ABSTRACT
Members of the family of small GTPases regulate a variety of important cellular functions. In order to accomplish this, tight temporal and spatial regulation is absolutely necessary. The two most important factors for this regulation are GTPase activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs), the latter being responsible for the activation of the GTPase downstream pathways at the correct location and time. Although a large number of exchange factors have been identified, it is likely that a similarly large number remains unidentified. We have therefore developed a procedure to specifically enrich GEF proteins from biological samples making use of the high affinity binding of GEFs to nucleotide-free GTPases. In order to verify the results of these pull-down experiments, we have additionally developed two simple validation procedures: An in vitro transcription/translation system coupled with a GEF activity assay and a yeast two-hybrid screen for detection of GEFs. Although the procedures were established and tested using the Rab protein Sec4, the similar basic principle of action of all nucleotide exchange factors will allow the method to be used for identification of unknown GEFs of small GTPases in general.
KEYWORDS: enrichment, GEF, in vitro transcription/translation, nucleotide exchange factor, pull-down, small GTPase, yeast two-hybrid
Introduction
Small GTPases of the Ras superfamily act as regulators of a variety of important functions such as cell proliferation (Ras),1,2 remodeling of the cytoskeleton (Rho, Rac and Cdc42),3 nuclear im- and export (Ran)4 or vesicular transport (Rab, Sar and Arf).5,6 They do so by switching between an inactive GDP-bound (guanosine-5′-diphosphate) and an active GTP-bound (guanosine-5′-triphosphate) state, only interacting with specific downstream effector proteins in the active conformation.7 The interconversion between these states is mediated by GTPase activating proteins (GAPs) that catalyze hydrolysis of GTP and guanine nucleotide exchange factors (GEFs) that catalyze GDP/GTP-exchange.8
A recent review giving a comprehensive overview of known GEFs and GAPs for small GTPases and their mechanisms of action can be found in Cherfils et al. (2013).9 The review illustrates the great diversity between the different families of structurally unrelated GEFs. For most small GTPase subfamilies, relative to the number of known members of the subfamily a similar or even higher number of GEFs have been identified to date (see Table 1). However, this is not true for the Rab-family, indicating that further GEFs still need to be identified for this family, but possibly also for other small GTPases. However, their great structural diversity hinders an easy identification of novel GEFs in silico based on primary sequences.
Table 1.
Small GTPase families and their known GEFs.
| GTPase family | Function | known GEFs | references |
|---|---|---|---|
| Ras (36 members) | Cell proliferation and differentiation | 3 families (SOS, RasGRP and RasGRF), all carrying a catalytic Cdc25-homology domain (27 members in humans) | 9,65-67 |
| Rho (20 members in human) | Regulation of the cytoskeleton | Dbl-homology (DH) domain GEFs (70 members in human) | 9,67,68 |
| Dock Homology Region (DHR) GEFs, 11 members | |||
| PRONE GEFs in plants | 69 | ||
| Rab (61 members in humans) | Vesicular trafficking | Vps9 domain GEFs (10 members) | 9,67,70,71 |
| DENN domain GEFs (18 members) | 72 | ||
| TRAPP complex | 73 | ||
| Mon1-Ccz1 | 74 | ||
| Rabin8/Sec2 | 47,60 | ||
| Ric1-Rgp1 | 75,76 | ||
| BLOC-3 | 77 | ||
| Mss4 | 43 | ||
| REI-1 | 22 | ||
| Arf (5 members + Arls), Sar (127 in total) | Vesicular trafficking | ArfGEFs (Sec7 catalytic domain, 16 members) | 9,67 |
| Sec12 | |||
| Ran | Nuclear im- and export | RCC1 | 9,67 |
| Importin β | 78 | ||
| RanBP1 | 79 |
In the early days of small GTPase research, the first GEF proteins were found by phenotypic observations related to their function (e.g. SOS,10 Dbl-GEFs11) and/or identified via standard fractionation approaches (e.g., RCC112) of cell extracts or overexpression of the target gene and subsequent analysis of its GEF activity.13-16 Similarly, Vps9-domain containing17 and DENN-domain18 GEFs were found via phenotypic defects associated with mutations or deletions and subsequent analysis of their interacting proteins and their function.19,20
In many cases, interactions of a protein with small GTPases containing the mutation corresponding to S17N in Ras (often termed the dominant inactive form of a small GTPase) served as an indication for GEF activity. Consequently, yeast two-hybrid approaches using small GTPase mutants with lowered nucleotide binding affinity (such as the S17N mutation in Ras, but also others as discussed later) have been used to identify GEFs.21-25 However, additional and efficient high throughput methodologies are still missing.
