Abstract
Constitutive heterochromatin, an essential structure that has been conserved throughout evolution, is required to maintain genome stability. Although heterochromatin is enriched for repressive traits, it can be actively transcribed to generate thousands of noncoding RNAs that are required for correct chromatin assembly. Despite the importance of this structure, how and why heterochromatin transcription is regulated, and the proteins responsible for this regulation, remain poorly understood. Here, we summarize recent findings in heterochromatin transcription regulation during different cellular processes with a focus on the epithelial–mesenchymal transition (EMT), which elicits important changes in cell behavior, has a key role in early development, and is involved in cancer progression.
Keywords: epithelial to mesenchymal transition, heterochromatin, histones
Abbreviations
- 3C
chromosome conformation capture
- ChIP
chromatin immunoprecitpitation
- CSC
cancer stem cells
- DGE
digital gene expression
- EMT
epithelial to mesenchymal transition
- EMT-TF
EMT-Transcription Factors
- ES
embryonic stem cells
- HP1
heteroprotein 1
- LINES
long interspersed elements
- LNA
locked-nucleic acid
- lncRNAs
long non-coding RNAs
- LOCKs
large organized chromatin domains enriched in H3K9m2
- MEFs
mouse embryonic fibroblasts
- MET
mesenchymal epithelial transition
- NMuMg
mouse epithelial mammary gland
- SINES
short interspersed elements
- TERRA
telomeric repeat-containing RNA
Chromatin Organization
DNA in the nucleus is folded into chromatin through the formation of a basic structural and functional unit called the nucleosome. Each nucleosome consists of 146 base pairs of DNA wrapped around a histone octamer composed of 2 molecules each of histones H3, H2A, H4, and H2B.1 Nucleosomes are kept in clusters by linker DNA that is associated with histone H1 or H5. These linker histones organize the nucleosome arrays into a more condensed 30-nm chromatin fiber, typically postulated as the second structural level of DNA organization.2 However, chromatin is additionally organized into a higher-order structure inside the nucleus. Although it is clear that this structure is biologically relevant, how the spatial organization of the genome is linked to its biological function is still poorly understood.3 Recent advances in genomic technologies, such as the development of chromosome conformation capture (3C)4 and, more recently, the Hi-C technique,5 have provided new information about trans and cis interactions of DNA and have led to rapid advances in the study of the 3-dimensional genome organization in vivo. For example, distant loci on the same chromosome have been found to be physically associated.6
Chromatin can be divided into 2 types, heterochromatin and euchromatin, based on its compaction status in interphase. Heterochromatin has been defined as a chromosomal region that remains condensed throughout the cell cycle and is gene-poor, contains heterochromatin protein 1 (HP1), and is enriched in hypoacetylated histones, methylated histone H3 at lysine 9 (H3K9), and methylated DNA.7,8 On the other hand, euchromatin is less condensed, contains most of the genes, and is enriched in acetylated histones H3 and H4 and methylated H3K4.9 Heterochromatin can be subdivided into constitutive and facultative heterochromatin. In higher eukaryotes, repetitive and noncoding sequences as well as regions important for genome integrity, such as telomeres and centromeres, are kept stably heterochromatinized; these regions are referred to as constitutive heterochromatin. In contrast, facultative heterochromatin is defined as condensed and transcriptionally silent chromatin that can decondense for transcription depending on the developmental state, cell cycle phase, or chromatin positioning. Therefore, although facultative heterochromatin is transcriptionally silent, it retains the potential to convert to euchromatin.10,11
The centromere region is characterized by the presence of 2 different types of chromatin: centric and pericentric heterochromatin. These 2 domains are enriched in repetitive sequences and specific epigenetic traits. Centric chromatin has a particular histone variant called cenH3, as well as specific proteins that are required for kinetochore formation. Pericentromeric heterochromatin contains hypoacetylated histones and methylated H3K9 and H4K20. Pericentromeric heterochromatin is also rich in HP1, a protein essential for maintaining the heterochromatin structure. In mouse somatic cells, these heterochromatin areas are organized into clusters called chromocenters12; the number and the size of these clusters are indicators for how the pericentromeric heterochromatin is organized.
