Abstract
The DNA-directed primase-polymerase PrimPol of the archaeo-eukaryotic primase superfamily represents an ancient solution to the many problems faced during genome duplication. This versatile enzyme is capable of initiating de novo DNA/RNA synthesis, DNA chain elongation, and has the capacity to bypass modifications that stall the replisome by trans-lesion synthesis or origin-independent re-priming, thus allowing discontinuous synthesis of the leading strand. Recent studies have shown that PrimPol is an important new player in replication fork progression in eukaryotic cells; this review summarizes our current understanding of PrimPol and highlights important questions that remain to be addressed.
Keywords: AEP, DNA, lesions, polymerase, primase, PrimPol, replication, repriming, restart, TLS
Abbreviations
- AEP
archaeo-eukaryotic primase
- CPD
cyclobutane pyrimidine dimer
- DSB
DNA double-strand break
- γH2AX
phosphorylated form of histone H2AX
- HR
homologous recombination
- HU
hydroxyurea
- IR
ionizing radiation
- LigD
ligase D
- MEF
mouse embryonic fibroblast
- MMS
methyl methanesulfonate
- NER
nucleotide excision repair
- NHEJ
non-homologous end joining
- Pol α
DNA polymerase α
- Pol η
DNA Polymerase eta
- PolDom
polymerase domain of bacterial LigD
- PPL
PrimPol-like
- PrimPol
primase-polymerase
- Prim1/PriS
primase small subunit 1
- Prim2/PriL
primase small subunit 2
- RPA
replication protein A
- TLS
trans-lesion synthesis
- UV
ultraviolet
- XP-V
xeroderma pigmentosum variant
- 4NQO
4-nitroquinoline 1-oxide
- 6–4PP
pyrimidine (6–4) pyrimidone photoproduct
Introduction
The integrity of the eukaryotic genome is constantly under threat. During DNA replication, if the template is damaged or DNA synthesis is disrupted, the replication fork can stall leading to the formation of pathologic DNA structures.1 This vulnerability of DNA during genome duplication is commonly exploited by many anticancer treatments currently in use.2 Thus, a complete understanding of the enzymes involved in processing and restarting stalled replication forks is of great importance. PrimPol is a recently identified DNA-directed primase-polymerase responsible for the efficient progression of replication forks, particularly under non-optimal conditions, such as when the DNA template is damaged or when the nucleotide precursor pool is depleted.3-10 This review article will summarize our current understanding of this newly discovered enzyme and outline important questions that remain to be addressed. For a broader overview of DNA replication and the additional mechanisms that exist to overcome replication perturbations, readers are directed to previous articles11,12 and references cited therein.
The AEP Superfamily—Much More than Primases
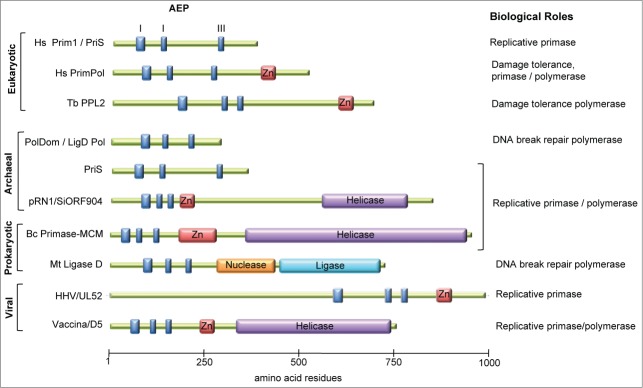
PrimPol is a member of the archaeo-eukaryotic primase (AEP) superfamily.13 AEPs are typically thought of as DNA-dependent RNA polymerases that specialize in the de novo synthesis of short RNA polymers called primers that provide the 3′ hydroxyl group that is absolutely required by DNA polymerases to begin synthesis of a new DNA chain.14,15 The defining member of the AEP superfamily is the DNA-dependent RNA polymerase Prim1 (or PriS).13 In eukaryotes, Prim1 binds to the non-catalytic primase large subunit Prim2 (or PriL) to form the heterodimeric DNA primase complex that is associated with DNA polymerase α (Pol α). Pol α-primase is required for the synthesis and initial extension of primers at replication origins on the leading strand and at each Okazaki fragment on the lagging strand, and is essential for the completion of genome duplication.16 Historically, the absence of well-characterized AEPs has resulted in the prevailing view that this class of enzymes are strictly DNA-dependent RNA polymerases responsible for primer synthesis. However, an increasing body of work published over the past decade, including the recent characterization of PrimPol, has established that this not the case.
The identification of AEP homologues in bacteria provided one of the first hints that AEPs could have additional biological roles, because prokaryotes already contain a dedicated replicative DnaG primase. Notably, the AEP homologues are often part of a multidomain protein called ligase D (LigD) that contains putative DNA ligase and nuclease domains and is encoded by a gene that is co-operonic with homologues of the eukaryotic DNA repair protein Ku.17-19 Ku and LigD were demonstrated to form a minimal non-homologous end-joining (NHEJ) complex in bacteria that is required for the repair of DNA double-strand breaks (DSBs).20 The AEP domain of LigD, called PolDom (or LigD POL), is capable of a multitude of nucleotidyl transferase activities and possesses all DNA/RNA synthetic activities that are possibly required at a DNA break; in eukaryotes these functions are shared among 3 family-X DNA polymerases. Notably, PolDom can perform template-dependent DNA/RNA extension and has gap-filling, strand displacement, template-dependent RNA priming, and template-independent terminal transferase activities.20-23 PolDom can also tolerate DNA lesions by catalyzing error-free gap-filling opposite a template 8-oxo-guanine, and can bypass abasic sites by template scrunching.23,24 Further demonstrating its specialization in DNA break repair, PolDom can bind to the termini of DSBs and mediate the synapsis of broken DNA ends.25-27 PolDom was the first AEP demonstrated to have a bona fide role outside of priming DNA synthesis, and a recent study suggests this utilization of the AEP family in DNA break repair is not restricted to prokaryotes, as archaeal homologues of PolDom have been identified and shown to co-operate with Ku to catalyze similar DSB repair activities in vitro.28
This catalytic versatility of AEPs is not restricted to distant AEP homologues either, as an archaeal Prim1 homologue from the hypothermophile Pyrococcus furiosus, PfuPriS, was demonstrated to be a competent DNA-dependent DNA polymerase in vitro. Additionally, PfuPriS was shown to initiate DNA synthesis de novo with dNTPs, thus generating DNA primers.29 The ability to synthesize DNA primers and extend DNA chains was shown to be a conserved feature of other archaeal PriS orthologues.30-32 PriS has also been demonstrated to catalyze DNA repair activities reminiscent of PolDom in vitro, such as terminal transferase, gap-filling, strand displacement, and DNA end-synapsis functions.31,33-35 As archaeal genomes encode no detectable Pol α homolog or family-X repair polymerases, it has been suggested that PriS might be responsible for these different roles;36 however, to date no biological roles have been attributed to these activities. Additional AEP primase-polymerases have also been characterized in archaeal genomes, the best example being the pRN1 plasmid-encoded AEP from Sulfolobus islandicus.37-39 AEP primase-polymerase homologues have also been characterized in phage, bacterial, and viral genomes.40-42 In summary, AEPs are not restricted to archaea and eukaryotes but are present in all domains of life, and the vast majority characterized to date are capable of catalyzing a wide variety of nucleotidyl transferase activities. Thus, it seems that AEPs have been functionally mis-annotated, and rather than being strictly DNA-dependent RNA polymerases specialized in priming DNA synthesis, they are in fact a group of versatile DNA/RNA primase-polymerases with additional biological roles (Fig. 1). The recent characterization of PrimPol extends this paradigm into eukaryotes.