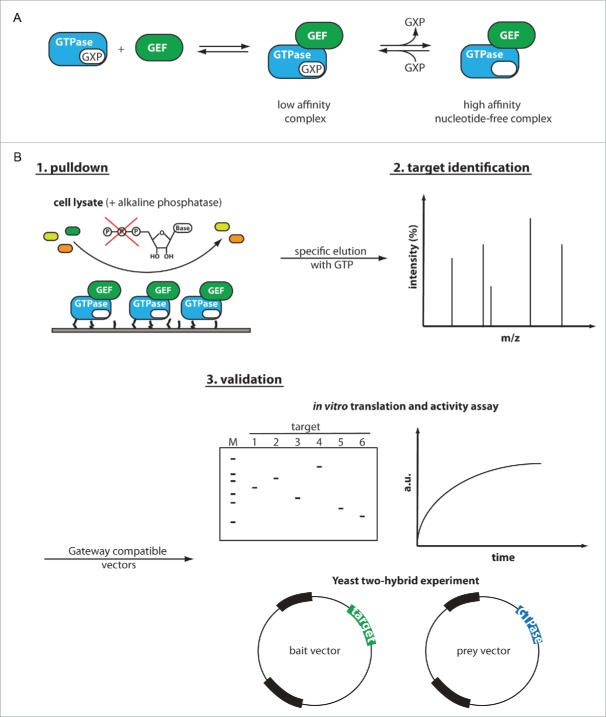
We have therefore established a procedure for specific enrichment of GEFs from biological samples making use of the known mechanistic principles of these exchange factors. In order to exchange one bound nucleotide for another, small GTPases have to pass through a nucleotide-free intermediate state. The basic mechanism of GEF action is the stabilization of this nucleotide-free intermediate. The mechanism catalyzed by GEFs can be described as a two-step process, with a first low-affinity encounter complex (Rab:GXP:GEF; GXP can be both GDP or GTP) in which the nucleotide is still bound, and a second high affinity nucleotide-free Rab:GEF complex (see Figure. 1A).26-28 In the reverse reaction, another nucleotide can bind, thereby releasing the Rab protein from the GEF. Any directionality of this reaction (i.e. from a GDP-bound toward a GTP-bound small GTPase) is achieved by the excess of GTP over GDP present in cells under physiological conditions29 and the relative affinities of the corresponding GTPase toward GDP and GTP. Because of the high concentrations of guanine nucleotides (˜0.5 mM GTP, ˜0.15 mM GDP) inside cells,29 the nucleotide-free Rab:GEF complex is only short-lived in vivo. However, the complex can be stabilized by artificially depleting the environment of guanine nucleotides as has been done in many cases to obtain stable complexes for X-ray crystallography (see for example references30-34).
Figure 1.
