Abstract
During autophagy, a double-membraned vesicle called the autophagosome is responsible for the degradation of long-lived proteins and damaged/old organelles, thus contributing to the maintenance of cellular homeostasis. Physiological stimuli and stressors enhance autophagy in order to accomplish important processes such as cell differentiation or as a cytoprotective response. In line with this, numerous studies have demonstrated the relevance of proper autophagy regulation to health. Autophagy defects are associated with the insurgence of neurological/neurodegenerative diseases and cancer. Moreover, the autophagy pathway is often potentiated in cancer cells to increase cell survival. Increased knowledge of the molecular mechanisms underlying autophagy regulation and their interplay with other cellular pathways would provide advances in cancer treatment. In this context, post-translational modifications, protein–protein interactions, and regulative feedback loops offer promising insights. In this review, we focus on AMBRA1, a proautophagic protein that was recently demonstrated to participate in numerous crucial regulative mechanisms of the autophagy process.
Keywords: apoptosis, cancer, cell survival, embryonic development, mitophagy
Abbreviations
- AD
Alzheimer's disease
- AMBRA1
activating molecule in Beclin 1-regulated autophagy
- ASD
autism-spectrum disorder
- ATG
autophagy related gene
- BCL2
B-cell lymphoma 2
- BH3
Bcl-2 homology3
- BIM
Bcl2-interacting mediator of cell death
- CB1R
Cannabinoid receptor type 1
- CUL4
Cullin 4
- DDA1
DET1- and DDB1-associated protein 1
- DDB1
Damage-specific DNA binding protein 1
- DFCP1
Double FYVE-containing protein 1
- DLC1/2
Dynein light chain 1/2
- E16.5
Embryonic day 16.5
- ER
Endoplasmic reticulum
- FIP200
FAK family kinase-interacting protein of 200 kDa
- GPCRs
G protein-coupled receptors
- ID
Intrinsically disordered
- IDR
intrinsically disordered region
- IDP
Intrinsically disordered protein
- JNK
c-Jun N-terminal kinase
- K48-ubiquitylation
Lys48-ubiquitilation
- K63-ubiquitylation
Lys63-ubiquitilation
- LC3
Light chain 3
- LIR
LC3-interacting region
- MIZ1
Myc-interacting zinc finger protein 1
- MLST8
Target of rapamycin complex subunit LST8
- mVPS
mammalian vacuolar protein sorting
- mTOR
mammalian target of rapamycin
- mTORC1
mammalian target of rapamycin complex1
- NeP
Neuropatic pain
- PcG
Polycomb group
- PI3KIII
Class III phosphatidyl inositol 3-Kinase
- PI3P
Phosphatidyl inositol 3-phosphate
- PINK1
PTEN-induced putative kinase 1
- RAPTOR
Regulatory-associated protein of TOR 1
- RNF2
Ring finger protein
- SCs
Shwann cells
- Shh
Sonic hedgehog protein
- TP53
Tumor suppressor p53
- TRAF6
TNFR-associated factor 6
- ULK1
unc-51 like autophagy activating kinase 1
- ULK2
Unc-51-like kinase 2
- WASH
SCAR homolog protein
- WASP
Whiscott-Aldrich syndrome protein
Introduction
In recent years, both physiological and pathological roles have been attributed to macroautophagy (hereafter referred to as autophagy).1 This cellular process is normally dedicated to the removal of damaged/old organelles, protein aggregates, and long-lived proteins, thus ensuring quality control of cellular components and maintaining cellular homeostasis. On the other hand, upon environmental stresses and in response to certain stimuli cells can undergo a dramatic increase in the level of autophagy (called ‘induced autophagy’). In this context, autophagy constitutes a crucial cytoprotective response and allows adaptation and survival in unfavorable conditions.1-3
In the last decade the proteins on which the autophagy pathway relies have been identified. Comprehensively, they are part of the ‘core autophagy machinery’ and mainly constitute 4 macromolecular complexes that have been already reviewed in detail.3 Briefly, these complexes are the Unc-51 like kinase 1 (ULK1) kinase complex, the Class III phosphatidyl inositol 3-kinase (PI3KIII) complex, the ubiquitin-like conjugation system (mainly composed of ATG12 and LC3), and transmembrane proteins such as mATG9.3
The ULK1 and PI3KIII complexes both participate in the early steps of autophagy leading to the formation of a double-membraned organelle called the autophagosome, which is unique to the autophagy process. The autophagosome swallows autophagy substrates such as entire organelles and long-lived proteins, which, after fusion of the autophagosome with a lysosome, are degraded by lytic enzymes.3
In recent years, our knowledge of autophagy regulation has grown exponentially along with our appreciation of the complexity of the autophagy pathway. The main reasons for this complexity are (i) a plethora of other signaling pathways that impact on autophagy, and vice versa; (ii) the large number of post-translational modifications occurring on autophagy-related proteins, which are being identified at a rapid rate and embrace various kinds of enzymatic reactions; (iii) positive- and negative-feedback loops that allow tight regulation of the autophagy pathway.
In 2007, a new positive regulator of autophagy was identified: activating molecule in beclin 1-regulated autophagy 1 (Ambra1).4 Within a few years, Ambra1 was demonstrated be a critical regulator of autophagy, undergoing and mediating post-translational modifications and regulative feedback loops. A large number of Ambra1 interacting proteins have also been identified (Table 1), accounting for crosstalk between autophagy and other cellular processes.5
Table 1.
Interaction networks and post-translational modifications of AMBRA1. Known AMBRA1 binding partners and functional interactors are listed. Where characterized, the binding sites, localization, and the functional relevance of the interaction are reported.
| Binding protein | Binding-site on AMBRA1 | Post translational modifications on AMBRA1 | Localisation | Function | Effect on autophagy | References |
|---|---|---|---|---|---|---|
| BECLIN 1 |
aa 533-751 |
none |
Cytosol, Cytoskeleton, ER |
Increase of mVPS34 kinase activity |
+ |
5, 18 |
| DLC1/2 |
aa 1075-1077, aa 1087-1089 |
none |
Cytoskeleton |
Prevention of PI3KIII complex localisation to the ER |
− |
18 |
| mTORC1 |
? |
Phosphorylation at Ser52 |
? |
Prevention of AMBRA1 activation |
− |
23 |
| ULK1 |
aa 1-532, aa 752-1269 |
Phosphorylation |
Cytosol |
AMBRA1 dissociation from DLC1 |
+ |
18, 23 |
| TRAF6 |
aa 618-623, aa 680-685 |
none |
Cytosol |
Substrate receptor for the ubiquitylation of ULK1 |
+ |
23 |
| RNF2 |
? |
K48-ubiquitylation at Lys45 |
? |
AMBRA1 degradation by the proteasome |
− |
26 |
| CULLIN4–DDB1–complex |
? |
none |
? |
As substrate receptor for ubiquitylation of BECLIN 1 |
+ |
11, 30, 35 |
| Parkin |
aa 1-532 |
none |
Mitochondria |
Increase of VPS34 kinase activity |
+ |
38 |
| LC3 |
aa 1013-1022 |
none |
Mitochondria |
Mitochondria clearance |
? |
39 |
| BCL2 |
aa 1-532, aa 752-1269 |
none |
Mitochondria |
Sequesters AMBRA1 at mitochondria |
− |
41 |
| BECLIN 2 |
? |
none |
? |
Endosomal trafficking and GPCRs degradation |
? |
77 |
| MIZ1 |
? |
none |
? |
Transcriptional regulation |
? |
59 |
| Calpain/ Caspases | ? | Cleavage at Asp484 | ? | Cleavage | − | 43 |
Ambra1 is a 1,300-amino acid protein with a predicted molecular weight of 130 kDa. The AMBRA1 gene is located on chromosome 11 and is composed of 18 exons.4 Of the large number of alternative transcript variants predicted to exist (Ensembl Database), it will be interesting to determine which encode functional products and the significance of these proteins.
