consequences of Telomere Uncapping
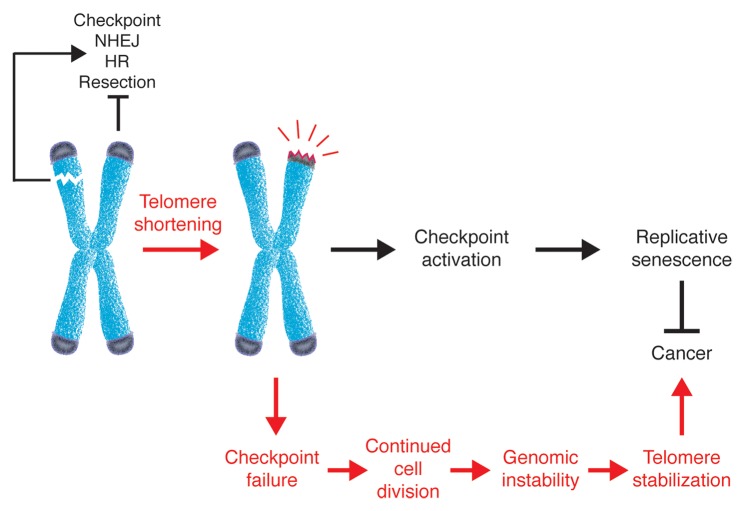
A major focus in telomere biology is to understand the structural changes occurring in the switch from a protected to a deprotected state. Telomere deprotection can occur by either altering the activities of proteins with capping functions or by erosion of the telomeric DNA that occurs spontaneously in human cells with each cell division. Both conditions lead to dramatic changes in the epigenetic pattern and the nucleosome organization.57 Furthermore, both elicit a DNA damage checkpoint response that leads to cell cycle arrest (Fig. 2).9-13,46 However, although depletion of shelterin leads to acute deprotection that results in telomere fusions and rampant genome instability, the checkpoint elicited by critically short telomeres in aged human cells initiates replicative senescence, during which telomeres accumulate γH2A and 53BP1 but do not undergo fusion events.58 To explain this difference, a three-state model of chromosome protection has been proposed.59 In this model, the “closed” state corresponds to a shelterin- and length-dependent structure that protects chromosome ends from eliciting the DDR. Telomere shortening disrupts this structure leading to an “intermediate” state, in which telomeres are recognized as DNA damage and induce the DDR, but retain sufficient shelterin proteins to prevent NHEJ-mediated fusions. Further shortening results in an “uncapped” state, in which the amount of shelterin is not sufficient to prevent chromosome end-to-end fusions. In agreement with the existence of an intermediate state, telomeres from senescent cells retain sufficient telomeric DNA to bind TRF2 and confer protection from NHEJ.58,60 In addition, telomere fusions only occur in cells that have bypassed senescence as a result of checkpoint deactivation.58 Interestingly, it has recently been shown that two distinct regions within TRF2 are required to inhibit ATM signaling and NHEJ, respectively.61 While ATM is inhibited by the TRFH dimerization domain, NHEJ repression depends on the C-terminal region of the hinge domain, which acts by inhibiting the ubiquitin ligase RNF168 that is necessary for the recruitment of 53BP1.
Figure 2. Telomere deprotection in carcinogenesis. Although intrachromomal double-strand breaks (DSBs) elicit a checkpoint response and can be repaired, the natural chromosome ends are protected from fusion and degradation and do not activate the checkpoint. This protective function, referred to as capping, is due to proteins that bind the telomeric DNA. Loss of capping due to either deficiences in capping proteins or loss of telomeric DNA induces a DNA damage checkpoint response that leads to cell cycle arrest and senescence, thus providing a potent anticancer mechanism. However, rare failure to activate the checkpoint may allow cells to undergo cell divisions during which uncapped telomeres can be subjected to unscheduled DNA repair events. The resulting genomic instability, coupled with activation of telomere restabilizing mechanisms, can drive the oncogenic process.