Heterochromatin Transcription
Constitutive heterochromatin accounts for more than 50% of the human genome13 and, although highly compacted, is transcribed.14,15 In stark contrast, protein-coding sequences represent approximately 2% of the human genome.16 Despite this huge difference, little is known about the function of most of these heterochromatin RNAs and their regulation. It has recently become clear that these regions are transcribed to generate a new subtype of long non-coding RNAs (lncRNAs); indeed, long pericentromeric transcripts of up to 8 kb have been detected.17 These heterochromatin transcripts are differentially expressed in different cell types and their levels are modulated by distinct stimuli, suggesting that this transcription is tightly regulated.
Although this review focuses on the transcription of pericentromeric and interspersed repeats, it is worth mentioning that telomeres of eukaryotic chromosomes are also transcribed by RNA Pol II to generate different non-coding RNAs, including the telomeric repeat-containing RNA (TERRA).18
Heterochromatin Regulation during Fertilization, Cell Differentiation, Cellular Stress, and Cancer
At the moment of fertilization the organization of pericentromeric maternal and paternal heterochromatin differs considerably,19 although both are equivalent by the 8-cell stage.20,21 During this early stage of development in mice a particular heterochromatin state is acquired that requires transcription of a pericentromeric repetitive sequence, known as major satellite RNA, that is initiated by a burst in synthesis of the forward major satellite strand. At the 4-cell stage, transcription of both strands is downregulated. Disruption of this transcriptional heterochromatin regulation using locked-nucleic acid (LNA)-DNA gapmers that target major transcripts leads to developmental arrest at the 2-cell stage.22 These repetitive transcripts are detected again in advanced stages of mouse development and are ubiquitously expressed in several tissues. However, the expression pattern in the adult is more restricted and tissue-specific.23
The expression levels of pericentromeric transcripts are also modified during several processes of differentiation. For example, in P19 cells the major satellites are downregulated by retinoic acid, which induces differentiation, suggesting a relationship between major satellite transcription and reprogramming.23 In contrast, an increase in transcription is observed during terminal muscle differentiation, resulting in global heterochromatin reorganization.24
Pluripotent stem cells express high amounts of HERV-H retrovirus, a heterochromatin sequence, and this expression decreases upon differentiation.25 In fact, transcription of satellite repeat sequences, including major, minor, long interspersed elements (LINES), and short interspersed elements (SINES), is significantly higher in embryonic stem (ES) cells than in ES-derived neural progenitor cells, and the transcriptional status of these sequences correlates with the degree of active chromatin marks such as H3K4me3 and H3K9/K14ac.26
Regulation of pericentromeric transcription is also important for the cellular stress response. In human cells, alpha-satellite repetitive sequences are located in the pericentromeric and centromeric regions in almost all chromosomes. The high degree of similarity between these sequences makes it difficult to discern the origin of specific sequences. However, other sequences within pericentromeric regions, such as LINEs, SINEs, and satellite III, can be more easily analyzed.27,28 SINEs are upregulated in stress conditions. The human Alu RNA transcribed from SINEs represses the transcription of 4 housekeeping genes in trans during the heat shock response.29,30 Heat shock also activates the formation of RNA Pol II factories, facilitating satellite III transcription.31-33
Senescence can be considered a stress-dependent exit of the cell cycle. Transcription of the major satellite increases during senescence in cardiac muscle.34 Heterochromatin transcription is also regulated during the cell cycle; the transcription of 2 different populations of major and minor satellite repeats from heterochromatin regions increases in late G1 and then decreases during mid-late S phase, when heterochromatin is replicated.35
The expression of repetitive sequences in human primary tumors has not been extensively analyzed, probably because of the bias of microarray platforms toward including annotated coding sequences and excluding repetitive sequences. Recent studies using a next-generation digital gene expression (DGE) method showed that pericentromeric satellites are overexpressed in human pancreatic adenocarcinomas, especially for the α satellite and HSTAIII transcripts; this pattern is also observed in other cancers (lung, kidney, ovary, colon, and prostate).36 Furthermore, epithelial primary tumors were found to overexpress LINE1 and aberrantly express neuroendocrine-associated genes located near LINE1 insertions. The authors suggest that this aberrant expression of heterochromatin transcripts may reflect global alterations in heterochromatin.