AMBRA1 is absent in lower eukaryotes but is highly conserved among vertebrates.4 The structure of the protein is characterized by WD40-domains in the N-terminal region. WD40 domains are highly abundant in eukaryotes and can participate in diverse cellular functions (including signal transduction, cell division, and RNA processing) by mediating protein–protein, protein–peptide, and protein–DNA interactions. These domains are probably the most pervasive interactors in the cell because they can provide platforms for the assembly of macromolecular complexes; for this reason WD40 domains are also peculiar to scaffold proteins.6
Intriguingly, no other protein domains have been assigned to the rest of the AMBRA1 protein sequence, thus labeling AMBRA1 as an ‘intrinsically disordered protein’ (IDP) (Pfam database).7,8 IDPs are characterized by long regions of protein sequence that lack recognizable domains. Such regions are thought to fold upon binding to their interaction partners, thus allowing different protein folding with different partners.9,10 Recently, it has been found that many autophagy proteins contain at least small regions that are intrinsically disordered (ID); functional analysis of these IDs revealed that they are enriched in protein–protein interaction sequences.7,8 This feature accounts for the great plasticity of AMBRA1 and the other ID autophagy proteins by increasing the number of possible protein–protein interactions that they can undergo. Such a characteristic is probably very useful within the complex interaction network of autophagy itself and for the extensive crosstalk between autophagy and other pathways.1,11,12
AMBRA1-Mediated Regulation of Autophagy
Autophagy regulation at the cytoskeleton
In the early steps of autophagy, a pool of phosphatidyl inositol 3-phosphate (PI3P) is required in order to form the autophagosome. The PI3P localizes to a specific site of the endoplasmic reticulum (ER) with a Ω-like shape, called the ‘omegasome’. The omegasome is a platform for recruitment of the autophagy machinery proteins to the autophagosome nucleation site.13,14 In mammals, the BECLIN 1/PI3KIII complex responsible for the production of PI3P consists of the kinase mVPS34, the membrane-associated protein mVPS15, and BECLIN 1. Diverse autophagy proteins have been found to bind BECLIN 1 in order to activate or inhibit PI3KIII or confer other functions on the BECLIN 1/PI3KIII complex.15 In particular, AMBRA1 directly binds BECLIN 1, increasing its interaction with mVPS34. This leads to stronger activation of mVPS34 and autophagosome formation, thus promoting autophagy.4
In recent years, various groups have described interactions of the BECLIN 1/PI3KIII complex with cytoskeletal components,16,17 and AMBRA1 has also been shown to play a role in this context.18 In fact, the interaction of AMBRA1 with BECLIN 1 is responsible for tethering the entire BECLIN 1/PI3KIII complex to the dynein motor complex. This localization is mediated by binding of AMBRA1 to the dynein light chain1/2 (DLC1/2) by two specific DLC-binding motifs (TQT) (Table 1) at the carboxy terminal region of AMBRA1.18 This tethering exerts negative regulation on autophagy by preventing localization of the PI3KIII complex to the ER, where it induces PI3P production and autophagosome formation. Upon autophagy induction, a phosphorylation event (see below) on AMBRA1 is responsible for release of the PI3KIII complex, allowing full autophagy induction.18
The BECLIN 1/PI3KIII complex can be recruited to the cytoskeleton by BECLIN 1 binding partners in 2 other ways. In the first case, BIM, a BH3-only protein, mediates the binding of BECLIN 1 to the dynein motor complex, thus inhibiting autophagy. Again, upon proautophagic stimuli, a JNK-mediated phosphorylation event on BIM protein is responsible for BECLIN 1/PI3KIII translocation to the ER and autophagy induction.13
Secondly, through AKT-mediated phosphorylation, BECLIN 1 is recruited to the intermediate filaments by the 14-3-3 proteins, which bind VIMENTIN.15 In this scenario, a dephosphorylation occurs to reactivate the BECLIN 1/PI3KIII complex.
It is evident that interactions of BECLIN 1 with the cytoskeleton are highly relevant to proper autophagy regulation but several open questions remain, for example: (i) Are these cytoskeleton-mediated regulation mechanisms coordinated and/or correlated? (ii) Do they capture distinct pools of BECLIN 1? (iii) Do their upstream signals work in a synergistic or an antagonist manner?
It has been known for decades that the cytoskeleton constitutes the structure along which the formed autophagosomes fuse with lysosomes.19 Taken together, the data reported here also highlight the role of the cytoskeleton as a crucial regulator of the first steps of autophagy induction.
Phosphorylation and ubiquitylation
It has long been established that phosphorylation events are crucial for the initiation of autophagy by regulating the activity of and interactions among key players of upstream signaling of autophagy. There is growing evidence that post-translational modifications, including phosphorylation and ubiquitylation of AMBRA1 protein and its binding partners, play an important role in autophagy induction and progression.
Two different kinases are known to regulate AMBRA1 by phosphorylation: the major nutrient/energy sensor, mTOR, and the proautophagic kinase ULK1.18,20
mTOR is a highly conserved serine/threonine kinase that coordinates signals to maintain metabolic homeostasis.21 It is part of a complex (mTORC1) consisting of mTOR, RAPTOR, and MLST8 and functions as a potent repressor of autophagy in eukaryotes. mTORC1 is able to directly phosphorylate the ULK1 complex to repress its activity in autophagy initiation.22 Recent studies demonstrate that under nutrient-rich conditions mTORC1 binds directly to AMBRA1 and is able to phosphorylate AMBRA1 at Serine 52; this mTOR-driven AMBRA1 phosphorylation correlates with autophagy inhibition (Fig. 1).20,23 How this phosphorylation event on AMBRA1 exerts its inhibitory effects remains to be characterized, although it could sequester or physically separate AMBRA1 from its binding partners. The identification of relevant phosphatases will also be important to gain a deeper understanding of how this complex is regulated. Moreover, it is still unclear whether mTOR-AMBRA1 association occurs in the cytosol or in the proximity of certain structures such as lysosomes or the cytoskeleton.
Figure 1.
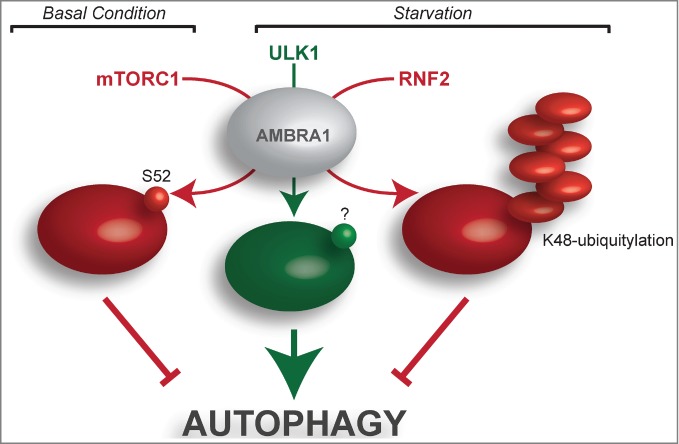
Major phosphorylation and ubiquitylation events targeting AMBRA1. In the presence of nutrients (basal condition), mTORC1 is responsible for AMBRA1 phosphorylation at Ser52 (S52), resulting in AMBRA1 inhibition and prevention of autophagy induction. Upon nutrient deprivation (starvation), the ULK1-mediated phosphorylation of AMBRA1 promotes the relocalization of the BECLIN 1/PI3KIII complex to the endoplasmic reticulum, where it participates in autophagosome formation. The AMBRA1 residue(s) affected by ULK1 phosphorylation (?) are still unknown. After prolonged starvation, AMBRA1 is targeted by K48-ubiquitylation, which is responsible for its proteasome-mediated degradation. This event constitutes a negative feedback loop upon autophagy induction. Pro- and antiautophagic molecules are represented in green and red, respectively.