Scheme for the specific enrichment and identification of unknown GEFs of small GTPases. (A) General reaction scheme of guanine nucleotide exchange factors for small GTPases. GEFs operate by transiting from a low affinity ternary GTPase:GXP:GEF intermediate to a high-affinity binary GTPase:GEF complex and back. In a first step, the GEF binds the GTPase:GXP complex with low affinity. GEF-mediated release of the nucleotide in the second step leads to a high affinity GEF:GTPase complex. In the reverse reaction, a different guanine nucleotide can bind, thereby completing the exchange reaction. (B) We envision to enrich the specific GEFs from cell lysates by exploiting the high-affinity of the intermediary nucleotide-free GTPase:GEF complex. The immobilized GTPase of interest is incubated with cell lysate containing its cognate GEF (green) among several other proteins (yellow and orange). Upon formation of a binary GTPase:GEF complex, the released nucleotide is degraded by exogenously added alkaline phosphatase to further stabilize the GTPase:GEF complex. After washing and removal of unbound proteins, the GEF can be specifically eluted by addition of GDP or GTP followed by subsequent identification via mass spectrometry. Since complex mixtures such as cell lysates may give rise to false positive targets, we designed the following procedures for GEF validation: I) An in vitro translation system coupled with an activity based GEF assay and II) a Y2H experiments with specifically designed GTPase mutants favoring GEF-interaction. The use of Gateway compatible vectors for both validation procedures greatly simplifies the cloning of different target proteins into expression vectors or vectors for Y2H experiments.
Because of the high-affinity binding of nucleotide-free GTPases and GEFs, the nucleotide-free GTPases should be a suitable bait for pull-down experiments in order to identify unknown GEFs for a given small GTPase from natural sources. We have established this procedure using the known Rab:GEF pair Sec4:Sec2, showing that the endogenous GEF protein can be specifically enriched from yeast cell lysate. Furthermore, we have established a straightforward and rapid procedure to validate putative targets from such pull-down experiments.
Results
Pull-down procedure and analysis by mass spectrometry
In order to specifically enrich GEF proteins from cell lysates, Sec4 was first biotinylated using the commercially available EZ-Link™ Maleimide-PEG2-Biotin and immobilized on streptavidin magnetic beads (SMBs). Subsequently, the immobilized Sec4 was incubated with yeast cell lysate spiked with exogenously added alkaline phosphatase to degrade all nucleoside tri-, di- and monophosphates present in the samples and to allow stable complex formation between the GTPase and GEFs. The idea behind this was that complex formation between the immobilized GTPase and the corresponding GEF would lead to release of the bound nucleotide and degradation by the alkaline phosphatase, thus yielding a high-affinity nucleotide-free GEF:GTPase complex. After several washing steps using standard buffer, bound GEFs were specifically eluted in 2 subsequent steps with buffer containing 500 µM GTP and 10 mM GTP, respectively. As negative controls, the same experiments were performed using SMBs without immobilized Sec4. The eluted samples from all experiments were digested with trypsin and the peptide pattern was subsequently analyzed via mass spectrometry (MS) in a nano-HPLC coupled Quadrupole-Orbitrap Mass Spectrometer (Fig. 1B, see materials and methods for the exact procedure). In order to allow statistical evaluation of the experiments, every experiment was repeated thrice with two experimental replicates (6 experiments in total) and only proteins that were specifically enriched in the Sec4 pull-down experiments, but not in the control experiments, were regarded as significant hits.
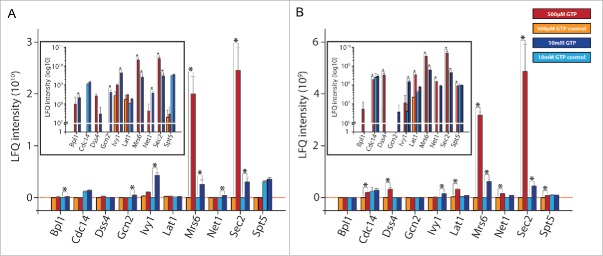
A total of 10 different proteins were significantly enriched in at least one of the triplicate experiments compared to the control experiments (Fig. 2 and Table S1). Besides several other proteins, both known GEFs of Sec4 (Sec2 and Dss435,36) were among these 10 proteins, indicating that the method will be suitable for identification of unknown GEFs for other GTPases as well. Most notably, Sec2 showed the highest label free quantification (LFQ) intensities in most experiments and was significantly enriched in all triplicate experiments (Fig. 2). In contrast, Dss4 was only significantly enriched in one triplicate. In addition to the known GEFs, two proteins known to interact with Rab proteins (Ivy137 and Mrs638) were enriched as well as several proteins not known to have a function in Rab-mediated vesicular trafficking.