Overall, these data show a relationship between the cellular state and global levels of heterochromatin transcripts. As indicated above, other results indicate the biological relevance of this regulation–for example, disruption of heterochromatin regulation in early mouse development induces 2-cell stage arrest22–thus highlighting the functional relevance of these transcripts. Indeed, we have recently shown that major satellite regulation plays a critical role during the epithelial–mesenchymal transition (EMT). Below, we further examine this phenomenon to exemplify how heterochromatin regulation helps control an important cellular transition.
The Epithelial–Mesenchymal Transition
The EMT program describes a series of events that affect epithelial cells, causing them to lose many of their epithelial characteristics and acquire mesenchymal properties. In physiological conditions, this process is critical in early embryonic morphogenesis37; for example, it is necessary for mesoderm formation, neural crest delamination, and secondary palate formation.37-39 EMT also takes place during progression of epithelial cancer, when carcinoma cells acquire a mesenchymal gene expression pattern accompanied by complex changes in cell architecture and behavior.39 These mesenchymal cells are characterized by their ability to migrate and invade surrounding tissue as well as by their increased resistance to cell death, allowing tumor cells to metastasize and establish secondary tumors at distant sites.40 Furthermore, EMT provides tumor cells with some cancer stem cell (CSC) traits.41
The EMT process displays a high degree of plasticity, and can be reversed through its conversion to the mesenchymal–epithelial transition (MET).42 It has been proposed that MET is essential for metastasis development in distal organs because secondary tumors typically show a similar morphology to that of primary tumors. Moreover, EMT is not always completed, such that in many tumors the epithelial cells only undergo a partial conversion to the mesenchymal state and present a metastable phenotype characterized by co-expression of mesenchymal and epithelial markers.43
The EMT process is driven by a set of specific transcription factors (including SNAIL1, ZEB1/2, and TWIST)44 and involves very extensive epigenetic reprogramming, similar to processes that occur during other cellular changes such as stem cell differentiation.45 EMT inducers promote general chromatin reorganization by recruiting chromatin remodelers, and this reorganization is required to convert the epithelial cell to a final mesenchymal state. Accordingly, it has recently been reported that SNAIL1 triggers sustained but reversible epigenetic changes.46 Several authors have demonstrated that SNAIL1 binds transiently to its repressed target promoters, concomitant with loss of histone activation marks and a gain of chromatin repressive marks.46
Distinct pieces of evidence now suggest that regulation and reprogramming of heterochromatin transcription are key events in the EMT process. When non-transformed mouse epithelial mammary gland (NMuMG) cells are treated with TGFβ to induce EMT, they exhibit a downregulation in major satellite expression at the same time that SNAIL1 is induced. Chromatin immunoprecipitation (ChIP) experiments show that SNAIL1 binds to major satellite DNA sequences at the start of EMT, correlating with a block in major satellite transcription. This effect on major satellite transcription promotes the release of HP1 from chromocenters. Importantly, this regulation is essential for the EMT process, since NMuMG cells that are unable to downregulate major satellites show impaired EMT.47 This model suggests that impairing heterochromatin transcription compromises the EMT program, leading to cells that are less able to migrate, less invasive, and with altered expression of mesenchymal genes47 (Fig. 1). Along the same lines, McDonald et al. have demonstrated that the nontransformed hepatocyte cell line AML12 also undergoes EMT after treatment with TGFβ, and that this occurs with global loss of electron-dense heterochromatin areas, suggesting global epigenetic changes during this process.48 Although the authors did not analyze constitutive heterochromatin transcription, they detected a decrease in the global levels of H3K9me2, specifically in large nonrepetitive heterochromatin domains enriched in H3K9m2 (LOCKs) (Fig. 1). They also observed that, although these domains are not actively transcribed, they present increased H3K4me3 levels, leading to the suggestion that such domains are in a “poised” state.48 These changes in the global levels of H3K9me2 and H3K4me3 seem to be governed by the histone demethylase LSD1; siRNA against LSD1 blocks this reprogramming and also impairs the EMT,48 suggesting that this histone mark reorganization is an essential prerequisite for EMT. Interestingly, LSD1 interacts with SNAIL1 and is required for repression of SNAIL1 target genes such as CDH1.49
Figure 1.