As stated above, the serine/threonine kinase ULK1 also regulates AMBRA1 activation in response to amino acid starvation.18 Regulation very upstream of the autophagy pathway focuses on the dynamic interaction between AMBRA1 and the dynein motor complex. Phosphorylation of AMBRA1 by ULK1 (Fig. 1) and the association of AMBRA1 with DLC1 are crucial steps in this subtle regulation. Indeed, this ULK1-mediated AMBRA1 phosphorylation has the potential to prime detachment of the AMBRA1–BECLIN 1–mVPS34 complex from cytoskeletal docking sites and mediate its translocation to the endoplasmic reticulum, where it regulates autophagosome nucleation; however, the ULK1 phosphorylation sites on AMBRA1 protein remain to be identified. Another important goal will be identification of the factors responsible for docking the AMBRA1–BECLIN 1 complex on the omegasome compartment, with DFCP1 (or other PI3P-binding proteins) or ATG14 being good candidates for such a role.
Although two of the kinases responsible for AMBRA1 phosphorylation have been identified, more than 20 different AMBRA1 phosphorylation sites have been reported, some of which are modulated in a cell-cycle-regulated manner.24,25 This suggests that further studies are necessary to define AMBRA1 regulation by phosphorylation.
The regulation of AMBRA1 is not limited to phosphorylation. Ubiquitylation is another important post-translational modification that regulates multiple autophagy proteins. Very recently, Xia and colleagues identified AMBRA1 as a novel substrate of the E3 ligase RNF2.26 RNF2, also known as RING1B, is an E3 ligase belonging to the Polycomb group (PcG) of proteins that alter chromatin conformation and mediate target gene repression.27 The PcG complex acts via chromatin remodeling and modification (such as ubiquitylation) of histones, leading to heritable changes in the expressibility of chromatin. A recent study shows that RNF2 also polyubiquitylates the tumor suppressor TP53 in selective cell types, leading to tumor formation.28
Use of RNF2 as a bait in a yeast 2-hybrid system identified AMBRA1 as an interactor of RNF2, suggesting that RNF2 might also play a role in autophagy.26 In particular, Xia et al. found that RNF2 ubiquitylates AMBRA1 at Lysine 45 by Lys48-linked ubiquitin chains (Fig. 1); this modification mediates AMBRA1 degradation by the proteasome. Importantly, AMBRA1 Lys48-ubiquitylation occurs after the induction of autophagy by starvation and leads to autophagy suppression. Specifically, the authors found that the recruitment of RNF2 for AMBRA1 degradation is mediated by Wiskott-Aldrich syndrome protein (WASP) and SCAR homolog protein (WASH), an endosomal protein present in the FAM21-containing multiprotein complex.29 The same group recently proposed WASH as an inhibitor of autophagy by mediating the suppression of BECLIN 1 ubiquitylation30 and, in this case, mediating AMBRA1 degradation. Of note, an independent paper reported that the WASH complex is instead necessary for autophagosome formation, acting as a positive regulator of autophagy.31 Further studies are therefore necessary to clarify the role of the WASH complex in autophagy.
It is also worth noting that other sites in AMBRA1 were found to be modified by ubiquitin in a study characterizing the ubiquitin-modified proteome (ubiquitinome)32 and that AMBRA1 has been also identified as a substrate for cullin-related ligase (CRL) complexes.33,34
AMBRA1 as an adaptor substrate for the E3 ligases
In addition to the above studies that clearly link ubiquitylation to the regulation of AMBRA1, some additional reports related to ubiquitylation could also be relevant when studying the regulation of AMBRA1. In fact, AMBRA1 should not just be considered a substrate for ubiquitylation as many papers propose AMBRA1 as an essential co-factor for the activity of different E3 ligases. AMBRA1, also known as DCAF3, has been found to be a binding partner of the CUL4–DDB1–DDA1 CRL complex.11,35 DCAF substrate receptors confer E3 substrate specificity36 and Xia et al. recently found that AMBRA1 is the substrate receptor for ubiquitylation of BECLIN 1 in starvation-induced autophagy.30 In particular, they found that AMBRA1 acts as a substrate receptor for the DDB1–Cullin 4 E3 ubiquitin ligase complex mediating Lys63-linked ubiquitylation of BECLIN 1. This modification on BECLIN 1 has been proven to be important in augmenting mVPS34 activity in starvation-induced autophagy (Fig. 2). The recent finding that mVPS34-associated proteins are direct targets for ubiquitin ligases leads to the model of ubiquitin-dependent regulation of the PI3KIII activity. As previously mentioned, the authors propose that the AMBRA1-mediated ubiquitylation of BECLIN 1 is regulated by WASH through competitive binding of AMBRA1.
Figure 2.

The crucial role of AMBRA1 in ubiquitin-mediated regulation of autophagy. Two different regulative ubiquitylation events are mediated by the adaptor molecule AMBRA1 during autophagy induction. First, the CULLIN4–DDB1–AMBRA1 complex mediates the K63-ubiquitylation of BECLIN 1, which results in increased activity of the lipid kinase mVPS34 and induces autophagy. Second, ULK1 is targeted by the TRAF6–AMBRA1 complex and, once ubiquitylated, self-associates and promotes autophagy. Similarly, BECLIN 1 is a substrate of TRAF6 in T cells, but it is not clear whether AMBRA1 plays any role in this regulation (dotted line). BECLIN 1 and ULK1 complexes are represented as individual complexes to simplify the scheme; however, their physical interaction cannot be ruled out. AMBRA1, together with the E3 ligase Parkin, also contributes to K48-ubiquitylation of mitochondria or other unknown substrates with a mitochondrial localization (?). In this case, the substrate (the entire mitochondrion) is selectively degraded by autophagy.
Recently, AMBRA1 was shown to promote Lys63-linked ubiquitylation of ULK1 by the tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6) E3 ligase upon autophagy induction by starvation and after rapamycin administration (Fig. 2).23
Two specific TRAF6-binding sites (P-X-E-X-X-aromatic/acidic) were identified on the AMBRA1 protein. Rendering AMBRA1 insensitive to TRAF6 binding (through mutation of the glutamic acid residue to alanine) impairs ULK1 kinase activity and stability and greatly reduces AMBRA1-mediated autophagy.23 The mechanism regulating AMBRA1–TRAF6 binding is unclear but AMBRA1 phosphorylation might have a priming function as certain E3 ligases have specific phosphorylation requirements. Moreover, like many other kinases, ULK1 is able to self-associate during autophagy and AMBRA1–TRAF6 binding is essential for ULK1 self-association. A key question remains as to whether ULK1-mediated phosphorylation on AMBRA1 takes place before or after AMBRA1–TRAF6 regulatory function on ULK1. One possibility is that the ULK1-dependent regulation of AMBRA1 localization represents a positive feedback loop regulating autophagy enhancement.