Figure 2.
Proteins that were specifically enriched in the pull-down experiments. (A) and (B) show the two independent experimental replicates of the pull-down experiments with each single experiment performed in triplicate. The average values of the label free quantification (LFQ) with their standard deviations are indicated in the graphs for all proteins that were significantly enriched in at least one triplicate (red: elution with 500 µM GTP; orange: control experiment not containing immobilized Sec4 and elution with 500 µM GTP; blue: elution with 10 mM GTP; cyan: control experiment not containing immobilized Sec4 and elution with 10 mM GTP; * indicates statistically relevant enrichment comparing the pull-down and the control experiment (t-test)). The insets show the same graphs with logarithmic scale of the LFQ intensities for better visualization of the differences between the experiments and the corresponding controls.
We wondered whether the established method might also be suitable to identify proteins that directly interact with the active (i.e., GTP-bound) GTPases, but not with GEF proteins. Therefore, in an additional experiment, biotinylated Sec4:GppNHp (GppNHp – Guanosine-5′-[β,γ-imido]triphosphate) was immobilized on the SMBs for similar pull-down experiments, but without the addition of alkaline phosphatase. Interestingly, both Ivy1 and Mrs6 were again specifically enriched in these pull-down experiments as well as the known Sec4 effector protein Sro739 (besides several other proteins; see Table S2 and Fig. S1) and were identified via MS. In contrast, neither Sec2 nor Dss4 were detected via MS, thus indicating that nucleotide-free GTPases need to be used to specifically enrich their corresponding GEFs.
Validation of putative targets
Since 10 different proteins were specifically enriched in the previous Sec4 pull-down experiments compared to the control experiments and pull-down experiments from complex samples generally contain many false positive results, we set up a strategy to validate these putative binding partners. The methods used aimed at detecting the activity of potential GEFs, but also to verify other potential interaction partners that were co-enriched in the pull-down experiments. Because commonly used recombinant expression and purification strategies are laborious and time-consuming when multiple different putative targets need to be tested, we used a commercially available in vitro transcription/translation (IVTT) system40,41 coupled with a fluorescence based exchange assay to validate the GEF(s) among the other identified targets. Independently, all putative targets were used in yeast two-hybrid (Y2H) experiment to allow validation of GEFs and other binding partners of the GTPase (Fig. 1B). To simplify the procedure, Gateway compatible vectors were used that allow easy shuttling of the genes coding the putative targets for the different validation steps into expression vectors as well as vectors for the Y2H screen.
Validation of putative targets via IVTT and GEF assay
For the IVTT system, all constructs were cloned into expression vectors containing N-terminal His6-tags via the Gateway cloning system.41 Subsequently, all constructs were produced in vitro in small scale IVTT-expressions (typically ∼40 µl) and expression of the different proteins was confirmed via western-blotting against the His6-tag with an α-His6-tag-antibody (Fig. S2a). Out of 10 proteins, 7 could be readily produced in detectable amounts and these were directly used for a fluorescence based GEF activity assay without further purification. For the activity assay, Sec4 was preparatively loaded with the fluorescent nucleotide mantGDP and displacement of mantGDP with GDP was monitored. In these experiments, only the known GEF Sec2 induced a significant increase in the rate of nucleotide exchange (Fig. 3 and Fig. S2). Dss4 did not increase the rate of nucleotide exchange in these experiments. However, keeping in mind the 23-fold lower activity of Mss4 toward Rab8 (Mss4 is the human homolog of Dss4; kcat/KM = 8.5·103 M−1 s−1) and the 285-fold lower activity of Dss4 toward Ypt1 (kcat/KM = 7.2·102 M−1s−1) compared to Sec2:Sec4 (kcat/KM = 2.0·105 M1 s−1) as well as the proposed function of Mss4 as a chaperone rather than a GEF,42,43 this is not surprising. Thus, a 23- to 285-fold higher expression level would be necessary for Dss4 compared to Sec2 to obtain a similar exchange activity.