Figure 1.
The domain organization of members of the archaeo-eukaryotic primase superfamily. Domain organization of various AEPs is depicted: motifs I, II, and III of the catalytic AEP domain (blue boxes), zinc finger (Zn) motifs (red boxes), and additional domains often associated with AEP enzymes are shown. The putative or characterized role of each AEP is indicated. Domain organizations were deduced from the article by Iyer et al.13
PrimPol—A Versatile DNA-Directed RNA/DNA Primase-Polymerase
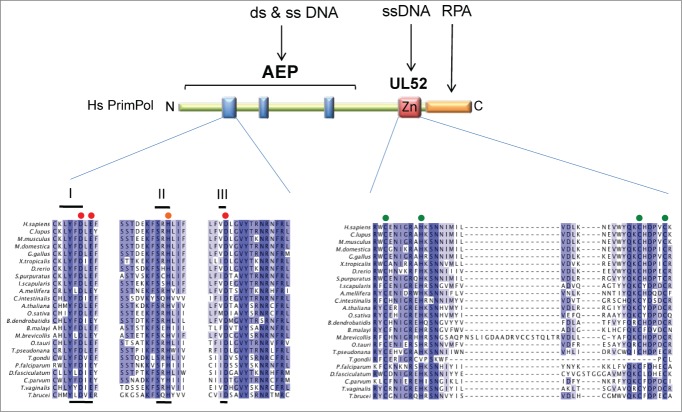
Human PrimPol (also referred to as hPrimPol13,9) is encoded by the PRIMPOL gene (alternative names are coiled-coil domain containing protein 111 [CCDC111] or FLJ33167) located on chromosome 4q35.1, and was originally identified as a putative AEP by in silico analyses.13 PrimPol was categorized as a member of the nucleo-cytoplasmic large DNA virus (NCLDV)-herpesvirus primase clade, which also contains herpes viral and kinetoplastid primases, some of which have been recently characterized.43,44 PrimPol homologues are conserved in a broad range of unicellular and multicellular eukaryotes, including animals, plants, and protists,4,5,13 and is notably duplicated in trypanosomatids.8 However, PrimPol is not conserved in all eukaryotes, being absent from Drosophila, Caenorhabditis elegans, and all fungal genomes sequenced to date with the exception of the parasitic Batrachochytrium dendrobatidis. This patchy phylogenetic distribution is consistent with the PRIMPOL gene being acquired early in eukaryotic evolution by horizontal gene transfer from viruses, and then lost independently on multiple occasions in some animals and fungi.13 Alignment of PrimPol homologues reveals several conserved regions that can be principally divided into 2 domains; an N-terminal catalytic AEP domain and a C-terminal CHC2 zinc finger motif (Fig. 2). The PrimPol AEP domain contains the 3 catalytic motifs conserved in all AEP-like enzymes. Motif I of PrimPol homologues contains the consensus LYFDLE with invariant DxE residues; this is unusual among members of the AEP superfamily, which usually have the sequence DxD. Motif II in PrimPol homologues is an invariant SxH and motif III is an invariant xD. Residues in motif I and III are predicted to be required for binding of divalent metal ions and motif II is required for nucleotide binding, consistent with these residues being essential for PrimPol activity in vitro.3-8 The second conserved region among PrimPol homologues is a C-terminal zinc finger motif with homology to the human herpesvirus UL52 primase, which has been shown to specifically bind zinc ions.7 Zinc fingers are known to have critical functions in primases15 and this is also the case with PrimPol, as discussed below.
Figure 2.
Conserved domains and motifs present in the PrimPol family. The catalytic archaeo-eukaryotic primase (AEP) domain containing 3 signature motifs (I, II, and III; blue boxes), the UL52-like zinc finger domain (Zn), and the replication protein A (RPA)-interaction site (orange box) are indicated for human PrimPol, including amino acid numbers. Multiple sequence alignment was generated for a selection of PrimPol homologues; blue shading indicates ≥40% sequence identity, red circles indicate residues required for metal ion binding, orange circles indicate those required for nucleotide binding, and green circles those required for chelation of zinc.
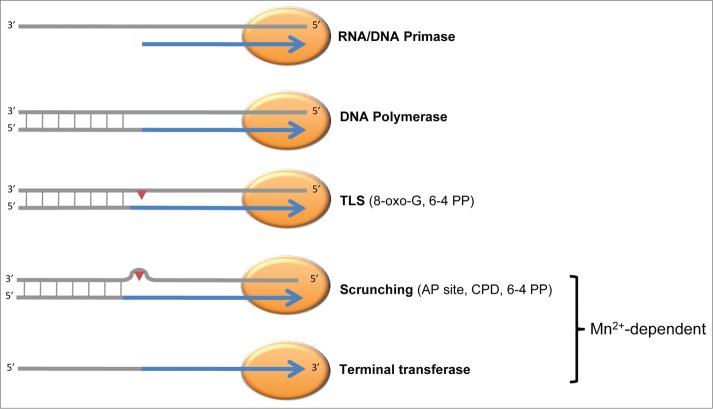
Recombinant human PrimPol has been purified by a number of groups, facilitating in-depth biochemical characterization. We and others found that, akin to prokaryotic and archaeal AEPs, human PrimPol is an extremely versatile nucleotidyl transferase in vitro3-7 (Fig. 3), and that this versatility is shared among 2 divergent PrimPol homologues from the African trypanosome.8 Human PrimPol is capable of synthesizing both RNA and DNA primers, which is unique for a eukaryotic enzyme. Preferential primer synthesis was repeatedly reported when dNTPs were used as substrates, with primers typically exceeding 50 nucleotides in length, whereas RNA primers typically contained 20 nucleotides, similar to the canonical RNA primers synthesized by Prim1. When manganese was the co-factor rather than magnesium, primers synthesized using dNTPs or NTPs exceeded 100 nucleotides in length and it was reported that PrimPol preferentially initiated synthesis with a NTP-dNTP di-nucleotide,4 akin to the archaeal primase-polymerase encoded on the archaeal pRN1 plasmid.39 Although increased activity has been reported with manganese as a co-factor;4 higher manganese concentrations can result in the catalysis of template-independent terminal transferase activity, indicating that this metal may promote additional activities.7 Which divalent metal is preferentially used in vivo remains to be established. To initiate di-nucleotide synthesis PrimPol requires templated pyrimidines,4-5 consistent with the template requirements of other eukaryotic primases,14 and primers synthesized by PrimPol are readily extended by replicative DNA polymerases. PrimPol is also a competent DNA-dependent DNA polymerase capable of extending already existing DNA/RNA chains (including its own primers). Mutation of the invariant residues in motif I of the AEP domain completely abolishes both primase and polymerase activities, confirming that they are intrinsic to the PrimPol enzyme.3-5,7,8 PrimPol has reasonably low fidelity, preferentially performing template-dependent synthesis, and is a distributive enzyme, polymerizing approximately 4 nucleotides in a single DNA binding event7 reminiscent of low-fidelity polymerases of the X-and Y-families.