Interestingly, TRAF6 E3 ligase activity is also involved in the Lys63-linked ubiquitylation of BECLIN 1.37 This modification is important for BECLIN 1 dissociation from BCL2 and for its self-association during inflammatory responses in macrophages. However, the involvement of AMBRA1 in this regulation remains to be addressed.
An interesting report has shown that AMBRA1 is also involved in a selective form of autophagy called mitophagy.38 The authors demonstrated binding of endogenous Ambra1 to the E3 ligase Parkin in adult mouse brain. A number of studies have revealed that, in the case of mitochondrial damage, Parkin is recruited from the cytosol to depolarized mitochondria by PTEN-induced putative kinase 1 (PINK1) and subsequently triggers the elimination of mitochondria through mitophagy. Van Humbeck and coworkers demonstrated that AMBRA1 protein translocates to perinuclear clusters of depolarized mitochondria in a Parkin-dependent manner; this event activates PI3KIII complex around these damaged mitochondria and contributes to their selective autophagic clearance (Fig. 2). Moreover, prolonged mitochondrial depolarization strongly enhances the interaction between endogenous Parkin and Ambra1 in neuronal cells. In this case there is no evidence for Parkin-mediated ubiquitylation of AMBRA1, supporting the hypothesis that AMBRA1 could act as an adaptor substrate for Parkin during mitophagy.38 Recently, Strappazzon and co-authors reported that AMBRA1 directly binds LC3 upon mitochondrial damage through an LC3-interacting region (LIR) and that this enhances Parkin-mediated autophagy. On the other hand, forced targeting of AMBRA1 to mitochondria is responsible for mitochondria damage and consequent mitophagy. Intriguingly, in this scenario Parkin is not essential but mitochondrial ubiquitylation occurs, suggesting that an unknown E3 ligase participates in this process.39
Taken together, these data strengthen the emerging view that AMBRA1 could also be involved in this selective form of autophagy. However, the mechanism underlying the increased binding of Parkin, as well as LC3 and AMBRA1, after mitochondrial depolarization remains unknown. There is also uncertainty over whether AMBRA1 is degraded together with the mitochondria or instead dissociates from them.
In summary, a number of studies have revealed that AMBRA1 is part of a larger complex that remains stable under basal and autophagic conditions. Post-translational modifications and protein–protein interactions are important in the regulation of, and by, the AMBRA1 complex; some complex members stabilize the others, whereas some are activated by post-translational modifications. Ultimately, identification of the phosphatases and deubiquitylases involved will be important in order to gain a deeper understanding of how this complex is regulated.
AMBRA1 in Apoptosis
Autophagy and apoptosis share common stimuli and signaling pathways, and exhibit some degree of mutual inhibition. In most cases, autophagy functions as an early induced cytoprotective response, favoring stress adaptation by removing damaged subcellular components. However, several reports suggest that autophagy also contributes to cell death execution when apoptotic signaling is compromised.40
As previously mentioned, Ambra1 protein is prematurely expressed in the nervous system during mouse development; in addition to neuronal abnormalities, Ambra1 mutant mice show excessive apoptosis of selective areas of the developing brain, supporting the idea that the apoptosis–autophagy equilibrium plays a crucial role during nervous system development.4
In addition to the prosurvival and prodeath roles of autophagy and apoptosis, it is clear that there is also a complex regulatory interplay between the two processes. In particular, proautophagic proteins such as AMBRA1 and BECLIN 1 appear to be regulated by the antiapoptotic protein BCL2; BCL2 and other members of this family exhibit antiautophagic activity via their interaction with these proteins.41,42 Whereas BCL2 inhibits BECLIN 1 when localized to the ER, AMBRA1 binds preferentially and dynamically to the mitochondrial pool of BCL2. This interaction is regulated by both autophagy and apoptosis stimuli. In normal conditions, a pool of AMBRA1 is docked by BCL2 at the mitochondria; after autophagy induction, mito-AMBRA1 dissociates from BCL2 and thus increases binding of BCL2 to BECLIN 1. AMBRA1 therefore has the potential to enhance BECLIN 1-dependent autophagy, highlighting the importance of the subcellular localization of AMBRA1 in its function. Further studies are required to understand how and why BCL2 binds AMBRA1, but phosphorylation events could be involved. Whether ULK1, which phosphorylates AMBRA1 to dissociate it from the Dynein motor complex, is also responsible for its dissociation from BCL2 is an intriguing possibility that should be investigated.
Another important finding is that AMBRA1 is proteolytically degraded during the early phases of apoptosis by caspases and calpains, contributing to abrogation of the prosurvival function of autophagy.43 The activation of calpains and caspases during apoptosis is required for cell demise, and other autophagic proteins such as BECLIN 1 and mVPS34 are also targets of these apoptotic proteases.44,45 In a large number of cell lines, a reduction in the level of AMBRA1 increases susceptibility to different apoptotic stimuli. Using an in vitro caspase cleavage assay, the authors found that AMBRA1 is a substrate of different caspases and identified D482 as a major caspase cleavage site in the AMBRA1 protein.18 Significantly, a D482→A AMBRA1 mutation prevents the formation of cleavage products in AMBRA1-overexpressing cells and protects cells from apoptosis more efficiently than the wild-type protein does. However, in contrast to other autophagic factors, the AMBRA1 cleavage products have no proapoptotic functions. Further investigation of these cleavage events will be important for greater understanding of the inter-relationship between autophagy and apoptosis.
AMBRA1: From Physiology to Pathology
Role of Ambra1 in embryonic morphogenesis
Autophagy is a powerful catabolic pathway that can be triggered by environmental and hormonal stimuli, making it a good candidate for driving the rapid cellular remodeling necessary for proper development.46,47 Accordingly, the impairment of basal autophagy may be sufficient to interfere with normal embryogenesis. In fact, severe phenotypes have been observed in both Ambra1 mutated mice and in mice deficient for the functional AMBRA1-binding proteins Beclin 1, Ulk1/2, and FIP200.4,48,49 In this context, the premature death of Beclin 1 knockout mice, occurring at embryonic day 7.5 (E7.5), constitutes a striking example of the crucial role of autophagy in development.15 Furthermore, an in vivo large-scale mutagenesis approach revealed that targeting the murine Ambra1 locus results in (i) hyperproliferation and an excess of apoptotic cells in the neuroepithelium, (ii) insurgence of neural tube defects (midbrain and hindbrain exencephaly) and, eventually, (iii) embryonic lethality at day E16.5.4 Here, a central role for Ambra1 in the development of the central nervous system arises, as recently confirmed by in vitro studies on neural stem cells and in the Zebrafish model.50-52 In addition, Ambra1 is required for proper development of skeletal muscle, with Ambra1 ablation resulting in hyperproliferation of the tissue and altered morphology.53
A function in neurodevelopment has also been attributed to the Ambra1-associated kinase Ulk1. Ulk1 is highly expressed in developing cerebellum, where it is crucial for axon outgrowth, neurite extension, and the formation of fibers.54 Whether the role of Ulk1 in neurodevelopment depends on its function in regulating autophagy remains to be clarified.