The signal(s) that trigger the checkpoint response in senescent cells are far from being fully understood. In mammals, senescence is not determined by the average telomere length, but by the presence of telomeres that are short enough to trigger the DDR.62 Analysis of dysfunctional telomeres with increased population doubling in a variety of human primary cells has shown that the threshold of DNA damage signaling corresponds to an average of five dysfunctional telomeres.58 Critically short telomeres are also the major determinant of senescence onset in S. cerevisiae, in which the artificial shortening of a single telomere is sufficient to accelerate the onset of senescence in cells lacking telomerase activity.63
Analysis of telomeres of different lengths in S. cerevisiae cells deprived of telomerase activity has shown that 5′-3′ resection is stimulated at the shortest telomeres,64 suggesting that an excessive amount of ssDNA at telomeres could be the signal triggering checkpoint activation and senescence initiation. Consistent with this hypothesis, replicative senescence is regulated by proteins that modulate generation of ssDNA at the DSB ends.64,65 In fact, the onset of senescence is delayed when the MRX complex, which is known to transiently increase the generation of ssDNA at telomeres, is defective.64-66 In contrast, senescence is accelerated in the absence of Rif2,65 which is known to inhibit MRX-mediated resection at telomeres by competing with Tel1 for binding to Xrs2.67,68 Finally, this severe senescence phenotype is reversed by the loss of MRX or Tel1.65 Together, these data suggest that short telomeres become more accessible to the action of nucleases and thus accumulate ssDNA that activates the checkpoint.
Moreover, telomeres might have a role in senescence establishment independent from their shortening/uncapping functions. Telomeric DNA has been shown to resist DNA damage repair and trigger persistent DDR, possibly through the inability to repair DNA lesions.60,69 These data raise the possibility that telomeres might act as sinks of DNA damage that are capable of triggering senescence in nonproliferating tissues.
Telomere Uncapping and Cancer
The occurrence of telomere erosion in cancer is evidenced by typically short telomeres in most human cancers relative to normal tissues.70 Although telomerase is constitutively expressed in unicellular eukaryotes, its expression is downregulated in most somatic human tissues, resulting in progressive reduction of telomere repeats at each DNA replication cycle. When telomeres become critically short, cells can undergo one of two opposing fates (Fig. 2). Dysfunctional telomeres can activate a DNA damage checkpoint response that causes cell cycle arrest and entry into senescence, thus preventing cell proliferation.11,13,49 This limit to the number of cell divisions represents an important barrier against cancer because it reduces the risk of accumulating mutations that could lead to malignant transformation. On the other hand, rare cells that have inactivated the checkpoint response overcome the senescence barrier and continue to divide, resulting in total loss of telomere function. Continued telomere erosion could result in high genomic instability that generally results in cell death and therefore constrains full malignant progression. However, the very few cells capable of surviving this period can display a procarcinogenic mutator phenotype.71-73 Thus, although telomere shortening and repression of telomerase can represent protective mechanisms against cancer development, the associated genome instability has a cancer promoting activity and the stepwise removal of telomere protection provides a mechanism to block cell division before excessive telomere shortening endangers genome stability.59
Indeed, experiments to measure genome stability during telomere shortening demonstrate that S. cerevisiae cells lacking telomerase exhibit increased chromosome rearrangements compared with telomerase-positive cells, with sequence losses being more frequent at the ends of the chromosomes than at internal sites.74 Furthermore, telomere dysfunction in aging telomerase-deficient p53 mutant mice promotes the development of epithelial cancers by a process of fusion-bridge breakage.71 Finally, high-resolution analysis of telomere length at different stages of the hematologic malignancy chronic lymphocytic leukemia have shown that telomere erosion and fusion events can be detected in the early stages of the disease and strongly correlate with disease progression.75
Although tumors may arise from cells with eroded telomeres, indicating that telomere dysfunction can drive early stages of cancer development, almost all human cancer cells acquire mechanisms to elongate their telomeres and overcome senescence, suggesting that subsequent restoration of telomere function is critical for malignant progression. Most human cancers maintain their telomeres through reactivation of telomerase but on rare occasions telomeres can be elongated through a homologous recombination-mediated process called alternative lengthening of telomeres (ALT).76 The existence of a telomerase-independent telomere maintenance mechanism was first demonstrated in telomerase-negative S. cerevisiae cells.77 This mechanism was found to be dependent on HR, although other mechanisms have been identified.78
Supporting the view that telomerase serves as an active driver of cancer progression in cells displaying telomere-based genome instability, expression of an engineered inducible version of telomerase in mouse models of prostate cancer and T-cell lymphoma leads to aggressive cancers with rearranged genomes and new tumor biologic capabilities (i.e., bone metastases).79,80 Upon subsequent telomerase extinction tumor growth eventually slows, suggesting that tumor progression is constrained by the lack of telomerase.80 However, growth can subsequently resume as tumors activate ALT and other adaptive responses,80 suggesting that therapies targeting telomere maintenance in cancer should encompass inhibitors of telomerase and inhibitors of ALT.