Figure 3.
PrimPol is a versatile DNA/RNA primase-polymerase in vitro. The reported in vitro activities of human PrimPol are represented schematically. Some activities are dependent on specific metal co-factors: for example, template scrunching to facilitate bypass and template-independent terminal transferase activity are only observed when manganese ions are used as co-factors. TLS, translesion synthesis; 8-oxo-guanine, 8-oxo-G; pyrimidine (6-4) pyrimidone photoproduct, 6-4 PP; apurinic/apyrimidinic site, AP site; cyclobutane pyrimidine dimer, CPD.
Like most primases characterized to date, the zinc finger motif of PrimPol plays an important role in its catalytic activities. Truncation of the UL52 domain or mutation of zinc chelating residues abolishes primase activity in vitro, providing an important biological tool as a ‘separation-of-function’ mutant.3,6,7 However, the UL52 domain also modulates the polymerase activity of PrimPol.7 For example, mutation of the zinc-chelating residues decreases the processivity of PrimPol, whereas truncating the carboxyl terminus (thereby removing the UL52 domain) increases processivity but at the cost of fidelity. Keen et al.7 demonstrated that whereas the AEP domain of PrimPol binds to both single-stranded and double-stranded DNA, the UL52 domain binds only single-stranded DNA and thus probably binds upstream of the primer-template junction in vivo.
PrimPol is also a competent trans-lesion synthesis (TLS) DNA polymerase that can bypass a number of replicase-blocking DNA lesions. PrimPol has been reported to bypass templated oxidative lesions such as 8-oxo-guanine and abasic sites, the ultraviolet (UV)-induced cyclobutane pyrimidine dimer (CPD), and pyrimidine (6–4) pyrimidone photoproducts. The reported mechanism and efficiency of bypass varies between reports, probably reflecting differences in the metal co-factor used. For example, when manganese is used as a cofactor, PrimPol can perform template re-arrangement to allow bypass of abasic sites, CPDs, and 6–4 photoproducts.4,6 However, in the presence of magnesium, PrimPol can completely bypass a 6–4 photoproduct but not an abasic site or CPD, and can only extend from 2 terminal dA residues opposite a template CPD.5 Notably, a C-terminal truncation of PrimPol lacking the UL52 domain can completely bypass a CPD in an error-free manner.7 Bypass of 8-oxo-G was equally error-free and error-prone, whereas bypass of a 6–4 photoproduct was error prone, with PrimPol incorporating dT opposite the 3′T, and dC or dG equally frequently opposite the 5′T.5
These studies demonstrate a catalytic flexibility of human PrimPol that is unprecedented for AEPs, and this flexibility is shared with other eukaryotic PrimPol orthologues.7,8 Notably, 2 PrimPol-like (PPL) proteins in the African trypanosome, one of the earliest diverging organisms from the eukaryotic tree and an important human pathogen, possess almost identical catalytic capabilities to those of human PrimPol.8 Trypanosoma brucei PPL1 and PPL2 can bypass a template 8-oxo-guanine and 6–4 photoproduct, and extend from mis-matched termini opposite template CPDs. Interestingly, although PPL1 is a primase-polymerase like human PrimPol, PPL2 does not appear to possess any detectable primase activity, being the first example of an AEP to have completely lost this activity, and probably plays a specialized role during genome replication in trypanosomes.
PrimPol—A New Player at Replication Forks
Elucidating the biological function of an enzyme capable of catalyzing a broad range of nucleotidyl transferase activities in vitro can be challenging. However, published reports on PrimPol from several groups all arrived at a similar conclusion: PrimPol is required to ensure replication fork progression during chromosomal DNA replication, particularly when DNA synthesis is perturbed.3,5,6 PrimPol is also present within mitochondria and was demonstrated to be important for maintenance of the small circular mitochondrial genome,4 probably performing similar roles as those undertaken during nuclear DNA synthesis.
Pathways that stabilize and restart disrupted replication forks
In eukaryotic cells, the DNA replication machinery faces many obstacles that can prevent efficient DNA polymerization by the replicative polymerases, and thus slow or stall replication forks. The best-characterized examples are perhaps when the nucleotide pool is depleted using the ribonucleotide reductase inhibitor hydroxyurea (HU), or when the DNA template contains damage such as base modifications or cross-linking of adjacent pyrimidines induced by UV radiation. Other obstacles include non-B form DNA, RNA-DNA hybrids, and replisome collisions with protein complexes, such as the transcription machinery. To overcome these obstacles and ensure timely and complete genome duplication, eukaryotic cells contain multiple distinct pathways to stabilize and restart disrupted replication forks. In the case of an obstacle such as a DNA lesion, alternative flexible DNA polymerases primarily of the of the Y- and X-families can be employed after stalling of the replicative DNA polymerase to extend the stalled primer terminus by TLS.45,46 Alternatively, the homologous recombination (HR) machinery can allow replication of an alternative sister template to facilitate lesion bypass.47 Additionally, prior to the beginning of DNA synthesis, an excess of replication origins are licensed and these can be fired to allow replication forks to converge on the damaged site, thus allowing completion of bulk DNA synthesis.48 Synthesis of the lagging strand is known to be tolerant of DNA damage because synthesis of a new Okazaki fragment can occur downstream of the lesion. Discontinuous synthesis on the leading strand by re-priming can also occur,49-51 and its mechanism in bacteria has been described.52-54 Discontinuous DNA synthesis will result in the damage being encompassed by single-stranded gaps left behind the replication fork, which can be filled using TLS or HR-mediated mechanisms.55 Additionally, stalled forks can be remodeled by multiple enzymes, facilitating removal of the damage and/or resumption of DNA synthesis.12 There are 2 non-mutually exclusive models for PrimPol-mediated fork progression (Fig. 4). First, PrimPol is able to bypass some DNA lesions using TLS in vitro, and therefore could directly extend the stalled primer terminus to facilitate resumption of DNA synthesis. Second, PrimPol is also a primase and could potentially catalyze origin-independent re-priming downstream of the lesion, allowing discontinuous synthesis of the newly growing DNA strand. Cellular studies supporting these 2 models will be discussed below.
Figure 4.