Figure 3.
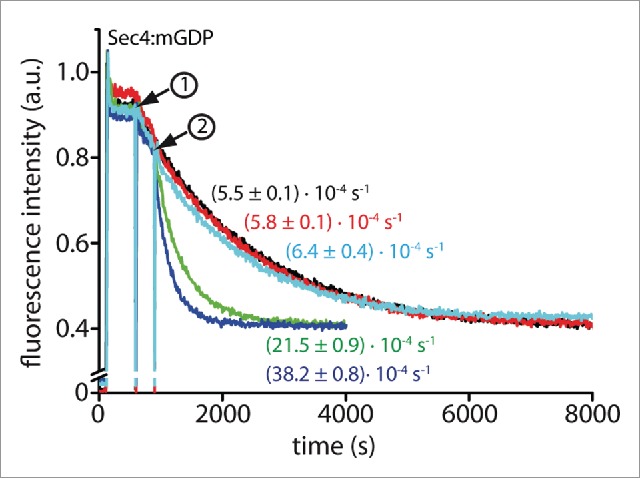
Nucleotide exchange assay using proteins from cell free expression. Sec4:mantGDP (1 µM) was incubated with 200 µM GDP (step 1) without addition of cell-free expressed protein showing the intrinsic rate of nucleotide exchange (black curve) and after addition (step 2) of 20 µl cell-free expression mixture (red curve) as control experiments. These experiments show that the cell-free expression mixture does not contain factors that accelerate nucleotide exchange. Additionally, similar experiments were repeated after expression of the putative targets (green and blue curves: 10 µl and 20 µl cell free expression mixture after expression of Sec2, respectively; cyan: 20 µl cell free expression mixture after expression of Dss4). Observed rate constants are indicated for each curve.
Validation of putative targets via yeast-two hybrid experiments
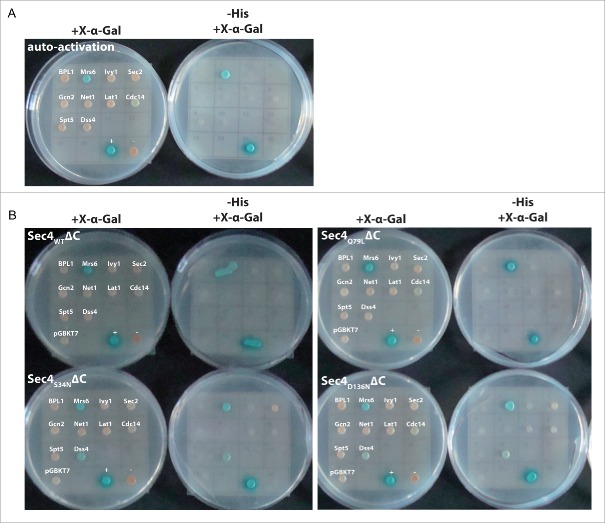
In a second validation step, all putative targets were cloned into a Gateway-compatible pGBKT7 bait-vector for Y2H-screening. In order to impede GTPase-prenylation that could interfere with targeting of Sec4 to the nucleus required for monitoring protein-protein interactions in Y2H, Sec4WT (WT – wild type) without the two C-terminal Cys-residues (Sec4WTΔC) was used as a prey. Additionally, we constructed different mutants of Sec4 mimicking the GDP-bound inactive state (Sec4S34NΔC), the GTP-bound active state (Sec4Q79LΔC) and a xanthosine 5′-triphosphate- (XTP-) binding mutant (Sec4D136NΔC).44 The D136N-substitution in Sec4D136NΔC changes the preference of the GTPase from guanine to xanthosine nucleotides. The latter mutant was designed since the environment of the small GTPase cannot be depleted of nucleotides in vivo (e.g. in a Y2H screen), hence not allowing the stable GEF:GTPaseWT complex formation that is necessary for the Y2H experiment. Since XTP occurs at extremely low abundance in cells45 we reasoned that this mutant might be effectively uncomplexed with any nucleotide and therefore faithfully represent a nucleotide-free Sec4 mimic. Expression of all bait and prey constructs in yeast were tested via western-blotting using Gal4 DNA-binding domain (Gal4 DBD) or Gal4 transcriptional activation domain (Gal4 TA) specific antibodies (Fig. S4) indicating that all except Net1 and Spt5 were well expressed.