Conclusions
There is mounting evidence for the existence of important links between telomeres and cancer. Uncapping of telomeres as a result of repressed telomerase activity in humans represents a protection mechanism against cancer because it activates the DDR that arrests the cell cycle and initiates senescence. However, proliferation beyond this limit may drive genomic instability that, coupled with the activation of telomere maintenance mechanisms, can lead to malignant transformation. An outstanding gap in our knowledge is the role of telomerase activation in the suppression of genome instability. If telomerase activation provides stability to the genome of a cancer cell, telomerase-based therapies might be effective strategies to limit tumor progression specifically in cancer cells. However, as tumor cells lacking telomerase might activate alternative pathways to maintain their telomeres and gain immortal growth capacity, development of successful therapeutic strategies targeting telomere maintenance in cancers requires a deep understanding of how these mechanisms are activated.
Glossary
Abbreviations:
- ALT
alternative lengthening of telomeres
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related protein
- DDR
DNA damage response
- DSB
double-strand break
- HR
homologous recombination
- NHEJ
non-homologous end joining
- RPA
replication protein A
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Giovanna Lucchini for critical reading of the manuscript. Work in the laboratory of M.P.L. is supported by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC) (grant IG11407) and Cofinanziamento 2010–2011 MIUR/Università di Milano-Bicocca. C.T. was supported by a fellowship from Fondazione Italiana Ricerca sul Cancro (FIRC). We apologize to all authors whose publications have not been cited because of length limitations.
References
- 1.de Lange T. . How telomeres solve the end-protection problem. Science 2009; 326:948 - 52; http://dx.doi.org/ 10.1126/science.1170633; PMID: 19965504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wellinger RJ, Zakian VA. . Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics 2012; 191:1073 - 105; http://dx.doi.org/ 10.1534/genetics.111.137851; PMID: 22879408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greider CW, Blackburn EH. . Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985; 43:405 - 13; http://dx.doi.org/ 10.1016/0092-8674(85)90170-9; PMID: 3907856 [DOI] [PubMed] [Google Scholar]
- 4.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. . Specific association of human telomerase activity with immortal cells and cancer. Science 1994; 266:2011 - 5; http://dx.doi.org/ 10.1126/science.7605428; PMID: 7605428 [DOI] [PubMed] [Google Scholar]
- 5.Harley CB, Futcher AB, Greider CW. . Telomeres shorten during ageing of human fibroblasts. Nature 1990; 345:458 - 60; http://dx.doi.org/ 10.1038/345458a0; PMID: 2342578 [DOI] [PubMed] [Google Scholar]
- 6.Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. . Telomere reduction in human colorectal carcinoma and with ageing. Nature 1990; 346:866 - 8; http://dx.doi.org/ 10.1038/346866a0; PMID: 2392154 [DOI] [PubMed] [Google Scholar]
- 7.Hayflick L, Moorhead PS. . The serial cultivation of human diploid cell strains. Exp Cell Res 1961; 25:585 - 621; http://dx.doi.org/ 10.1016/0014-4827(61)90192-6; PMID: 13905658 [DOI] [PubMed] [Google Scholar]
- 8.Lundblad V, Szostak JW. . A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 1989; 57:633 - 43; http://dx.doi.org/ 10.1016/0092-8674(89)90132-3; PMID: 2655926 [DOI] [PubMed] [Google Scholar]
- 9.Shay JW, Pereira-Smith OM, Wright WE. . A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res 1991; 196:33 - 9; http://dx.doi.org/ 10.1016/0014-4827(91)90453-2; PMID: 1652450 [DOI] [PubMed] [Google Scholar]