A lethal phenotype has also been associated with depletion of an Ulk1-interacting protein, substrate focal adhesion kinase family interacting protein of Mr 200K (FIP200). In this case, fetal hematopoiesis is abolished and defects in the development of heart and liver are likely responsible for the embryonic death.49,55
In contrast to the examples reported above, the majority of Atg genes (such as Atg3−/−, Atg5−/−, Atg7−/−) do not seem to be essential for embryogenesis,56-58 with the respective knockout embryos surviving the entire embryonic period. There is a clear difference between the phenotype associated with depletion of upstream regulators of autophagy (Beclin 1, Ambra1, Ulk1/2, FIP200) and those involved in the later stages of autophagy pathway (such as Atg5 and 7). Several functions and interacting partners that affect cell pathways other than autophagy have been reported for the Beclin 1-core complex and FIP200, which could account for their strong impact on embryogenesis.15 Moreover, in this context a functional redundancy between the Atg proteins might explain the mild phenotypes associated with their depletion.
Importantly, it is also clear that depletion of Ambra1 during embryogenesis interferes with autophagy, cell proliferation, and apoptosis,4 suggesting that these processes are coordinately regulated in order to prevent uncontrolled cell death and cell growth.
AMBRA1 in pathologies
A striking role for Ambra1 in neurogenesis is highlighted by the consequences of its depletion in vivo.4 In line with this, Ambra1 is highly expressed in the cortex, hippocampus, and striatum of the postnatal brain, where it is differentially regulated in an age-dependent manner; during aging, a progressive decrease in the expression of Ambra1 is observed in the neocortex and in the hippocampus.4 In addition, Ambra1 has emerged as one of the key targets of the transcription factor Miz1, which prevents loss of Purkinje cells and consequent neurodegeneration during aging.59 Moreover, Ambra1 expression in the brain was shown to be altered in a mouse model of Alzheimer's Disease (AD).60 These data allow us to hypothesize that Ambra1, as already demonstrated for autophagy in toto, might play a role in neural aging and the etiology of AD.
More recently, psychiatric disorders have been associated with AMBRA1 variations. A genome-wide association study carried out on patients with schizophrenia revealed a genetic risk variation in a region on chromosome 11 containing the AMBRA1 locus.61,62 Interestingly, some personality traits of patients with schizophrenia (such as a deficit in social cognition and communication) are common to patients with autism-spectrum disorders (ASDs). In line with this, a recent study found that Ambra1 haploinsufficiency (Ambra1+/gt) was a genetic trait associated with autism-like behavior in mice.63 Despite the fact that ASDs preferentially affect males,64 Ambra1+/gt-associated ASD behavior predominantly impacted females. In this context, Dere and co-authors observed major expression of Ambra1 protein in the brain cortex of female mice compared to male mice.63 This finding opens the possibility of sexual dimorphism regarding the role of autophagy in the central nervous system.
In fact, the role of AMBRA1 in neural disorders is not limited to the central nervous system as a recent paper reported a correlation between autophagy and neuropathic pain (NeP).65 After nerve injury, axonal and myelin debris are removed from the Schwann cells (SCs) and the autophagy process plays a key role in this clearance during the first days after the injury.66 Degeneration/regeneration promoted by SCs and the inflammatory reaction represent early events in the onset and maintenance of NeP. Using Ambra1-deficient mice (Ambra1+/gt), the same authors found that NeP is dramatically enhanced and prolonged when the autophagy process is impaired, demonstrating that Ambra1 haploinsufficiency reflects a reduction of SC autophagy after nerve injury and that this effect is associated with persistent neuropathic pain.65
In recent years, it has been shown that both deficient and excessive autophagy play a role in cancer insurgence, progression, and resistance to therapy by preventing tumor initiation or sustaining tumor growth.67-69 AMBRA1, together with other autophagy proteins, seems to function in tumor survival in colon cancer cells and its increased expression is associated with poor prognosis in pancreatic ductal adenocarcinoma.70,71 On the other hand, numerous AMBRA1-interacting proteins (such as PI3KIII complex components and BCL2) are protagonists in this scenario.16,72-75 In keeping with this, a defect in AMBRA1 could also be relevant to cancer insurgence. For example, binding of AMBRA1 to BCL2 (see above) could be crucial for the shift from autophagy to apoptosis, and vice versa, thus regulating both cell death and cell survival in cancer cells.
Intriguingly, deregulation of the morphogen Sonic hedgehog (Shh) is observed in Ambra1 mutated embryos,4 suggesting a possible involvement of Ambra1 in the insurgence of Shh-dependent tumors such as medulloblastoma.76
The hyperproliferative phenotype characterizing AMBRA1-deficient systems both in vitro and in vivo4 is another indirect proof supporting a possible role of AMBRA1 in tumorigenesis. Understanding the molecular mechanisms accounting for AMBRA1-mediated control of cell proliferation, and their possible link to regulation of the autophagy pathway, would be highly relevant.
New Perspectives from New Interactors
As discussed above, numerous interactions occur in autophagy pathways, both between autophagy proteins and with proteins from other cellular processes. With increasing data from mass spectrometry, the list of such interactions is constantly growing.11
In addition, many new intriguing binding partners for the AMBRA1 protein have been described and deserve further functional characterization.
BECLIN 2
Beth Levine's group has identified a previously unknown mammalian protein, BECLIN 2, as a member of the BECLIN 1 protein family. BECLIN 1 and 2 share a large number of protein interactors including AMBRA1, which is unique in this context because of its tighter binding to BECLIN 2 than to BECLIN 1.77 These authors described a dual role for BECLIN 2, which can bind the PI3KIII complex thus promoting autophagy but also participates in the ligand-induced endosomal degradation of several G protein-coupled receptors (GPCRs). Strikingly, these 2 functions of BECLIN 2 are mechanistically independent. Moreover, as a result of deregulated degradation of the cannabinoid receptor type 1 (CB1R), Beclin 2-deficient mice are affected by increased food intake, obesity, and insulin resistance.77 It would be worth testing whether AMBRA1 functions in the role of BECLIN 2 in GPCR degradation as such a finding would make AMBRA1 a candidate risk allele for obesity and diabetes.
MIZ1
A role for the transcription factor Miz1 in regulation of the autophagy flux has been recently described, with Ambra1 as the main autophagy-related target of this transcription factor.59 A functional role for Miz1-mediated regulation of Ambra1 has previously been described in the central nervous system (see above) and in relation to autophagy regulation. Notably, Miz1 can also transcribe Ambra1 in other tissue types and in proliferating cells, suggesting additional physiological roles for the Ambra1-Miz1 interaction.59 Moreover, Miz1 plays roles in cellular processes other than autophagy, impacting skin papilloma and B- and T-cell development.78-80 The relevance of Ambra1 in these contexts should be investigated.
The primary cilium
The primary cilium is an organelle present in most resting mammalian cells that protrudes from the cell membrane and constitutes a hub for sensing and transmitting signals.81 The laboratories of Cuervo and Zhong82,83 independently showed a reciprocal regulation between autophagy and the primary cilium, paving the way for numerous physiological and pathological implications.84 AMBRA1, and some other autophagy proteins, localize to the cilium, probably through direct interaction with the cytoskeleton. A possible functional relevance for AMBRA1 in the regulation of cilium length or in the pathologies resulting from cilium defects (named ciliopathies) should be considered.81
Concluding Remarks
The relevance of autophagy in human diseases is clear and the central issue in the field of autophagy is now the design of therapeutic strategies targeting autophagy in order to properly modulate it in a convenient manner.85
In the era of mass spectrometry, comprehensive characterization of the interaction networks and post-translational modifications of autophagy proteins is already available.11,86 In many instances, molecular studies confirming the validity and the functional relevance of these data have also been reported. These data pave the way to a complex scenario in which (i) autophagy proteins perform functions other than their autophagic role; (ii) autophagy has a large impact on other cellular processes and vice versa; (iii) not only protein–protein interactions, but also a large number of different kinds of post-translational modifications, are involved in autophagy regulation and have a striking relevance.