All 10 putative targets were first tested against an empty prey-vector (Fig. 4A), and this showed that only Mrs6 caused auto-activation in the Y2H screen. In a second step, the experiments were repeated against the prey-vectors containing the different Sec4 mutants. Growth on selective media lacking histidine indicated that both Dss4 and Sec2 interact with the dominant negative mutant Sec4S34N as well as the supposedly nucleotide-free (and xanthosine nucleotide specific) Sec4D136N variant, but not with Sec4WT or the constitutively active Sec4Q79L (Fig. 4B and Table 2). We also observed a very weak positive signal for the non-GEF protein Cdc14 in the Y2H experiments using Sec4D136N. In addition to growth on the selective medium we also observed turnover of the chromogenic substrate X-α-Gal in the case of Dss4, but not Sec2. For the latter this is presumably a consequence of the low-affinity interaction between GEFs and small GTPases in the presence of high concentrations of nucleotides present in cells. Furthermore these results suggest that Sec4D136NΔC still binds endogenous nucleotides under these conditions in vivo, and this is in keeping with the observation that for the corresponding Ras mutant (H-RasD119N) the affinity of GDP (and presumably GTP), while much weaker than that of the wild-type protein, is still in the submicromolar range.46 We consequently also tested a double mutant (Sec4N133I, D136N) since both mutations have been reported to strongly reduce guanine nucleotide binding affinity.44 However Sec4N133I, D136N was not expressed in yeast (Fig. S4) probably because it was toxic toward yeast arising from sequestering the complete cellular pool of endogenous cognate nucleotide exchange factors (as already suggested in the original publication on these mutations44). Despite this, for human GTPases the corresponding double mutant might be a better mimic of a nucleotide-free protein without having the toxic side effects seen in yeast.
Figure 4.
Results of the yeast two-hybrid screens with Sec4. (A) All 10 putative targets from the pull-down experiments were cloned into bait-vectors and tested for auto-activation against empty prey-vectors. In these experiments, only Mrs6 displayed auto-activation. (B) The Y2H experiments were repeated using the prey-vectors containing Sec4WTΔC, Sec4S34NΔC, Sec4Q79LΔC or Sec4D136NΔC to test for interaction with putative binding partners. Growth on the selective medium lacking histidine (-His) indicates an interaction of both known GEFs (Sec2 and Dss4) with Sec4S34NΔC and Sec4D136NΔC, but not with Sec4WTΔC and Sec4Q79LΔC. However the interaction is probably weak (as expected for an enzymatic interaction) and turnover of the chromogenic substrate X-α-Gal could only be observed for Dss4, but not Sec2 ((+) and (−): positive (pGBKT7–53/pGADT7-T) and negative (pGBKT7-Lam/pGADT7-T) controls).
Table 2.