- 10.Enomoto S, Glowczewski L, Berman J. . MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae.. Mol Biol Cell 2002; 13:2626 - 38; http://dx.doi.org/ 10.1091/mbc.02-02-0012; PMID: 12181334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. . A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003; 426:194 - 8; http://dx.doi.org/ 10.1038/nature02118; PMID: 14608368 [DOI] [PubMed] [Google Scholar]
- 12.IJpma AS, Greider CW. . Short telomeres induce a DNA damage response in Saccharomyces cerevisiae.. Mol Biol Cell 2003; 14:987 - 1001; http://dx.doi.org/ 10.1091/mbc.02-04-0057; PMID: 12631718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. . Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol Cell 2004; 14:501 - 13; http://dx.doi.org/ 10.1016/S1097-2765(04)00256-4; PMID: 15149599 [DOI] [PubMed] [Google Scholar]
- 14.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. . Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J 1992; 11:1921 - 9; PMID: 1582420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. . Extension of life-span by introduction of telomerase into normal human cells. Science 1998; 279:349 - 52; http://dx.doi.org/ 10.1126/science.279.5349.349; PMID: 9454332 [DOI] [PubMed] [Google Scholar]
- 16.Ciccia A, Elledge SJ. . The DNA damage response: making it safe to play with knives. Mol Cell 2010; 40:179 - 204; http://dx.doi.org/ 10.1016/j.molcel.2010.09.019; PMID: 20965415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. . DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 2004; 431:1011 - 7; http://dx.doi.org/ 10.1038/nature02964; PMID: 15496928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu D, Topper LM, Wilson TE. . Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae.. Genetics 2008; 178:1237 - 49; http://dx.doi.org/ 10.1534/genetics.107.083535; PMID: 18245831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clerici M, Mantiero D, Guerini I, Lucchini G, Longhese MP. . The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep 2008; 9:810 - 8; http://dx.doi.org/ 10.1038/embor.2008.121; PMID: 18600234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longhese MP, Bonetti D, Manfrini N, Clerici M. . Mechanisms and regulation of DNA end resection. EMBO J 2010; 29:2864 - 74; http://dx.doi.org/ 10.1038/emboj.2010.165; PMID: 20647996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mimitou EP, Symington LS. . Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 2008; 455:770 - 4; http://dx.doi.org/ 10.1038/nature07312; PMID: 18806779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. . Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 2008; 134:981 - 94; http://dx.doi.org/ 10.1016/j.cell.2008.08.037; PMID: 18805091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakada D, Matsumoto K, Sugimoto K. . ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev 2003; 17:1957 - 62; http://dx.doi.org/ 10.1101/gad.1099003; PMID: 12923051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou L, Elledge SJ. . Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003; 300:1542 - 8; http://dx.doi.org/ 10.1126/science.1083430; PMID: 12791985 [DOI] [PubMed] [Google Scholar]
- 25.Lydall D, Weinert T. . Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 1995; 270:1488 - 91; http://dx.doi.org/ 10.1126/science.270.5241.1488; PMID: 7491494 [DOI] [PubMed] [Google Scholar]
- 26.Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, Muzi-Falconi M. . Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J 2008; 27:1502 - 12; PMID: 18418382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen HT, Bosque D, Callen E, et al. . Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell 2011; 42:319 - 29; http://dx.doi.org/ 10.1016/j.molcel.2011.03.019; PMID: 21549309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams KE, Medhurst AL, Dart DA, Lakin ND. . Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene 2006; 25:3894 - 904; http://dx.doi.org/ 10.1038/sj.onc.1209426; PMID: 16474843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. . ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 2006; 8:37 - 45; http://dx.doi.org/ 10.1038/ncb1337; PMID: 16327781 [DOI] [PubMed] [Google Scholar]