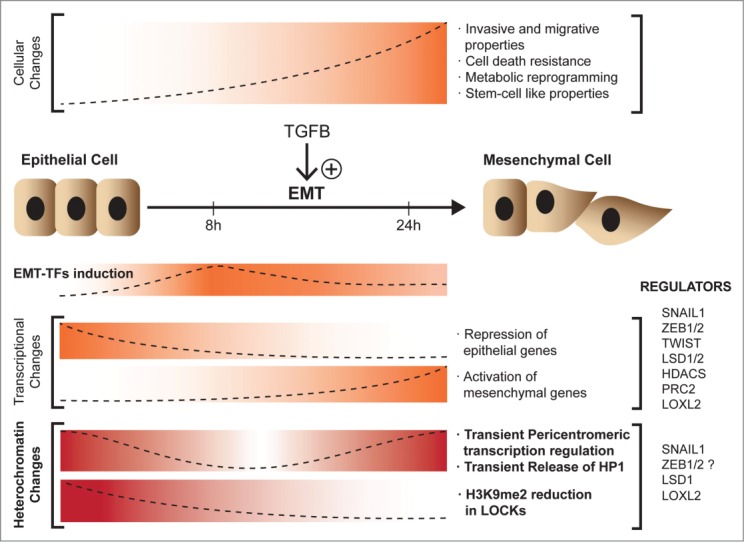
Chromatin dynamics during epithelial-mesenchymal transition (EMT). Epithelial cells undergo EMT upon treatment with TGFβ to acquire mesenchymal properties. During this process EMT-transcription factors (EMT-TFs) are upregulated and transcriptional and heterochromatin changes take place.
Heterochromatin Regulators
Transcription factors and histone modifications
Although it is now clear that the transcription of non-coding heterochromatin RNAs has to be regulated, little is known about the sequences and specific transcription factors involved in its control. Some data suggest a partial dependence on several zinc finger transcription factors. For example, the transcription factors YY1 and Ikaros directly bind to the major satellite (γ-satellite).50 Pax3 and Pax9 repress pericentric repeats in MEFs and ES cells, and depletion of these transcription factors results in an increase of major satellite transcripts that leads to an impairment of the heterochromatin structure and defects in chromosome segregation.51
The tumor suppressor gene BRCA1 is also involved in the regulation of heterochromatin transcription. Knockout mice for BRCA1 present disorganized heterochromatin that is associated with upregulation of major satellite transcripts.52 The BRCA1 protein contains a RING finger domain with ubiquitin ligase E3 activity that (mono) ubiquitinylates histone H2A in major and minor satellite regions to repress transcription. In the absence of BRCA1 the presence of ubiquityl-H2A in these sequences is abolished.52
Likewise, the phosphatase WIP1 is implicated in regulating heterochromatin-associated sequences. Knockdown of WIP1 in the cancer cell lines HCT116 and MCF7 leads to downregulation of the L1 LINEs and HERV-H sequences.53 In the absence of WIP1, levels of the repressive marks H3K9me3 and H3K20me3, as well as 5-methyl-cytosine, are increased, while those of H3K9ac are significantly reduced. This increase in H3K9me3 correlates with enrichment in BRCA1, HP1γ, and DNMT3B, although no ubiquityl-H2A was detected in these heterochromatin regions. The authors also found a role for ATM beyond its function at double-strand breaks. In L1 LINEs and HERV-H sequences, ATM is both necessary and sufficient to establish global DNA methylation; activation of ATM results in enhanced BRCA1 binding to HP1γ, which correlates with H3K9 methylation and transcriptional repression.53
H3K9me3 governs pericentromeric heterochromatin and is required for genome stability.54 Double knockout of the histone methyltransferases SUV39 1 and 2, which are required for H3K9 methylation, in ES cells leads to upregulation of pericentromeric transcription. In this system, SUV39 also mediates DNA methylation of major satellite regions through the action of DNMT3.55
Transcription factors and histone modifications in EMT
SNAIL1, the main inducer of the EMT program, also binds major satellite sequences in mouse embryonic fibroblasts (MEFs).47 Snail1 knockout cells show an increase in major satellite transcription and an altered heterochromatin structure. Moreover, in NMuMG cells, entry into EMT after treatment with TGFβ induces the expression of SNAIL1, which then represses major satellite transcription; this step is necessary for the cells to fully acquire mesenchymal properties.47 The repressive function of SNAIL1 requires the histone-modifying enzyme LOXL2,56 which oxidizes H3K4 in heterochromatin major satellite regions, a modification associated with the repression of major satellite transcription during EMT.47 Interestingly, Zeb1, another zinc finger transcription factor and EMT inducer, has also been shown to bind to major satellite regions.51