Results of the Y2H experiments shown in Figure 4.
| Bpl1 | Mrs6 | Ivy1 | Sec2 | Gcn2 | Net1 | Lat1 | Cdc14 | Spt5 | Dss4 | |
|---|---|---|---|---|---|---|---|---|---|---|
| autoactivation | − | +++ | − | − | − | − | − | − | − | − |
| Sec4WTΔC | − | +++ | − | − | − | − | − | − | − | − |
| Sec4S34NΔC | − | +++ | − | + | − | − | − | − | − | ++ |
| Sec4Q79LΔC | − | +++ | − | − | − | − | − | − | − | − |
| Sec4D136NΔC | − | +++ | − | + | − | − | − | (+) | − | ++ |
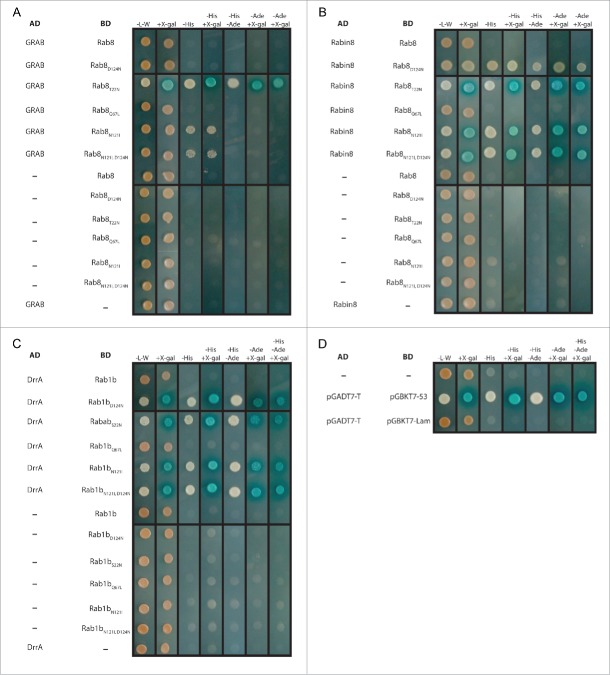
We subsequently used other known Rab:GEF pairs with different mutations of the Rab proteins in a similar experiment to verify the general applicability of this approach. The pairs tested included Rab8:Rabin8 and Rab8:GRAB (Rabin8 and GRAB are Sec2 homologues from human)47 as well as Rab1b:DrrA30 (Fig. 5). All experiments indicated an interaction between the GEFs and most of those Rab mutants impaired in nucleotide binding, but not with wildtype Rabs or the constitutively active (Q67L in Rab1b and Rab8) mutant. However we observed notable differences in the strength of the observed signals and the possible stringency of selection in the Y2H experiments. Notably, the Rab proteins containing the mutations N121I and D124N only showed interactions at low stringency of selection for Rab8N121I:GRAB (Fig. 5A) and Rab8D124N:Rabin8 (Fig. 5B) and completely missing interaction for Rab8D124N:GRAB (Fig. 5A). In contrast, the mutation corresponding to S22N in Rab1b and T22N in Rab8 showed strong interaction with all GEFs tested and under maximum possible stringency of selection (Fig. 5A-C), thus making this mutation the preferable one for identification of interacting GEFs.
Figure 5.
Results of the yeast two-hybrid screens with known Rab:GEF pairs. (A) The GEF domains of GRAB (aa 73–154 cloned into the vector pGADT7) and (B) Rabin8 (aa 153–237 in vector pGADT7) were tested for interaction with Rab8A (aa 1–203 in vector pGBKT7) and different mutants of Rab8 (Rab8T22N, Rab8Q67L, Rab8N121I, Rab8D124N, Rab8N121I, D124N). (C) In a similar experiment, the GEF domain of the Legionella pneumophila GEF DrrA (aa 340–533) was tested for interaction with Rab1B (aa 1–199) and the corresponding mutants of Rab1B (Rab1S22N, Rab1Q67L, Rab1N121I, Rab1D124N, Rab1N121I, D124N). All experiments were also performed with the corresponding empty vectors (−) to exclude auto-activation. (D) Shown are the positive (pGBKT7–53/pGADT7-T) and negative (pGBKT7-Lam/pGADT7-T) controls as well as the control with empty vectors (−/−).