- 30.Mantiero D, Clerici M, Lucchini G, Longhese MP. . Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep 2007; 8:380 - 7; http://dx.doi.org/ 10.1038/sj.embor.7400911; PMID: 17347674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiotani B, Zou L. . Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell 2009; 33:547 - 58; http://dx.doi.org/ 10.1016/j.molcel.2009.01.024; PMID: 19285939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia X, Weinert T, Lydall D. . Mec1 and Rad53 inhibit formation of single-stranded DNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics 2004; 166:753 - 64; http://dx.doi.org/ 10.1534/genetics.166.2.753; PMID: 15020465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clerici M, Trovesi C, Galbiati A, Lucchini G, Longhese MP. . Mec1/ATR regulates the generation of single-stranded DNA that attenuates Tel1/ATM signaling at DNA ends. EMBO J 2014; 33:198 - 216; PMID: 24357557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longhese MP. . DNA damage response at functional and dysfunctional telomeres. Genes Dev 2008; 22:125 - 40; http://dx.doi.org/ 10.1101/gad.1626908; PMID: 18198332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maringele L, Lydall D. . EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev 2002; 16:1919 - 33; http://dx.doi.org/ 10.1101/gad.225102; PMID: 12154123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirano Y, Sugimoto K. . Cdc13 telomere capping decreases Mec1 association but does not affect Tel1 association with DNA ends. Mol Biol Cell 2007; 18:2026 - 36; http://dx.doi.org/ 10.1091/mbc.E06-12-1074; PMID: 17377065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. . RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol 2007; 14:208 - 14; http://dx.doi.org/ 10.1038/nsmb1205; PMID: 17293872 [DOI] [PubMed] [Google Scholar]
- 38.Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE. . Evolution of CST function in telomere maintenance. Cell Cycle 2010; 9:3157 - 65; http://dx.doi.org/ 10.4161/cc.9.16.12547; PMID: 20697207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcand S, Pardo B, Gratias A, Cahun S, Callebaut I. . Multiple pathways inhibit NHEJ at telomeres. Genes Dev 2008; 22:1153 - 8; http://dx.doi.org/ 10.1101/gad.455108; PMID: 18451106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonetti D, Clerici M, Anbalagan S, Martina M, Lucchini G, Longhese MP. . Shelterin-like proteins and Yku inhibit nucleolytic processing of Saccharomyces cerevisiae telomeres. PLoS Genet 2010; 6:e1000966; http://dx.doi.org/ 10.1371/journal.pgen.1000966; PMID: 20523746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vodenicharov MD, Laterreur N, Wellinger RJ. . Telomere capping in non-dividing yeast cells requires Yku and Rap1. EMBO J 2010; 29:3007 - 19; http://dx.doi.org/ 10.1038/emboj.2010.155; PMID: 20628356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribeyre C, Shore D. . Anticheckpoint pathways at telomeres in yeast. Nat Struct Mol Biol 2012; 19:307 - 13; http://dx.doi.org/ 10.1038/nsmb.2225; PMID: 22343724 [DOI] [PubMed] [Google Scholar]
- 43.Xue Y, Rushton MD, Maringele L. . A novel checkpoint and RPA inhibitory pathway regulated by Rif1. PLoS Genet 2011; 7:e1002417; http://dx.doi.org/ 10.1371/journal.pgen.1002417; PMID: 22194703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anbalagan S, Bonetti D, Lucchini G, Longhese MP. . Rif1 supports the function of the CST complex in yeast telomere capping. PLoS Genet 2011; 7:e1002024; http://dx.doi.org/ 10.1371/journal.pgen.1002024; PMID: 21437267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qi H, Zakian VA. . The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated est1 protein. Genes Dev 2000; 14:1777 - 88; PMID: 10898792 [PMC free article] [PubMed] [Google Scholar]
- 46.Denchi EL, de Lange T. . Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 2007; 448:1068 - 71; http://dx.doi.org/ 10.1038/nature06065; PMID: 17687332 [DOI] [PubMed] [Google Scholar]