AML12 cells that are treated with TFGβ to induce EMT show a reduced global level of H3K9me2 and increased levels of the active marks H3K4me3/K36me3 with no variation in the DNA methylation pattern, correlating with a general increase in transcription.48 This increase in global transcription fits well with the acquisition of a “stem-like” phenotype by cells that are undergoing the EMT program. The authors show a global reprogramming in terms of histone marks associated specifically with large heterochromatin domains (LOCKs) that is LSD1-dependent. The levels of H3K36me2 increase in gene-rich regions between LOCKs and the gene ontology of the corresponding genes indicates that they have EMT-related functions. The authors also demonstrated that the increase in H3K4me3 is located in 190 LOCKs but is not associated with an increase in gene transcription.48
The histone-modifying enzyme implicated in this process is LSD1. Considering that LSD1 can demethylate H3K9 and H3K4, the fact that it interacts with a distinct set of proteins at the onset of EMT presents the intriguing possibility that LSD1 has distinct functions in cells undergoing EMT and in differentiated cells. Indeed, H3K9me2 LOCKs are absent in ES cells and are acquired during differentiation, contributing to a stable cell phenotype. As mentioned above, LOCK reprogramming that is acquired during EMT resembles that existing in ES cells, with higher levels of H3K4m3 than H3K9me2.
Concluding Remarks
We still have only a poor understanding of the function of the majority of lncRNAs, in particular those arising from pericentromeric regions. Some of these lncRNAs, transcribed mostly from LINE sequences, seem to have the potential to form intermolecular structures such as C0T-1 RNA.57 When sumoylated, HP1α binds forward major satellite transcripts at the periphery of pericentromeric regions, an interaction that guides HP1α to these domains.58 Moreover, these pericentromeric transcripts are essential for both nuclear reorganization and developmental progression.59 Further possibilities to be investigated are that these transcripts require processing analogous to RNAi-mediated degradation or, alternatively, that heterochromatin transcripts function in cis or in trans on homologous target sequences. Future studies will clarify the role of these RNA-based structures in genome packaging, gene regulation and, ultimately, overall genome function.
It is obvious that a process that instigates important changes in cell behavior, shape, and responses, such as the EMT, should be linked to not only modifications in gene expression but also to heterochromatin transcription regulation and global epigenetic changes. Recent evidence suggests that this link could involve rearrangements of local 3-dimensional chromatin architecture. Epithelial cells treated with TGFβ showed an increase in the number of HP1α foci strongly suggestive of chromatin rearrangement.47 We have to keep in mind that this transition requires a massive and finely-tuned regulation of heterochromatin and gene transcription.49 It is clear that a true understanding of genome function requires us to integrate what we have learned about the genome sequence with what are still discovering about heterochromatin transcription and the function of these noncoding RNAs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Work in our laboratory is supported by grants from Instituto de Salud Carlos III (ISCIII) FIS/FEDER (PI12/01250; CP08/00223), MINECO (SAF2013-40922-R), Association for International Cancer Research (AICR), Red Temática de Investigación Cooperativa en Cáncer (RTICC, RD012/0036/005), Fundación Científica de la Asociación Española contra el Cáncer (AECC), Fundació Marató de TV3 and Fundació Fero. SP was a recipient of a Miguel Servet contract (ISCIII/FIS); JPC is supported by a contract from Fundació Marató de TV3.