The new challenge will be to consider all of these regulative pathways and modifications when we want to modulate autophagy for therapeutic purposes. Regarding AMBRA1 in this context, specific binding sites for protein–protein interactions and post-translational modifications have been identified (Table 1) and all of these sites would be considered good candidates for tight modulation of autophagy without interfering with the entire autophagy machinery. Such an approach would be an alternative or synergic option to the drugs already in clinical trials for the modulation of autophagy.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We wish to thank M.W. Bennett for editorial work.
Funding
Our laboratory is supported in part by grants from KBVU (R72-A4408), Lundbeck Foundation (R167-2013-16100), Novo Nordisk Foundation (7559), AIRC (IG2012), FISM (2012), the Italian Ministry of University and Research (PRIN 2009 and FIRB Accordi di Programma 2011) and the Italian Ministry of Health (RF 2009). VC is supported by the Lundbeck Foundation R165-2013-15982). The Unit of Cell Stress and Survival is part of the Center for Autophagy, Recycling and Disease (CARD), funded by the Danish National Research Foundation.
References
- 1. Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol 2013; 15:713-20; PMID:23817233; http://dx.doi.org/ 10.1038/ncb2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. New Engl J Med 2013; 368:651-62; PMID:23406030; http://dx.doi.org/ 10.1056/NEJMra1205406 [DOI] [PubMed] [Google Scholar]
- 3. Abada A, Elazar Z. Getting ready for building: signaling and autophagosome biogenesis. EMBO Rep 2014; 15:839-52; PMID:25027988; http://dx.doi.org/ 10.15252/embr.201439076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. . Ambra1 regulates autophagy and development of the nervous system. Nature 2007; 447:1121-5; PMID:17589504 [DOI] [PubMed] [Google Scholar]
- 5. Fimia GM, Corazzari M, Antonioli M, Piacentini M. Ambra1 at the crossroad between autophagy and cell death. Oncogene 2013; 32:3311-8; PMID:23069654; http://dx.doi.org/ 10.1038/onc.2012.455 [DOI] [PubMed] [Google Scholar]
- 6. Stirnimann CU, Petsalaki E, Russell RB, Muller CW. WD40 proteins propel cellular networks. Trends Biochem Sci 2010; 35:565-74; PMID:20451393; http://dx.doi.org/ 10.1016/j.tibs.2010.04.003 [DOI] [PubMed] [Google Scholar]
- 7. Mei Y, Su M, Soni G, Salem S, Colbert CL, Sinha SC. Intrinsically disordered regions in autophagy proteins. Proteins 2014; 82:565-78; PMID:24115198; http://dx.doi.org/ 10.1002/prot.24424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peng Z, Xue B, Kurgan L, Uversky VN. Resilience of death: intrinsic disorder in proteins involved in the programmed cell death. Cell Death Differ 2013; 20:1257-67; PMID:23764774; http://dx.doi.org/ 10.1038/cdd.2013.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 2005; 6:197-208; PMID:15738986; http://dx.doi.org/ 10.1038/nrm1589 [DOI] [PubMed] [Google Scholar]
- 10. Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics 2008; 9 Suppl 2:S1; PMID:18831774; http://dx.doi.org/ 10.1186/1471-2164-9-S2-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466:68-76; PMID:20562859; http://dx.doi.org/ 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N. Crosstalk between apoptosis, necrosis and autophagy. Biochimica et Biophysica Acta 2013; 1833:3448-59; PMID:23770045; http://dx.doi.org/ 10.1016/j.bbamcr.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 13. Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685-701; PMID:18725538; http://dx.doi.org/ 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roberts R, Ktistakis NT. Omegasomes: PI3P platforms that manufacture autophagosomes. Essays Biochem 2013; 55:17-27; PMID:24070468; http://dx.doi.org/ 10.1042/bse0550017 [DOI] [PubMed] [Google Scholar]
- 15. Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol 2010; 20:355-62; PMID:20356743; http://dx.doi.org/ 10.1016/j.tcb.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 2012; 338:956-9; PMID:23112296; http://dx.doi.org/ 10.1126/science.1225967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luo S, Garcia-Arencibia M, Zhao R, Puri C, Toh PP, Sadiq O, Rubinsztein DC. Bim inhibits autophagy by recruiting Beclin 1 to microtubules. Mol Cell 2012; 47:359-70; PMID:22742832; http://dx.doi.org/ 10.1016/j.molcel.2012.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al. . The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol 2010; 191:155-68; PMID:20921139; http://dx.doi.org/ 10.1083/jcb.201002100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mackeh R, Perdiz D, Lorin S, Codogno P, Pous C. Autophagy and microtubules - new story, old players. J Cell Sci 2013; 126:1071-80; PMID:23620510; http://dx.doi.org/ 10.1242/jcs.115626 [DOI] [PubMed] [Google Scholar]
- 20. Nazio F, Cecconi F. mTOR, AMBRA1, and autophagy: an intricate relationship. Cell Cycle 2013; 12:2524-5; PMID:23907135; http://dx.doi.org/ 10.4161/cc.25835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149:274-93; PMID:22500797; http://dx.doi.org/ 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. . Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 2009; 20:1981-91; PMID:19211835; http://dx.doi.org/ 10.1091/mbc.E08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. . mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15:406-16; PMID:23524951; http://dx.doi.org/ 10.1038/ncb2708 [DOI] [PubMed] [Google Scholar]
- 24. Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Nat Acad Sci U S A 2008; 105:10762-7; PMID:18669648; http://dx.doi.org/ 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell 2008; 31:438-48; PMID:18691976; http://dx.doi.org/ 10.1016/j.molcel.2008.07.007 [DOI] [PubMed] [Google Scholar]
- 26. Xia P, Wang S, Huang G, Du Y, Zhu P, Li M, Fan Z. RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy. Cell Res 2014; 24:943-58; PMID:24980959; http://dx.doi.org/ 10.1038/cr.2014.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J 2006; 25:2465-74; PMID:16710298; http://dx.doi.org/ 10.1038/sj.emboj.7601144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Su WJ, Fang JS, Cheng F, Liu C, Zhou F, Zhang J. RNF2Ring1b negatively regulates p53 expression in selective cancer cell types to promote tumor development. Proc Nat Acad Sci U S A 2013; 110:1720-5; PMID:23319651; http://dx.doi.org/ 10.1073/pnas.1211604110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp23 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell 2009; 17:712-23; PMID:19922875; http://dx.doi.org/ 10.1016/j.devcel.2009.09.010 [DOI] [PubMed] [Google Scholar]