- 47.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. . p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 1999; 283:1321 - 5; http://dx.doi.org/ 10.1126/science.283.5406.1321; PMID: 10037601 [DOI] [PubMed] [Google Scholar]
- 48.Celli GB, de Lange T. . DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol 2005; 7:712 - 8; http://dx.doi.org/ 10.1038/ncb1275; PMID: 15968270 [DOI] [PubMed] [Google Scholar]
- 49.Takai H, Smogorzewska A, de Lange T. . DNA damage foci at dysfunctional telomeres. Curr Biol 2003; 13:1549 - 56; http://dx.doi.org/ 10.1016/S0960-9822(03)00542-6; PMID: 12956959 [DOI] [PubMed] [Google Scholar]
- 50.Doksani Y, Wu JY, de Lange T, Zhuang X. . Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 2013; 155:345 - 56; http://dx.doi.org/ 10.1016/j.cell.2013.09.048; PMID: 24120135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T. . POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J 2005; 24:2667 - 78; http://dx.doi.org/ 10.1038/sj.emboj.7600733; PMID: 15973431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, et al. . Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 2006; 126:49 - 62; http://dx.doi.org/ 10.1016/j.cell.2006.05.037; PMID: 16839876 [DOI] [PubMed] [Google Scholar]
- 53.Gong Y, de Lange T. . A Shld1-controlled POT1a provides support for repression of ATR signaling at telomeres through RPA exclusion. Mol Cell 2010; 40:377 - 87; http://dx.doi.org/ 10.1016/j.molcel.2010.10.016; PMID: 21070964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. . TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 2011; 471:532 - 6; http://dx.doi.org/ 10.1038/nature09772; PMID: 21399625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. . Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 2010; 327:1657 - 61; http://dx.doi.org/ 10.1126/science.1185100; PMID: 20339076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sfeir A, de Lange T. . Removal of shelterin reveals the telomere end-protection problem. Science 2012; 336:593 - 7; http://dx.doi.org/ 10.1126/science.1218498; PMID: 22556254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galati A, Micheli E, Cacchione S. . Chromatin structure in telomere dynamics. Front Oncol 2013; 3:46; http://dx.doi.org/ 10.3389/fonc.2013.00046; PMID: 23471416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaul Z, Cesare AJ, Huschtscha LI, Neumann AA, Reddel RR. . Five dysfunctional telomeres predict onset of senescence in human cells. EMBO Rep 2012; 13:52 - 9; http://dx.doi.org/ 10.1038/embor.2011.227; PMID: 22157895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cesare AJ, Karlseder J. . A three-state model of telomere control over human proliferative boundaries. Curr Opin Cell Biol 2012; 24:731 - 8; http://dx.doi.org/ 10.1016/j.ceb.2012.08.007; PMID: 22947495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, Kaplunov JM, Bucci G, Dobreva M, Matti V, Beausejour CM, et al. . Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol 2012; 14:355 - 65; http://dx.doi.org/ 10.1038/ncb2466; PMID: 22426077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR 3rd, Denchi EL. . A two-step mechanism for TRF2-mediated chromosome-end protection. Nature 2013; 494:502 - 5; http://dx.doi.org/ 10.1038/nature11873; PMID: 23389450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hemann MT, Strong MA, Hao LY, Greider CW. . The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 2001; 107:67 - 77; http://dx.doi.org/ 10.1016/S0092-8674(01)00504-9; PMID: 11595186 [DOI] [PubMed] [Google Scholar]