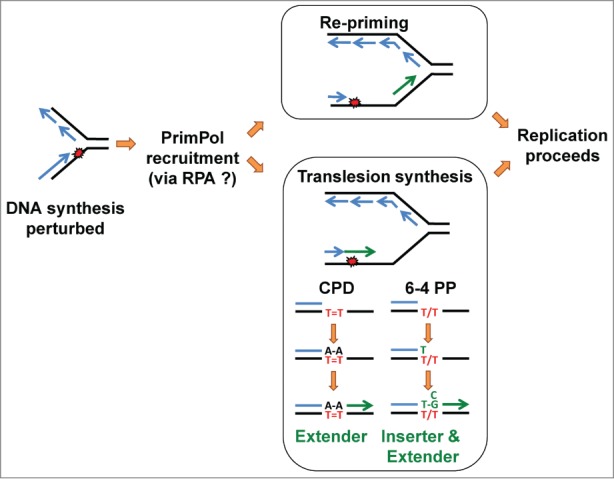
Model of PrimPol-mediated replication fork progression. Distortion of DNA and base modification can be induced by various environmental insults and endogenous processes and, if not corrected prior to replication, can disrupt DNA synthesis by the cellular replicases (blue lines). A DNA modification on the leading strand is shown, which in this example has caused uncoupling of leading and lagging strand synthesis. This generates stretches of single-stranded DNA that will be coated by replication protein A (RPA), which in turn recruits PrimPol. PrimPol-dependent DNA or RNA synthesis (green lines) then facilitates restart of DNA replication. PrimPol may re-prime DNA synthesis downstream of the lesion leaving a daughter strand gap that can be subsequently filled by translesion synthesis (TLS) or homologous recombination (HR)-mediated processes. Alternatively, in the case of DNA lesions such as UV photoproducts (depicted in red lettering), PrimPol can use its TLS activity and directly extend the stalled primer terminus to synthesize DNA opposite the lesion, either alone or by cooperating with another DNA polymerase. For example, in the case of a template cyclobutane pyrimidine dimer (CPD), after incorporation of 2 terminal dA residues opposite the lesion, PrimPol can catalyze the extension of this mismatched terminus. In the case of a pyrimidine (6–4) pyrimidone photoproduct (6–4 PP), PrimPol can catalyze both the insertion of nucleotides opposite the damaged bases and the subsequent extension from the mismatched terminus, and thus could possibly catalyze complete bypass of this lesion. PrimPol misincorporates a dT opposite the 5′T of the lesion and either dG or dC opposite the 3′T, as shown in green.
PrimPol is required for progression of disrupted replication forks
The biological role of PrimPol is most apparent after perturbation of DNA replication. PrimPol assembles into multiple subnuclear foci upon treatment of cells with DNA damage inducing agents; focal accumulation of PrimPol was observed following UV-C irradiation5 and HU-treatment,3,5 and PrimPol relocalized to sites of microirradiation by UV-A laser.6 A common feature of these DNA damaging treatments is their ability to stall cellular replicases. In accordance with this, PrimPol focal accumulation was also observed after treatment with the UV-mimetic 4-nitroquinoline 1-oxide (4NQO) or the alkylating agent methyl methanesulfonate (MMS) (JB, SGR, AJD, unpublished). However, Wan et al.3 reported PrimPol focal accumulation after ionizing radiation (IR), which is known to induce strand breaks and prevent overall DNA synthesis,56 whereas we observed no IR-induced foci.5 DNA damage-induced PrimPol foci were also shown to be sites of chromatin association.5,6 Focal accumulation of a particular enzyme is not always an indicator of enzyme function; however, the DNA damage sensitivities of PrimPol knockdown or knockout cell lines are largely in accordance with this notion. PrimPol depletion using shRNA sensitized HeLa cells to the cytotoxic effects of HU, but not IR,3 and disruption of the Gallus gallus PRIMPOL gene sensitized avian DT40 cells to UV-C and 4NQO, but not IR.5 However, RNAi-depletion of PrimPol in human fibroblasts did not result in UV-C sensitivity,5 which may be explained by the relatively rapid doubling times and larger S-phase population of chicken DT40 cells compared to human cells. Analysis of spread DNA fibers in PrimPol knockdown or knockout (DT40 and MEF) cells that were treated with UV-C or HU indicated a specific role for PrimPol in the resumption of DNA synthesis after replication perturbations,3,5-7 placing PrimPol directly at stalled replication forks in accordance with the chromatin association and DNA damage-induced sensitivities discussed above. Furthermore, PrimPol knockdown cells treated with HU or UV display an increase in replication stress markers, such as chromatin-associated replication protein A (RPA) and phosphorylation (S345) of the intra-S phase checkpoint kinase Chk1.3,5 This indicates that generation of single-stranded DNA is most likely the result of fork stalling and uncoupling of the replicative helicase and polymerase, which occurs more frequently in the absence of PrimPol. Following UV-C irradiation, knockdown cells also showed an increase in chromatin-bound Rad51 recombinase, suggesting HR-mediated rescue of stalled replication forks in the absence of PrimPol,5 and a further increase in origin firing was also reported,6 highlighting the pathways that compensate for the loss of PrimPol.
The defect in the resumption of replication after UV-C irradiation or nucleotide deprivation could be consistent with either the in vitro primase or TLS polymerase activity of PrimPol. In this regard, an important tool is the ‘separation-of-function’ zinc finger mutant that has been used in a number of reports.3,6,7 As discussed above, inactivation of the PrimPol zinc finger abolishes primase activity while leaving polymerase activity intact, although not unaffected. Although the fork-restart defect observed in PrimPol knockdown or knockout cell lines could be rescued by ectopic expression of wild-type PrimPol, it could not be rescued by expression of a zinc finger mutant.3,6,7 This demonstrates that the replication resumption role of PrimPol is dependent on an intact zinc finger, which would be consistent with PrimPol mediating the fork restart by re-priming. However, it should be noted that the zinc finger mutant does modulate polymerase activity in vitro,7 therefore further work will be required to pinpoint the precise role of PrimPol at stalled replication forks. The recruitment of PrimPol to sites of DNA damage is independent of checkpoint signaling by ataxia telangiectasia mutated (ATM) and ATM Rad3 related (ATR) kinases.6 Wan et al.3 demonstrated that PrimPol interacts with RPA1 via a conserved C-terminal region, and that this interaction is required for its localization to sites of damage and its replication restart role. When the replisome encounters a DNA lesion, the helicase and polymerase can functionally uncouple, producing long stretches of single-stranded DNA that are presumably the sites of PrimPol re-priming. It therefore makes sense that PrimPol's role is mediated by RPA interaction and further suggests that this is an immediate response that could be likened to re-priming in prokaryotes as it is an inherent property of the replisome (although catalyzed by the replicative primase).53 The replication restart role of PrimPol following UV-C irradiation is independent of the well-characterized role of the Y-family polymerase Pol η, which is specialized in bypassing CPDs, the most commonly occurring UV-induced lesions. PrimPol depletion renders human cells sensitive to UV-C induced killing only in the absence of functional Pol η, as in cells of xeroderma pigmentosum variant (XP-V) patients,5 and in accordance, a synergistic defect in fork-restart was observed in cells depleted of both PrimPol and Pol η6 and constitutive activation of the intra-S phase checkpoint was observed.5 This could be consistent with both the re-priming and TLS models of PrimPol-mediated fork restart. The in vitro TLS capabilities of PrimPol are complementary to the function of Pol η: whereas Pol η can completely bypass CPDs and only insert opposite 6–4 photoproducts, PrimPol can completely bypass 6–4 photoproducts and only extend from CPDs (Fig. 4).5,57,58 The mutagenic signature of PrimPol activity in vitro, the incorporation of a dT opposite the 3′T of a 6–4 photoproduct, has been observed in UV-irradiated cells, with the polymerase responsible so far remaining unidentified.59,60 Future studies into the role of PrimPol in UV-C–induced mutagenesis will prove insightful, especially for patients with diseases like XP-V in whom UV-induced mutagenesis is known to promote carcinogenesis.