- 30. Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, Huang G, Ye B, Li C, Dai Z, et al. . WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J 2013; 32:2685-96; PMID:23974797; http://dx.doi.org/ 10.1038/emboj.2013.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zavodszky E, Seaman MN, Moreau K, Jimenez-Sanchez M, Breusegem SY, Harbour ME, Rubinsztein DC. Mutation in VPS35 associated with Parkinson's disease impairs WASH complex association and inhibits autophagy. Nat Commun 2014; 5:3828; PMID:24819384; http://dx.doi.org/ 10.1038/ncomms4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. . Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 2011; 44:325-40; PMID:21906983; http://dx.doi.org/ 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 2010; 143:951-65; PMID:21145461; http://dx.doi.org/ 10.1016/j.cell.2010.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. . Global identification of modular cullin-RING ligase substrates. Cell 2011; 147:459-74; PMID:21963094; http://dx.doi.org/ 10.1016/j.cell.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 2006; 23:709-21; PMID:16949367; http://dx.doi.org/ 10.1016/j.molcel.2006.08.010 [DOI] [PubMed] [Google Scholar]
- 36. Wertz IE, O’Rourke KM, Zhang Z, Dornan D, Arnott D, Deshaies RJ, Dixit VM. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 2004; 303:1371-4; PMID:14739464; http://dx.doi.org/ 10.1126/science.1093549 [DOI] [PubMed] [Google Scholar]
- 37. Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signaling 2010; 3:ra42; PMID:20501938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Van Humbeeck C, Cornelissen T, Hofkens H, Mandemakers W, Gevaert K, De Strooper B, Vandenberghe W. Parkin interacts with Ambra1 to induce mitophagy. J Neurosci: Off J Soc Neurosci 2011; 31:10249-61; PMID:21753002; http://dx.doi.org/ 10.1523/JNEUROSCI.1917-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Strappazzon F NF, Corrado M, Cianfanelli V, Romagnoli A, Fimia GM, Campello S, Piacentini M, Campanella M, Cecconi F. AMBRA1 is able to induce mitophagy via LC3 binding and independently of PARKIN and p62. Cell Death Differ 2014; PMID:25215947; http://dx.doi.org/ 10.1038/cdd.2014.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rubinstein AD, Kimchi A. Life in the balance - a mechanistic view of the crosstalk between autophagy and apoptosis. J Cell Sci 2012; 125:5259-68; PMID:23377657; http://dx.doi.org/ 10.1242/jcs.115865 [DOI] [PubMed] [Google Scholar]
- 41. Strappazzon F, Vietri-Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM, Piacentini M, Levine B, Cecconi F. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J 2011; 30:1195-208; PMID:21358617; http://dx.doi.org/ 10.1038/emboj.2011.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol 2010; 22:140-9; PMID:20097051; http://dx.doi.org/ 10.1016/j.ceb.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pagliarini V, Wirawan E, Romagnoli A, Ciccosanti F, Lisi G, Lippens S, Cecconi F, Fimia GM, Vandenabeele P, Corazzari M, et al. . Proteolysis of Ambra1 during apoptosis has a role in the inhibition of the autophagic pro-survival response. Cell Death Differ 2012; 19:1495-504; PMID:22441670; http://dx.doi.org/ 10.1038/cdd.2012.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Das A, Banik NL, Ray SK. Mechanism of apoptosis with the involvement of calpain and caspase cascades in human malignant neuroblastoma SH-SY5Y cells exposed to flavonoids. Int J Cancer J Int du Cancer 2006; 119:2575-85; PMID:16988947; http://dx.doi.org/ 10.1002/ijc.22228 [DOI] [PubMed] [Google Scholar]
- 45. Cho DH, Jo YK, Hwang JJ, Lee YM, Roh SA, Kim JC. Caspase-mediated cleavage of ATG6Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett 2009; 274:95-100; PMID:18842334; http://dx.doi.org/ 10.1016/j.canlet.2008.09.004 [DOI] [PubMed] [Google Scholar]
- 46. Sato M, Sato K. Dynamic regulation of autophagy and endocytosis for cell remodeling during early development. Traffic 2013; 14:479-86; PMID:23356349; http://dx.doi.org/ 10.1111/tra.12050 [DOI] [PubMed] [Google Scholar]
- 47. Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 2008; 15:344-57; PMID:18804433; http://dx.doi.org/ 10.1016/j.devcel.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Nat Acad Sci U S A 2003; 100:15077-82; PMID:14657337; http://dx.doi.org/ 10.1073/pnas.2436255100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL. Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J Cell Biol 2006; 175:121-33; PMID:17015619; http://dx.doi.org/ 10.1083/jcb.200604129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Benato F, Dalla Valle L, Skobo T, Alibardi L. Biomolecular identification of beta-defensin-like peptides from the skin of the soft-shelled turtle Apalone spinifera. J Exp Zool Part B, Mol Dev Evol 2013; 320:210-7; PMID:23554337; http://dx.doi.org/ 10.1002/jez.b.22495 [DOI] [PubMed] [Google Scholar]
- 51. Vazquez P, Arroba AI, Cecconi F, de la Rosa EJ, Boya P, de Pablo F. Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. Autophagy 2012; 8:187-99; PMID:22240590; http://dx.doi.org/ 10.4161/auto.8.2.18535 [DOI] [PubMed] [Google Scholar]
- 52. Yazdankhah M F-VS, Tonchev AB, Stoykova A, Cecconi F. The autophagy regulator Ambra1 and Beclin 1 are required for adult neurogenesis in the brain subventricular zone. Cell Death Dis 2014; 5:e1403; In press; PMID:25188513; http://dx.doi.org/ 10.1038/cddis.2014.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Skobo T, Benato F, Grumati P, Meneghetti G, Cianfanelli V, Castagnaro S, Chrisam M, Di Bartolomeo S, Bonaldo P, Cecconi F, et al. . Zebrafish ambra1a and ambra1b knockdown impairs skeletal muscle development. PloS One 2014; 9:e99210; PMID:24922546; http://dx.doi.org/ 10.1371/journal.pone.0099210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tomoda T, Bhatt RS, Kuroyanagi H, Shirasawa T, Hatten ME. A mouse serinethreonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron 1999; 24:833-46; PMID:10624947; http://dx.doi.org/ 10.1016/S0896-6273(00)81031-4 [DOI] [PubMed] [Google Scholar]
- 55. Liu F, Guan JL. FIP200, an essential component of mammalian autophagy is indispensible for fetal hematopoiesis. Autophagy 2011; 7:229-30; PMID:21088496; http://dx.doi.org/ 10.4161/auto.7.2.14125 [DOI] [PubMed] [Google Scholar]
- 56. Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature 2004; 432:1032-6; PMID:15525940; http://dx.doi.org/ 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- 57. Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. . Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 2005; 169:425-34; PMID:15866887; http://dx.doi.org/ 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, et al. . The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell 2008; 19:4762-75; PMID:18768753; http://dx.doi.org/ 10.1091/mbc.E08-03-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wolf E, Gebhardt A, Kawauchi D, Walz S, von Eyss B, Wagner N, Renninger C, Krohne G, Asan E, Roussel MF, et al. . Miz1 is required to maintain autophagic flux. Nat Commun 2013; 4:2535; PMID:24088869; http://dx.doi.org/ 10.1038/ncomms3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sepe S, Nardacci R, Fanelli F, Rosso P, Bernardi C, Cecconi F, Mastroberardino PG, Piacentini M, Moreno S. Expression of Ambra1 in mouse brain during physiological and Alzheimer type aging. Neurobiol Aging 2014; 35:96-108; PMID:23910655; http://dx.doi.org/ 10.1016/j.neurobiolaging.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 61. Heinrich A, Nees F, Lourdusamy A, Tzschoppe J, Meier S, Vollstadt-Klein S, Fauth-Buhler M, Steiner S, Bach C, Poustka L, et al. . From gene to brain to behavior: schizophrenia-associated variation in AMBRA1 alters impulsivity-related traits. Eur J Neurosci 2013; 38:2941-5; PMID:23551272 [DOI] [PubMed] [Google Scholar]