References
- 1. Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997; 389:251-60; PMID:9305837; http://dx.doi.org/ 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- 2. Robinson PJ, Rhodes D. Structure of the '30 nm' chromatin fibre: a key role for the linker histone. Curr Opin Struct Biol 2006; 16:336-43; PMID:16714106; http://dx.doi.org/ 10.1016/j.sbi.2006.05.007 [DOI] [PubMed] [Google Scholar]
- 3. Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev 2011; 21:175-86; PMID:21342762; http://dx.doi.org/ 10.1016/j.gde.2011.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science 2002; 295:1306-11; PMID:11847345; http://dx.doi.org/ 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- 5. Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods 2012; 58:268-76; PMID:22652625; http://dx.doi.org/ 10.1016/j.ymeth.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009; 326:289-93; PMID:19815776; http://dx.doi.org/ 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huisinga KL, Brower-Toland B, Elgin SC. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma 2006; 115:110-22; PMID:16506022; http://dx.doi.org/ 10.1007/s00412-006-0052-x [DOI] [PubMed] [Google Scholar]
- 8. Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293:1074-80; PMID:11498575; http://dx.doi.org/ 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- 9. Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 2001; 293:2453-5; PMID:11498546; http://dx.doi.org/ 10.1126/science.1064413 [DOI] [PubMed] [Google Scholar]
- 10. Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet 2007; 8:35-46; PMID:17173056; http://dx.doi.org/ 10.1038/nrg2008 [DOI] [PubMed] [Google Scholar]
- 11. Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature 2007; 447:399-406; PMID:17522672; http://dx.doi.org/ 10.1038/nature05914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol 2004; 166:493-505; PMID:15302854; http://dx.doi.org/ 10.1083/jcb.200403109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 2011; 7:e1002384; PMID:22144907; http://dx.doi.org/ 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aalto AP, Pasquinelli AE. Small non-coding RNAs mount a silent revolution in gene expression. Curr Opin Cell Biol 2012; 24:333-40; PMID:22464106; http://dx.doi.org/ 10.1016/j.ceb.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81:145-66; PMID:22663078; http://dx.doi.org/ 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature 2001; 409:860-921; PMID:11237011; http://dx.doi.org/ 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- 17. Eymery A, Callanan M, Vourc'h C. The secret message of heterochromatin: new insights into the mechanisms and function of centromeric and pericentric repeat sequence transcription. Int J Dev Biol 2009; 53:259-68; PMID:19412885; http://dx.doi.org/ 10.1387/ijdb.082673ae [DOI] [PubMed] [Google Scholar]
- 18. Bah A, Azzalin CM. The telomeric transcriptome: from fission yeast to mammals. Int J Biochem Cell Biol 2012; 44:1055-9; PMID:22504286; http://dx.doi.org/ 10.1016/j.biocel.2012.03.021 [DOI] [PubMed] [Google Scholar]
- 19. Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol 2005; 280:225-36; PMID:15766761; http://dx.doi.org/ 10.1016/j.ydbio.2005.01.025 [DOI] [PubMed] [Google Scholar]
- 20. Merico V, Barbieri J, Zuccotti M, Joffe B, Cremer T, Redi CA, Solovei I, Garagna S. Epigenomic differentiation in mouse preimplantation nuclei of biparental, parthenote and cloned embryos. Chromosome Res 2007; 15:341-60; PMID:17447149 [DOI] [PubMed] [Google Scholar]
- 21. Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U, Kolb C, Otte AP, Koseki H, Orkin SH, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet 2008; 40:411-20; PMID:18311137; http://dx.doi.org/ 10.1038/ng.99 [DOI] [PubMed] [Google Scholar]