The role of DNA polymerases in tolerance of UV lesions by TLS is well documented;45 however, the role of polymerases after nucleotide deprivation is less well established. TLS polymerases also accumulate into foci following HU treatment61 and it has been demonstrated that Pol η and E. coli Y-family polymerases (Pol IV and Pol V) are required for DNA replication during HU treatment.61,62 In the case of Y-family polymerases, it was suggested that because these polymerases have much lower binding affinities for dNTPs than replicative polymerases, they could take over DNA replication at times of nucleotide deprivation to prevent replication fork collapse,62 which could also be a possible function of PrimPol.
PrimPol is required during an unperturbed S phase
Although the consequences of loss of PrimPol function are most apparent in cells in which DNA replication has been challenged, they can also be observed in otherwise unperturbed cells. PrimPol knockout DT40 cells present minor proliferative defects, such as an increased G2/M transit and an overall reduction of replication fork speeds.5 Proliferative defects and reduced fork speeds were also observed in PrimPol knockdown HeLa cells.6 This reduced efficiency of replication fork progression probably leads to the reported genomic instability, as suggested by an increase in replication stress markers (RPA foci) and DSB markers (γH2AX and 53BP1 foci).3,5,6 An RPA-interaction mutant of PrimPol could not rescue the increase in γH2AX foci in PrimPol knockdown cells, suggesting that interaction with RPA is required for its normal role in S phase.3 Interestingly, the PrimPol zinc finger mutant could complement the reduced replication fork speed in unperturbed conditions, suggesting that the polymerase activity of PrimPol is biologically relevant.7 PrimPol knockdown cells and knockout MEFs also displayed increased chromosome aberrations consistent with S-phase defects, which were further increased after mild replication stress induced by low doses of aphidicolin, an inhibitor of replicative polymerases.3,5,6 To compensate for the loss of PrimPol function, knockdown cells fire dormant origins,6 which could explain the higher G2 population but slower fork speeds observed in PrimPol knockout DT40 cells.5 In support of a function in normal S phase, PrimPol associated with chromatin in a replication-dependent manner in unchallenged cells.5,6 Together, these studies demonstrate that PrimPol is required for replication fork progression during unperturbed S phase and functions to prevent genome instability. This could very well represent a role in bypassing naturally occurring replication obstacles, as re-priming would be a simple and elegant method to bypass any obstacles that do not block the replicative helicase but block the replicative polymerase. TLS polymerases are also known to play important roles during an unchallenged S phase, such as the bypass of non-B form structures,63 replication of chromosomal fragile sites,64,65 and possibly bypass of naturally occurring oxidative damage66 or misincorporated ribonucleotides,67 therefore a role for the TLS polymerase activity of PrimPol should not be excluded.
Underlining the importance of AEPs during unperturbed DNA synthesis, a PrimPol-like protein (PPL2) in Trypanosoma brucei is essential for the completion of genome duplication.8 This is probably a trypanosome-specific PrimPol, because trypanosomes encode another PrimPol-like protein (PPL1) that, like human PrimPol, is a primase-polymerase and dispensable for cell viability. It is likely that the PrimPol gene has been duplicated in trypanosomes and used for a trypanosome-specific DNA metabolic problem. Knockdown of PPL2 leads to the arrest of cells after bulk DNA synthesis with an abundance of irreparable DNA damage. We hypothesized that PPL2 might be responsible for the post-replication bypass of endogenously occurring DNA replication obstacles. This could be a consequence of the unorthodox transcription mechanism, an abundance of structural barriers, or the lack of active origins for replication of the bulk of the DNA,68 which render these parasites more reliant on damage tolerance pathways. Future work is required to pinpoint the precise biological role of PPL2, although at this early stage it appears to represent an attractive target for anti-trypanosomal drugs given that human PrimPol is not an essential gene and PPL2 knockdown results in a lethal phenotype.
Future questions
An interesting avenue for future study would be to determine the functional interplay/redundancy between PrimPol and Pol α-primase, although these experiments would be difficult given that Pol α-primase function is essential for cell viability. Mouron et al.6 observed a small but significant defect in fork restart after UV treatment following partial knockdown of Prim1, which increased after depletion of PrimPol. Yeast lack a detectable PrimPol homolog yet single-stranded gaps in the DNA behind replication forks in UV-irradiated NER-deficient yeast have been observed,50 suggesting that Pol α-primase is responsible for re-priming in yeast. Primers synthesized by Pol α-primase at stalled replication forks play an important role in generating DNA structures responsible for checkpoint signaling.69 Primed single-stranded DNA is required for activation of the intra-S checkpoint,70 and the accumulation of newly synthesized primers at stalled replication forks by Pol α-Primase and their elongation by DNA polymerases contributes to the activation of Chk1.71,72 It is interesting that UV-C irradiated or HU-treated PrimPol knockdown cells show no defect in Chk1 activation,3,5 suggesting that PrimPol plays no significant role in generating these structures. It is possible that checkpoint-activating DNA structures are primarily generated on the lagging strand, therefore if PrimPol is required for leading-strand re-priming its contribution to checkpoint activation would be less pronounced. However, arguing against this, Pol κ is hypothesized to function in leading-strand checkpoint activation and knockdown of Pol κ reduces Chk1 phosphorylation.72 It is also possible that PrimPol is primarily involved in ensuring continuation of DNA replication, while Pol α-primase generates structures for checkpoint activation at stalled forks. It has been suggested that generation of these checkpoint-activating DNA structures might be linked to replication restart,73 in which PrimPol clearly plays an important role. Elucidating the function, if any, of PrimPol in checkpoint activation would be an interesting topic for future research.
Another avenue of future study is the involvement of PrimPol, and its mutations, in human disease. A mutation of PrimPol (Y89D), close to the active site, has been identified in individuals with high myopia.74 A recent report has shown that this mutation significantly reduces the polymerase activity of PrimPol and is associated with defective DNA replication in vivo.75 A number of PrimPol mutations have also been identified in cancers, with the most common being the missense mutation R417L/W (http://www.cbioportal.org/public-portal/).76-79This residue is located in the UL52 zinc-finger and would likely impact upon the biological function of PrimPol. Over-expression of alternate DNA polymerases has been identified in a number of cancers, and has been hypothesised to be a source of the mutator phenotype and replicative stress observed in some cancer cells.80 PrimPol is up-regulated in some cancers, such as glioma81 and so determining the biological effects of PrimPol over-expression would be relevant. Given PrimPol's central role in maintaining replication fork progression, particularly when DNA synthesis is perturbed, and as it is not an essential gene, developing small molecule inhibitors of PrimPol could be a fruitful avenue for future studies. Such inhibitors have the potential to be combined with current genotoxic agents that perturb DNA synthesis to treat a range of cancers, particularly those that up-regulate expression of PrimPol.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Research in the AJD laboratory is supported by grants from Biotechnology and Biological Sciences Research Council (AJD and JB), and a center grant and DTA PhD studentship (SGR) from the Medical Research Council.