- 63.Abdallah P, Luciano P, Runge KW, Lisby M, Géli V, Gilson E, Teixeira MT. . A two-step model for senescence triggered by a single critically short telomere. Nat Cell Biol 2009; 11:988 - 93; http://dx.doi.org/ 10.1038/ncb1911; PMID: 19597486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fallet E, Jolivet P, Soudet J, Lisby M, Gilson E, Teixeira MT. . Length-dependent processing of telomeres in the absence of telomerase. Nucleic Acids Res 2014; 42:3648 - 65; http://dx.doi.org/ 10.1093/nar/gkt1328; PMID: 24393774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ballew BJ, Lundblad V. . Multiple genetic pathways regulate replicative senescence in telomerase-deficient yeast. Aging Cell 2013; 12:719 - 27; http://dx.doi.org/ 10.1111/acel.12099; PMID: 23672410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Larrivée M, LeBel C, Wellinger RJ. . The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev 2004; 18:1391 - 6; http://dx.doi.org/ 10.1101/gad.1199404; PMID: 15198981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hirano Y, Fukunaga K, Sugimoto K. . Rif1 and rif2 inhibit localization of tel1 to DNA ends. Mol Cell 2009; 33:312 - 22; http://dx.doi.org/ 10.1016/j.molcel.2008.12.027; PMID: 19217405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martina M, Clerici M, Baldo V, Bonetti D, Lucchini G, Longhese MP. . A balance between Tel1 and Rif2 activities regulates nucleolytic processing and elongation at telomeres. Mol Cell Biol 2012; 32:1604 - 17; http://dx.doi.org/ 10.1128/MCB.06547-11; PMID: 22354991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hewitt G, Jurk D, Marques FD, Correia-Melo C, Hardy T, Gackowska A, Anderson R, Taschuk M, Mann J, Passos JF. . Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat Commun 2012; 3:708; http://dx.doi.org/ 10.1038/ncomms1708; PMID: 22426229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Artandi SE, DePinho RA. . Telomeres and telomerase in cancer. Carcinogenesis 2010; 31:9 - 18; http://dx.doi.org/ 10.1093/carcin/bgp268; PMID: 19887512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. . Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 2000; 406:641 - 5; http://dx.doi.org/ 10.1038/35020592; PMID: 10949306 [DOI] [PubMed] [Google Scholar]
- 72.Rudolph KL, Millard M, Bosenberg MW, DePinho RA. . Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat Genet 2001; 28:155 - 9; http://dx.doi.org/ 10.1038/88871; PMID: 11381263 [DOI] [PubMed] [Google Scholar]
- 73.Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. . p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 1999; 97:527 - 38; http://dx.doi.org/ 10.1016/S0092-8674(00)80762-X; PMID: 10338216 [DOI] [PubMed] [Google Scholar]
- 74.Hackett JA, Feldser DM, Greider CW. . Telomere dysfunction increases mutation rate and genomic instability. Cell 2001; 106:275 - 86; http://dx.doi.org/ 10.1016/S0092-8674(01)00457-3; PMID: 11509177 [DOI] [PubMed] [Google Scholar]
- 75.Lin TT, Letsolo BT, Jones RE, Rowson J, Pratt G, Hewamana S, Fegan C, Pepper C, Baird DM. . Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood 2010; 116:1899 - 907; http://dx.doi.org/ 10.1182/blood-2010-02-272104; PMID: 20538793 [DOI] [PubMed] [Google Scholar]
- 76.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. . Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med 1997; 3:1271 - 4; http://dx.doi.org/ 10.1038/nm1197-1271; PMID: 9359704 [DOI] [PubMed] [Google Scholar]
- 77.Lundblad V, Blackburn EH. . An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 1993; 73:347 - 60; http://dx.doi.org/ 10.1016/0092-8674(93)90234-H; PMID: 8477448 [DOI] [PubMed] [Google Scholar]
- 78.Wellinger RJ. . When the caps fall off: responses to telomere uncapping in yeast. FEBS Lett 2010; 584:3734 - 40; http://dx.doi.org/ 10.1016/j.febslet.2010.06.031; PMID: 20600003 [DOI] [PubMed] [Google Scholar]
- 79.Ding Z, Wu CJ, Jaskelioff M, Ivanova E, Kost-Alimova M, Protopopov A, Chu GC, Wang G, Lu X, Labrot ES, et al. . Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell 2012; 148:896 - 907; http://dx.doi.org/ 10.1016/j.cell.2012.01.039; PMID: 22341455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu J, Hwang SS, Liesa M, Gan B, Sahin E, Jaskelioff M, Ding Z, Ying H, Boutin AT, Zhang H, et al. . Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell 2012; 148:651 - 63; http://dx.doi.org/ 10.1016/j.cell.2011.12.028; PMID: 22341440 [DOI] [PMC free article] [PubMed] [Google Scholar]