- 22. Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell 2010; 19:625-38; PMID:20951352; http://dx.doi.org/ 10.1016/j.devcel.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 23. Rudert F, Bronner S, Garnier JM, Dolle P. Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm Genome 1995; 6:76-83; PMID:7767009; http://dx.doi.org/ 10.1007/BF00303248 [DOI] [PubMed] [Google Scholar]
- 24. Terranova R, Sauer S, Merkenschlager M, Fisher AG. The reorganisation of constitutive heterochromatin in differentiating muscle requires HDAC activity. Exp Cell Res 2005; 310:344-56; PMID:16182285; http://dx.doi.org/ 10.1016/j.yexcr.2005.07.031 [DOI] [PubMed] [Google Scholar]
- 25. Santoni FA, Guerra J, Luban J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology 2012; 9:111; PMID:23253934; http://dx.doi.org/ 10.1186/1742-4690-9-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2008; 2:437-47; PMID:18462694; http://dx.doi.org/ 10.1016/j.stem.2008.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prades C, Laurent AM, Puechberty J, Yurov Y, Roizes G. SINE and LINE within human centromeres. J Mol Evol 1996; 42:37-43; PMID:8576962; http://dx.doi.org/ 10.1007/BF00163209 [DOI] [PubMed] [Google Scholar]
- 28. Tagarro I, Fernandez-Peralta AM, Gonzalez-Aguilera JJ. Chromosomal localization of human satellites 2 and 3 by a FISH method using oligonucleotides as probes. Hum Genet 1994; 93:383-8; PMID:8168808 [DOI] [PubMed] [Google Scholar]
- 29. Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 2008; 29:499-509; PMID:18313387; http://dx.doi.org/ 10.1016/j.molcel.2007.12.013 [DOI] [PubMed] [Google Scholar]
- 30. Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol 2004; 11:822-9; PMID:15300239; http://dx.doi.org/ 10.1038/nsmb812 [DOI] [PubMed] [Google Scholar]
- 31. Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, Riva S, Biamonti G. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell 2004; 15:543-51; PMID:14617804; http://dx.doi.org/ 10.1091/mbc.E03-07-0487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc'h C. Stress-induced transcription of satellite III repeats. J Cell Biol 2004; 164:25-33; PMID:14699086; http://dx.doi.org/ 10.1083/jcb.200306104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Valgardsdottir R, Chiodi I, Giordano M, Rossi A, Bazzini S, Ghigna C, Riva S, Biamonti G. Transcription of Satellite III non-coding RNAs is a general stress response in human cells. Nucleic Acids Res 2008; 36:423-34; PMID:18039709; http://dx.doi.org/ 10.1093/nar/gkm1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gaubatz JW, Cutler RG. Mouse satellite DNA is transcribed in senescent cardiac muscle. J Biol Chem 1990; 265:17753-8; PMID:1698780 [PubMed] [Google Scholar]
- 35. Lu J, Gilbert DM. Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J Cell Biol 2007; 179:411-21; PMID:17984319; http://dx.doi.org/ 10.1083/jcb.200706176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 2011; 331:593-6; PMID:21233348; http://dx.doi.org/ 10.1126/science.1200801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009; 139:871-90; PMID:19945376; http://dx.doi.org/ 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 38. Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 2008; 14:818-29; PMID:18539112; http://dx.doi.org/ 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 39. Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol 2011; 27:347-76; PMID:21740232; http://dx.doi.org/ 10.1146/annurev-cellbio-092910-154036 [DOI] [PubMed] [Google Scholar]
- 40. Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 2006; 7:131-42; PMID:16493418; http://dx.doi.org/ 10.1038/nrm1835 [DOI] [PubMed] [Google Scholar]
- 41. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008; 133:704-15; PMID:18485877; http://dx.doi.org/ 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science 2013; 342:1234850; PMID:24202173; http://dx.doi.org/ 10.1126/science.1234850 [DOI] [PubMed] [Google Scholar]
- 43. Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol 2006; 172:973-81; PMID:16567498; http://dx.doi.org/ 10.1083/jcb.200601018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013; 13:97-110; PMID:23344542; http://dx.doi.org/ 10.1038/nrc3447 [DOI] [PubMed] [Google Scholar]