References
- 1. Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 2008; 9:204-17; PMID:18227811; http://dx.doi.org/ 10.1038/nrg2268 [DOI] [PubMed] [Google Scholar]
- 2. Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer 2008; 8:193-204; PMID:18256616; http://dx.doi.org/ 10.1038/nrc2342 [DOI] [PubMed] [Google Scholar]
- 3. Wan L, Lou J, Xia Y, Su B, Liu T, Cui J, Sun Y, Lou H, Huang J. hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep 2013; 14:1104-12; PMID:24126761; http://dx.doi.org/ 10.1038/embor.2013.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. García-Gómez S, Reyes A, Martínez-Jiménez MI, Chocrón ES, Mourón S, Terrados G, Powell C, Salido E, Méndez J, Holt IJ, et al. . PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell 2013; 52:541-553; PMID:24207056; http://dx.doi.org/ 10.1016/j.molcel.2013.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bianchi J, Rudd SG, Jozwiakowski SK, Bailey LJ, Soura V, Taylor E, Stevanovic I, Green AJ, Stracker TH, Lindsay HD, et al. . PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol Cell 2013; 52:566-73; PMID:24267451; http://dx.doi.org/ 10.1016/j.molcel.2013.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mourón S, Rodriguez-Acebes S, Martínez-Jiménez MI, García-Gómez S, Chocrón S, Blanco L, Méndez J. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat Struct Mol Biol 2013; 20:1383-9; PMID:24240614; http://dx.doi.org/ 10.1038/nsmb.2719 [DOI] [PubMed] [Google Scholar]
- 7. Keen BA, Jozwiakowski SK, Bailey LJ, Bianchi J, Doherty AJ. Molecular dissection of the domain architecture and catalytic activities of human PrimPol. Nucleic Acids Res 2014; 42:5830-45; PMID:24682820; http://dx.doi.org/ 10.1093/nar/gku214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rudd SG, Glover L, Jozwiakowski SK, Horn D, Doherty AJ. PPL2 translesion polymerase is essential for the completion of chromosomal DNA replication in the African trypanosome. Mol Cell 2013; 52:554-65; PMID:24267450; http://dx.doi.org/ 10.1016/j.molcel.2013.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Im JS, Lee KY, Dillon LW, Dutta A. Human Primpol1: a novel guardian of stalled replication forks. EMBO Rep 2013; 14:1032-3; PMID:24189099; http://dx.doi.org/ 10.1038/embor.2013.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Helleday T. PrimPol breaks replication barriers. Nat Struct Mol Biol 2013; 20:1348-50; PMID:24304914; http://dx.doi.org/ 10.1038/nsmb.2727 [DOI] [PubMed] [Google Scholar]
- 11. Jones RM, Petermann E. Replication fork dynamics and the DNA damage response. Biochem J 2012; 443:13-26; PMID:22417748; http://dx.doi.org/ 10.1042/BJ20112100 [DOI] [PubMed] [Google Scholar]
- 12. Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol 2013; 5:a012815; PMID:23637285; http://dx.doi.org/ 10.1101/cshperspect.a012815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res 2005; 33:3875-96; PMID:16027112; http://dx.doi.org/ 10.1093/nar/gki702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frick DN, Richardson CC. DNA primases. Annu Rev Biochem 2001; 70:39-80; PMID:11395402; http://dx.doi.org/ 10.1146/annurev.biochem.70.1.39 [DOI] [PubMed] [Google Scholar]
- 15. Kuchta RD, Stengel G. Mechanism and evolution of DNA primases. Biochim Biophys Acta 2010; 1804:1180-9; PMID:19540940; http://dx.doi.org/ 10.1016/j.bbapap.2009.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Muzi-Falconi M, Giannattasio M, Foiani M, Plevani P. The DNA polymerase alpha-primase complex: multiple functions and interactions. ScientificWorldJournal 2003; 3:21-33; PMID:12806117; http://dx.doi.org/ 10.1100/tsw.2003.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koonin EV, Wolf YI, Kondrashov AS, Aravind L. Bacterial homologs of the small subunit of eukaryotic DNA primase. J Mol Microbiol Biotechnol 2000; 2:509-12; PMID:11075926 [PubMed] [Google Scholar]
- 18. Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res 2001; 11:1365-74; PMID:11483577; http://dx.doi.org/ 10.1101/gr.181001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weller GR, Doherty AJ. A family of DNA repair ligases in bacteria? FEBS Lett 2001; 505:340-2; PMID:11566200; http://dx.doi.org/ 10.1016/S0014-5793(01)02831-9 [DOI] [PubMed] [Google Scholar]
- 20. Della M, Palmbos PL, Tseng HM, Tonkin LM, Daley JM, Topper LM, Pitcher RS, Tomkinson AE, Wilson TE, Doherty AJ. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science 2004; 306:683-5; PMID:15499016; http://dx.doi.org/ 10.1126/science.1099824 [DOI] [PubMed] [Google Scholar]
- 21. Gong C, Bongiorno P, Martins A, Stephanou NC, Zhu H, Shuman S, Glickman MS. Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat Struct Mol Biol 2005; 12:304-12; PMID:15778718; http://dx.doi.org/ 10.1038/nsmb915 [DOI] [PubMed] [Google Scholar]
- 22. Pitcher RS, Tonkin LM, Green AJ, Doherty AJ. Domain structure of a NHEJ DNA repair ligase from Mycobacterium tuberculosis. J Mol Biol 2005; 351:531-44; PMID:16023671; http://dx.doi.org/ 10.1016/j.jmb.2005.06.038 [DOI] [PubMed] [Google Scholar]
- 23. Pitcher RS, Brissett NC, Picher AJ, Andrade P, Juarez R, Thompson D, Fox GC, Blanco L, Doherty AJ. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J Mol Biol 2007; 366:391-405; PMID:17174332; http://dx.doi.org/ 10.1016/j.jmb.2006.10.046 [DOI] [PubMed] [Google Scholar]
- 24. Yakovleva L, Shuman S. Nucleotide misincorporation, 3'-mismatch extension, and responses to abasic sites and DNA adducts by the polymerase component of bacterial DNA ligase D. J Biol Chem 2006; 281:25026-40; PMID:16816388; http://dx.doi.org/ 10.1074/jbc.M603302200 [DOI] [PubMed] [Google Scholar]
- 25. Brissett NC, Pitcher RS, Juarez R, Picher AJ, Green AJ, Dafforn TR, Fox GC, Blanco L, Doherty AJ. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science 2007; 318:456-9; PMID:17947582; http://dx.doi.org/ 10.1126/science.1145112 [DOI] [PubMed] [Google Scholar]
- 26. Brissett NC, Martin MJ, Pitcher RS, Bianchi J, Juarez R, Green AJ, Fox GC, Blanco L, Doherty AJ. Structure of a preternary complex involving a prokaryotic NHEJ DNA polymerase. Mol Cell 2011; 41:221-31; PMID:21255731; http://dx.doi.org/ 10.1016/j.molcel.2010.12.026 [DOI] [PubMed] [Google Scholar]
- 27. Brissett NC, Martin MJ, Bartlett EJ, Bianchi J, Blanco L, Doherty AJ. Molecular basis for DNA double-strand break annealing and primer extension by an NHEJ DNA polymerase. Cell Rep 2013; 5:1108-20; PMID:24239356; http://dx.doi.org/ 10.1016/j.celrep.2013.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bartlett EJ, Brissett NC, Doherty AJ. Ribonucleolytic resection is required for repair of strand displaced nonhomologous end-joining intermediates. Proc Natl Acad Sci U S A 2013; 110:E1984-91; PMID:23671117; http://dx.doi.org/ 10.1073/pnas.1302616110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bocquier AA, Liu L, Cann IK, Komori K, Kohda D, Ishino Y. Archaeal primase: bridging the gap between RNA and DNA polymerases. Curr Biol 2001; 11:452-6; PMID:11301257; http://dx.doi.org/ 10.1016/S0960-9822(01)00119-1 [DOI] [PubMed] [Google Scholar]