- 62. Rietschel M, Mattheisen M, Degenhardt F, Genetic R, Outcome P, Muhleisen TW, Kirsch P, Esslinger C, Herms S, Demontis D, et al. . Association between genetic variation in a region on chromosome 11 and schizophrenia in large samples from Europe. Mol Psychiat 2012; 17:906-17; PMID:21747397; http://dx.doi.org/ 10.1038/mp.2011.80 [DOI] [PubMed] [Google Scholar]
- 63. Dere E, Dahm L, Lu D, Hammerschmidt K, Ju A, Tantra M, Kastner A, Chowdhury K, Ehrenreich H. Heterozygous ambra1 deficiency in mice: a genetic trait with autism-like behavior restricted to the female gender. Front Behav Neurosci 2014; 8:181; PMID:24904333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Baron-Cohen S, Lombardo MV, Auyeung B, Ashwin E, Chakrabarti B, Knickmeyer R. Why are autism spectrum conditions more prevalent in males? PLoS Biol 2011; 9:e1001081; PMID:21695109; http://dx.doi.org/ 10.1371/journal.pbio.1001081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Marinelli S, Nazio F, Tinari A, Ciarlo L, D’Amelio M, Pieroni L, Vacca V, Urbani A, Cecconi F, Malorni W, et al. . Schwann cell autophagy counteracts the onset and chronification of neuropathic pain. Pain 2014; 155:93-107; PMID:24041962; http://dx.doi.org/ 10.1016/j.pain.2013.09.013 [DOI] [PubMed] [Google Scholar]
- 66. Yang Y, Coleman M, Zhang L, Zheng X, Yue Z. Autophagy in axonal and dendritic degeneration. Trends Neurosci 2013; 36:418-28; PMID:23639383; http://dx.doi.org/ 10.1016/j.tins.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell 2013; 155:1216-9; PMID:24315093; http://dx.doi.org/ 10.1016/j.cell.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Helgason GV, Holyoake TL, Ryan KM. Role of autophagy in cancer prevention, development and therapy. Essays Biochem 2013; 55:133-51; PMID:24070477; http://dx.doi.org/ 10.1042/bse0550133 [DOI] [PubMed] [Google Scholar]
- 69. Lorin S, Hamai A, Mehrpour M, Codogno P. Autophagy regulation and its role in cancer. Semin Cancer Biol 2013; 23:361-79; PMID:23811268; http://dx.doi.org/ 10.1016/j.semcancer.2013.06.007 [DOI] [PubMed] [Google Scholar]
- 70. Ko YH, Cho YS, Won HS, Jeon EK, An HJ, Hong SU, Park JH, Lee MA. Prognostic significance of autophagy-related protein expression in resected pancreatic ductal adenocarcinoma. Pancreas 2013; 42:829-35; PMID:23429496; http://dx.doi.org/ 10.1097/MPA.0b013e318279d0dc [DOI] [PubMed] [Google Scholar]
- 71. Gu W, Wan D, Qian Q, Yi B, He Z, Gu Y, Wang L, He S. Ambra1 is an essential regulator of autophagy and apoptosis in SW620 cells: pro-survival role of Ambra1. PloS One 2014; 9:e90151; PMID:24587252; http://dx.doi.org/ 10.1371/journal.pone.0090151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Abrahamsen H, Stenmark H, Platta HW. Ubiquitination and phosphorylation of Beclin 1 and its binding partners: tuning class III phosphatidylinositol 3-kinase activity and tumor suppression. FEBS Lett 2012; 586:1584-91; PMID:22673570; http://dx.doi.org/ 10.1016/j.febslet.2012.04.046 [DOI] [PubMed] [Google Scholar]
- 73. Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, Kinch L, Koduru P, Christudass CS, Veltri RW, et al. . EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 2013; 154:1269-84; PMID:24034250; http://dx.doi.org/ 10.1016/j.cell.2013.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sasi N, Hwang M, Jaboin J, Csiki I, Lu B. Regulated cell death pathways: new twists in modulation of BCL2 family function. Mol Cancer Ther 2009; 8:1421-9; PMID:19509269; http://dx.doi.org/ 10.1158/1535-7163.MCT-08-0895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. . Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003; 112:1809-20; PMID:14638851; http://dx.doi.org/ 10.1172/JCI20039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 1997; 277:1109-13; PMID:9262482; http://dx.doi.org/ 10.1126/science.277.5329.1109 [DOI] [PubMed] [Google Scholar]
- 77. He C, Wei Y, Sun K, Li B, Dong X, Zou Z, Liu Y, Kinch LN, Khan S, Sinha S, et al. . Beclin 2 functions in autophagy, degradation of G protein-coupled receptors, and metabolism. Cell 2013; 154:1085-99; PMID:23954414; http://dx.doi.org/ 10.1016/j.cell.2013.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol 2005; 6:1054-60; PMID:16142238; http://dx.doi.org/ 10.1038/ni1245 [DOI] [PubMed] [Google Scholar]
- 79. Saito M, Novak U, Piovan E, Basso K, Sumazin P, Schneider C, Crespo M, Shen Q, Bhagat G, Califano A, et al. . BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse large B cell lymphoma. Proc Nat Acad Sci U S A 2009; 106:11294-9; PMID:19549844; http://dx.doi.org/ 10.1073/pnas.0903854106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Basu D, Liu Y, Wu A, Yarlagadda S, Gorelik GJ, Kaplan MJ, Hewagama A, Hinderer RC, Strickland FM, Richardson BC. Stimulatory and inhibitory killer Ig-like receptor molecules are expressed and functional on lupus T cells. J Immunol 2009; 183:3481-7; PMID:19675166; http://dx.doi.org/ 10.4049/jimmunol.0900034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Satir P, Pedersen LB, Christensen ST. The primary cilium at a glance. J Cell Sci 2010; 123:499-503; PMID:20144997; http://dx.doi.org/ 10.1242/jcs.050377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pampliega O, Orhon I, Patel B, Sridhar S, Diaz-Carretero A, Beau I, Codogno P, Satir BH, Satir P, Cuervo AM. Functional interaction between autophagy and ciliogenesis. Nature 2013; 502:194-200; PMID:24089209; http://dx.doi.org/ 10.1038/nature12639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tang Z, Lin MG, Stowe TR, Chen S, Zhu M, Stearns T, Franco B, Zhong Q. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature 2013; 502:254-7; PMID:24089205; http://dx.doi.org/ 10.1038/nature12606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cianfanelli V, Cecconi F. Cell biology: molecular clearance at the cell's antenna. Nature 2013; 502:180-1; PMID:24089206; http://dx.doi.org/ 10.1038/nature12693 [DOI] [PubMed] [Google Scholar]
- 85. Gewirtz DA. The four faces of autophagy: implications for cancer therapy. Cancer Res 2014; 74:647-51; PMID:24459182; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-2966 [DOI] [PubMed] [Google Scholar]
- 86. Rigbolt KT, Zarei M, Sprenger A, Becker AC, Diedrich B, Huang X, Eiselein S, Kristensen AR, Gretzmeier C, Andersen JS, et al. . Characterization of early autophagy signaling by quantitative phosphoproteomics. Autophagy 2014; 10:356-71; PMID:24275748; http://dx.doi.org/ 10.4161/auto.26864 [DOI] [PMC free article] [PubMed] [Google Scholar]