- 45. Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science 2013; 339:1567-70; PMID:23539597; http://dx.doi.org/ 10.1126/science.1230184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Javaid S, Zhang J, Anderssen E, Black JC, Wittner BS, Tajima K, Ting DT, Smolen GA, Zubrowski M, Desai R, et al. Dynamic chromatin modification sustains epithelial-mesenchymal transition following inducible expression of Snail-1. Cell Rep 2013; 5:1679-89; PMID:24360956; http://dx.doi.org/ 10.1016/j.celrep.2013.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Millanes-Romero A, Herranz N, Perrera V, Iturbide A, Loubat-Casanovas J, Gil J, Jenuwein T, Garcia de Herreros A, Peiro S. Regulation of heterochromatin transcription by Snail1/LOXL2 during epithelial-to-mesenchymal transition. Mol Cell 2013; 52:746-57; PMID:24239292; http://dx.doi.org/ 10.1016/j.molcel.2013.10.015 [DOI] [PubMed] [Google Scholar]
- 48. McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol 2011; 18:867-74; PMID:21725293; http://dx.doi.org/ 10.1038/nsmb.2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. Embo J 2010; 29:1803-16; PMID:20389281; http://dx.doi.org/ 10.1038/emboj.2010.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shestakova EA, Mansuroglu Z, Mokrani H, Ghinea N, Bonnefoy E. Transcription factor YY1 associates with pericentromeric gamma-satellite DNA in cycling but not in quiescent (G0) cells. Nucleic Acids Res 2004; 32:4390-9; PMID:15316102; http://dx.doi.org/ 10.1093/nar/gkh737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bulut-Karslioglu A, Perrera V, Scaranaro M, de la Rosa-Velazquez IA, van de Nobelen S, Shukeir N, Popow J, Gerle B, Opravil S, Pagani M, et al. A transcription factor-based mechanism for mouse heterochromatin formation. Nat Struct Mol Biol 2012; 19:1023-30; PMID:22983563; http://dx.doi.org/ 10.1038/nsmb.2382 [DOI] [PubMed] [Google Scholar]
- 52. Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 2011; 477:179-84; PMID:21901007; http://dx.doi.org/ 10.1038/nature10371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Filipponi D, Muller J, Emelyanov A, Bulavin DV. Wip1 controls global heterochromatin silencing via ATM/BRCA1-dependent DNA methylation. Cancer Cell 2013; 24:528-41; PMID:24135283; http://dx.doi.org/ 10.1016/j.ccr.2013.08.022 [DOI] [PubMed] [Google Scholar]
- 54. Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 2001; 107:323-37; PMID:11701123; http://dx.doi.org/ 10.1016/S0092-8674(01)00542-6 [DOI] [PubMed] [Google Scholar]
- 55. Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 2003; 13:1192-200; PMID:12867029; http://dx.doi.org/ 10.1016/S0960-9822(03)00432-9 [DOI] [PubMed] [Google Scholar]
- 56. Herranz N, Dave N, Millanes-Romero A, Morey L, Diaz VM, Lorenz-Fonfria V, Gutierrez-Gallego R, Jeronimo C, Di Croce L, Garcia de Herreros A, et al. Lysyl Oxidase-like 2 Deaminates Lysine 4 in Histone H3. Mol Cell 2012; 46:369-76; PMID:22483618; http://dx.doi.org/ 10.1016/j.molcel.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 57. Hall LL, Carone DM, Gomez AV, Kolpa HJ, Byron M, Mehta N, Fackelmayer FO, Lawrence JB. Stable C0T-1 Repeat RNA Is Abundant and Is Associated with Euchromatic Interphase Chromosomes. Cell 2014; 156:907-19; PMID:24581492; http://dx.doi.org/ 10.1016/j.cell.2014.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, et al. SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat Genet 2011; 43:220-7; PMID:21317888; http://dx.doi.org/ 10.1038/ng.765 [DOI] [PubMed] [Google Scholar]
- 59. Casanova M, Pasternak M, El Marjou F, Le Baccon P, Probst AV, Almouzni G. Heterochromatin reorganization during early mouse development requires a single-stranded noncoding transcript. Cell Rep 2013; 4:1156-67; PMID:24055057; http://dx.doi.org/ 10.1016/j.celrep.2013.08.015 [DOI] [PubMed] [Google Scholar]