- 30. Matsui E, Nishio M, Yokoyama H, Harata K, Darnis S, Matsui I. Distinct domain functions regulating de novo DNA synthesis of thermostable DNA primase from hyperthermophile Pyrococcus horikoshii. Biochemistry 2003; 42:14968-76; PMID:14674773; http://dx.doi.org/ 10.1021/bi035556o [DOI] [PubMed] [Google Scholar]
- 31. Lao-Sirieix SH, Bell SD. The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase and 3′-terminal nucleotidyl transferase activities. J Mol Biol 2004; 344:1251-63; PMID:15561142; http://dx.doi.org/ 10.1016/j.jmb.2004.10.018 [DOI] [PubMed] [Google Scholar]
- 32. Chemnitz Galal W, Pan M, Kelman Z, Hurwitz J. Characterization of DNA primase complex isolated from the archaeon, Thermococcus kodakaraensis. J Biol Chem 2012; 287:16209-19; PMID:22351771; http://dx.doi.org/ 10.1074/jbc.M111.338145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. De Falco M, Fusco A, De Felice M, Rossi M, Pisani FM. The DNA primase of Sulfolobus solfataricus is activated by substrates containing a thymine-rich bubble and has a 3'-terminal nucleotidyl-transferase activity. Nucleic Acids Res 2004; 32:5223-30; PMID:15459292; http://dx.doi.org/ 10.1093/nar/gkh865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Le Breton M, Henneke G, Norais C, Flament D, Myllykallio H, Querellou J, Raffin JP. The heterodimeric primase from the euryarchaeon Pyrococcus abyssi: a multifunctional enzyme for initiation and repair? J Mol Biol 2007; 374:1172-85; PMID:17991487; http://dx.doi.org/ 10.1016/j.jmb.2007.10.015 [DOI] [PubMed] [Google Scholar]
- 35. Hu J, Guo L, Wu K, Liu B, Lang S, Huang L. Template-dependent polymerization across discontinuous templates by the heterodimeric primase from the hyperthermophilic archaeon Sulfolobus solfataricus. Nucleic Acids Res 2012; 40:3470-83; PMID:22189102; http://dx.doi.org/ 10.1093/nar/gkr1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lao-Sirieix SH, Pellegrini L, Bell SD. The promiscuous primase. Trends Genet 2005; 21:568-72; PMID:16095750; http://dx.doi.org/ 10.1016/j.tig.2005.07.010 [DOI] [PubMed] [Google Scholar]
- 37. Lipps G, Röther S, Hart C, Krauss G. A novel type of replicative enzyme harbouring ATPase, primase and DNA polymerase activity. EMBO J 2003; 22:2516-25; PMID:12743045; http://dx.doi.org/ 10.1093/emboj/cdg246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lipps G, Weinzierl AO, von Scheven G, Buchen C, Cramer P. Structure of a bifunctional DNA primase-polymerase. Nat Struct Mol Biol 2004; 11:157-62; PMID:14730355; http://dx.doi.org/ 10.1038/nsmb723 [DOI] [PubMed] [Google Scholar]
- 39. Beck K, Lipps G. Properties of an unusual DNA primase from an archaeal plasmid. Nucleic Acids Res 2007; 35:5635-45; PMID:17709343; http://dx.doi.org/ 10.1093/nar/gkm625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Halgasova N, Mesarosova I, Bukovska G. Identification of a bifunctional primase-polymerase domain of corynephage BFK20 replication protein gp43. Virus Res 2012; 163:454-60; PMID:22108584; http://dx.doi.org/ 10.1016/j.virusres.2011.11.005 [DOI] [PubMed] [Google Scholar]
- 41. Sanchez-Berrondo J, Mesa P, Ibarra A, Martínez-Jiménez MI, Blanco L, Méndez J, Boskovic J, Montoya G. Molecular architecture of a multifunctional MCM complex. Nucleic Acids Res 2012; 40:1366-80; PMID:21984415; http://dx.doi.org/ 10.1093/nar/gkr831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. De Silva FS, Paran N, Moss B. Products and substrate/template usage of vaccinia virus DNA primase. Virology 2009; 383:136-41; PMID:19007959; http://dx.doi.org/ 10.1016/j.virol.2008.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hines JC, Ray DS. A mitochondrial DNA primase is essential for cell growth and kinetoplast DNA replication in Trypanosoma brucei. Mol Cell Biol 2010; 30:1319-28; PMID:20065037; http://dx.doi.org/ 10.1128/MCB.01231-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hines JC, Ray DS. A second mitochondrial DNA primase is essential for cell growth and kinetoplast minicircle DNA replication in Trypanosoma brucei. Eukaryot Cell 2011; 10:445-54; PMID:21257796; http://dx.doi.org/ 10.1128/EC.00308-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol 2012; 13:141-52; PMID:22358330; http://dx.doi.org/ 10.1038/nrm3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hübscher U, Maga G. DNA replication and repair bypass machines. Curr Opin Chem Biol 2011; 15:627-35; PMID:21889903; http://dx.doi.org/ 10.1016/j.cbpa.2011.08.009 [DOI] [PubMed] [Google Scholar]
- 47. Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 2008; 18:99-113; PMID:18166982; http://dx.doi.org/ 10.1038/cr.2008.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yekezare M, Gómez-González B, Diffley JF. Controlling DNA replication origins in response to DNA damage - inhibit globally, activate locally. J Cell Sci 2013; 126:1297-306; PMID:23645160; http://dx.doi.org/ 10.1242/jcs.096701 [DOI] [PubMed] [Google Scholar]
- 49. Lehmann AR. Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J Mol Biol 1972; 66:319-37; PMID:5037019; http://dx.doi.org/ 10.1016/0022-2836(72)90418-4 [DOI] [PubMed] [Google Scholar]
- 50. Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 2006; 21:15-27; PMID:16387650; http://dx.doi.org/ 10.1016/j.molcel.2005.11.015 [DOI] [PubMed] [Google Scholar]
- 51. Elvers I, Johansson F, Groth P, Erixon K, Helleday T. UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res 2011; 39:7049-57; PMID:21646340; http://dx.doi.org/ 10.1093/nar/gkr420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature 2006; 439:557-62; PMID:16452972; http://dx.doi.org/ 10.1038/nature04329 [DOI] [PubMed] [Google Scholar]
- 53. Yeeles JT, Marians KJ. The Escherichia coli replisome is inherently DNA damage tolerant. Science 2011; 334:235-8; PMID:21998391; http://dx.doi.org/ 10.1126/science.1209111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yeeles JT, Marians KJ. Dynamics of leading-strand lesion skipping by the replisome. Mol Cell 2013; 52:855-65; PMID:24268579; http://dx.doi.org/ 10.1016/j.molcel.2013.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ulrich HD. Timing and spacing of ubiquitin-dependent DNA damage bypass. FEBS Lett 2011; 585:2861-7; PMID:21605556; http://dx.doi.org/ 10.1016/j.febslet.2011.05.028 [DOI] [PubMed] [Google Scholar]
- 56. Lamb JR, Petit-Frère C, Broughton BC, Lehmann AR, Green MH. Inhibition of DNA replication by ionizing radiation is mediated by a trans-acting factor. Int J Radiat Biol 1989; 56:125-30; PMID:2569014; http://dx.doi.org/ 10.1080/09553008914551271 [DOI] [PubMed] [Google Scholar]
- 57. Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol eta. Science 1999; 283:1001-4; PMID:9974380; http://dx.doi.org/ 10.1126/science.283.5404.1001 [DOI] [PubMed] [Google Scholar]
- 58. Johnson RE, Haracska L, Prakash S, Prakash L. Role of DNA polymerase eta in the bypass of a (6-4) TT photoproduct. Mol Cell Biol 2001; 21:3558-63; PMID:11313481; http://dx.doi.org/ 10.1128/MCB.21.10.3558-3563.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Szüts D, Marcus AP, Himoto M, Iwai S, Sale JE. REV1 restrains DNA polymerase zeta to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo. Nucleic Acids Res 2008; 36:6767-80; PMID:18953031; http://dx.doi.org/ 10.1093/nar/gkn651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hirota K, Sonoda E, Kawamoto T, Motegi A, Masutani C, Hanaoka F, Szüts D, Iwai S, Sale JE, Lehmann A, et al. . Simultaneous disruption of two DNA polymerases, Polη and Polζ, in avian DT40 cells unmasks the role of Polη in cellular response to various DNA lesions. PLoS Genet 2010; 6:e1001151; PMID:20949111; http://dx.doi.org/ 10.1371/journal.pgen.1001151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. de Feraudy S, Limoli CL, Giedzinski E, Karentz D, Marti TM, Feeney L, Cleaver JE. Pol eta is required for DNA replication during nucleotide deprivation by hydroxyurea. Oncogene 2007; 26:5713-21; PMID:17369853; http://dx.doi.org/ 10.1038/sj.onc.1210385 [DOI] [PubMed] [Google Scholar]
- 62. Godoy VG, Jarosz DF, Walker FL, Simmons LA, Walker GC. Y-family DNA polymerases respond to DNA damage-independent inhibition of replication fork progression. EMBO J 2006; 25:868-79; PMID:16482223; http://dx.doi.org/ 10.1038/sj.emboj.7600986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bétous R, Rey L, Wang G, Pillaire MJ, Puget N, Selves J, Biard DS, Shin-ya K, Vasquez KM, Cazaux C, Hoffmann JS. Role of TLS DNA polymerases eta and kappa in processing naturally occurring structured DNA in human cells. Mol Carcinog 2009; 48:369-78; PMID:19117014; http://dx.doi.org/ 10.1002/mc.20509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rey L, Sidorova JM, Puget N, Boudsocq F, Biard DS, Monnat RJ, Jr, Cazaux C, Hoffmann JS. Human DNA polymerase eta is required for common fragile site stability during unperturbed DNA replication. Mol Cell Biol 2009; 29:3344-54; PMID:19380493; http://dx.doi.org/ 10.1128/MCB.00115-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bergoglio V, Boyer AS, Walsh E, Naim V, Legube G, Lee MY, Rey L, Rosselli F, Cazaux C, Eckert KA, et al. . DNA synthesis by Pol η promotes fragile site stability by preventing under-replicated DNA in mitosis. J Cell Biol 2013; 201:395-408; PMID:23609533; http://dx.doi.org/ 10.1083/jcb.201207066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lange SS, Wittschieben JP, Wood RD. DNA polymerase zeta is required for proliferation of normal mammalian cells. Nucleic Acids Res 2012; 40:4473-82; PMID:22319213; http://dx.doi.org/ 10.1093/nar/gks054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lazzaro F, Novarina D, Amara F, Watt DL, Stone JE, Costanzo V, Burgers PM, Kunkel TA, Plevani P, Muzi-Falconi M. RNase H and postreplication repair protect cells from ribonucleotides incorporated in DNA. Mol Cell 2012; 45:99-110; PMID:22244334; http://dx.doi.org/ 10.1016/j.molcel.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tiengwe C, Marques CA, McCulloch R. Nuclear DNA replication initiation in kinetoplastid parasites: new insights into an ancient process. Trends Parasitol 2014; 30:27-36; PMID:24287149; http://dx.doi.org/ 10.1016/j.pt.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 69. Michael WM, Ott R, Fanning E, Newport J. Activation of the DNA replication checkpoint through RNA synthesis by primase. Science 2000; 289:2133-7; PMID:11000117; http://dx.doi.org/ 10.1126/science.289.5487.2133 [DOI] [PubMed] [Google Scholar]
- 70. MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev 2007; 21:898-903; PMID:17437996; http://dx.doi.org/ 10.1101/gad.1522607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Van C, Yan S, Michael WM, Waga S, Cimprich KA. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J Cell Biol 2010; 189:233-46; PMID:20385778; http://dx.doi.org/ 10.1083/jcb.200909105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bétous R, Pillaire MJ, Pierini L, van der Laan S, Recolin B, Ohl-Séguy E, Guo C, Niimi N, Grúz P, Nohmi T, et al. . DNA polymerase κ-dependent DNA synthesis at stalled replication forks is important for CHK1 activation. EMBO J 2013; 32:2172-85; PMID:23799366; http://dx.doi.org/ 10.1038/emboj.2013.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yan S, Michael WM. TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation. Cell Cycle 2009; 8:2877-84; PMID:19652550; http://dx.doi.org/ 10.4161/cc.8.18.9485 [DOI] [PubMed] [Google Scholar]
- 74. Zhao F, Wu J, Xue A, Su Y, Wang X, Lu X, Zhou Z, Qu J, Zhou X. Exome sequencing reveals CCDC111 mutation associated with high myopia. Hum Genet 2013; 132:913-21; PMID:23579484; http://dx.doi.org/ 10.1007/s00439-013-1303-6 [DOI] [PubMed] [Google Scholar]
- 75. Keen BA, Bailey LJ, Jozwiakowski SK, Doherty AJ Human PrimPol mutation associated with high myopia has a DNA replication defect. Nucleic Acids Res. 2014; http://dx.doi.org/ 10.1093/nar/gku879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cancer Genome Atlas Research Network . Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489:519-25; PMID:22960745; http://dx.doi.org/ 10.1038/nature11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cancer Genome Atlas Research Network . Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014; 507:315-22; PMID:24476821; http://dx.doi.org/ 10.1038/nature12965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. . Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012; 150:1107-20; PMID:22980975; http://dx.doi.org/ 10.1016/j.cell.2012.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. . The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2:401-4; PMID:22588877; http://dx.doi.org/ 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hoffmann JS, Cazaux C. Aberrant expression of alternative DNA polymerases: a source of mutator phenotype as well as replicative stress in cancer. Semin Cancer Biol 2010; 20:312-9; PMID:20934518; http://dx.doi.org/ 10.1016/j.semcancer.2010.10.001 [DOI] [PubMed] [Google Scholar]
- 81. Yan X, Ma L, Yi D, Yoon JG, Diercks A, Foltz G, Price ND, Hood LE, Tian Q. A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc Natl Acad Sci U S A 2011; 108:1591-6; PMID:21220328; http://dx.doi.org/ 10.1073/pnas.1018696108 [DOI] [PMC free article] [PubMed] [Google Scholar]