Discussion
GEFs play a crucial role for correct spatial and temporal activation of small GTPases. For Rab-proteins, it has emerged over the last few years that GEFs have an important function in localization of the Rab-proteins to the correct intracellular membranes.48 Furthermore, many intracellularly surviving pathogens provide GEFs to manipulate small GTPase signaling and regulate events in infected host cells.49 However, despite the apparent importance of GEF molecules, for many small GTPases the corresponding natural or pathogenic GEF(s) have yet to be identified.
In contrast to Rab GAPs, where systematic analyses of their functional roles and their corresponding Rab substrates have been performed based on the strong sequence similarity among the GAPs,50 a similar approach is not possible for GEFs due to their great structural diversity.9 We have therefore shown in this publication that a pull-down procedure can be used to specifically enrich GEFs for small GTPases by depleting their environment of nucleotides and thus making use of the high affinity of GEFs for nucleotide-free GTPases. A similar procedure has previously been applied to specifically enrich GEFs for Rho proteins51 making use of a mutation corresponding to G15A in Ras, a mutation that causes a dramatic decrease of nucleotide binding affinity.52 To the best of our knowledge, this is the first time that a pull-down procedure for small GTPases has been set up for specific enrichment of nucleotide exchange factors among the different GTPase interacting proteins by making use of the basic mechanism of GEFs and using the wildtype form of a GTPase. Since nucleotide-free GTPases (and GTPases carrying mutations that render them effectively nucleotide-free) are often inherently unstable and difficult to handle, this greatly simplifies the pull-down procedure and makes it more generally applicable. Testing the principle for the Rab protein Sec4 showed that the procedure can be applied successfully and both known exchange factors (Sec2 and Dss4) were specifically enriched and identified by this approach. However, even though the method worked very well for these Rab:GEF pairs, one has to keep in mind that missing factors in positive or negative feedback loops as observed in Rab cascades,53 the absence of post-translational modifications8 or putative GEFs containing transmembrane regions might interfere with an easy identification via this method.
Since pull-down experiments from complex samples always give rise to many false positive targets, we furthermore established validation procedures to identify the GEFs among the putative targets. One further development of this procedure could be the use of stable-isotope labeling of amino acids in cell culture (SILAC) to further reduce the number of false positives detected via mass spectrometry and consequently minimize the effort needed to validate the putative targets.54
The validation procedures reported in this publication include detection of the activity of potential GEFs as well as Y2H screens to allow identification and verification of GEFs and other general binding partners of small GTPases. The results using activity-based detection of GEF proteins presented above indicate that only potent GEFs can be easily identified by this approach (as shown for Sec2), while for less potent GEFs, more elaborate experiments will have to be performed. However the catalytic efficiencies (kcat/KM) of yet identified GEFs for small GTPases cover the whole range from 102 M−1 s−1 to 107 M−1 s−1 (kcat/KM = 2.0·105 M−1 s−1 for Sec2),55 thus making the procedure generally applicable for many of them.
A second approach for validation and identification of GEFs is provided by the Y2H experiments used in this publication. The possibility to identify GEFs via this approach has previously been shown for other exchange factors in several publications (see e.g., refs. 21-25) by using mutants of small GTPases with a lowered nucleotide affinity. Also, a modified yeast two-hybrid approach making use of the fact that small GTPases interact with their effectors only after activation by a GEF has been used in the past to characterize GEFs.56
Binding of both Dss4 and Sec2 to Sec4S34N and Sec4D136N observed in the Y2H experiments can be easily explained by the preferential binding of GEFs to nucleotide-free GTPases and more generally by the preferential binding of GEFs to the disordered open conformation of the switch regions of small GTPases.9 Several effects contribute to this. First, the S34N mutation in GTPases leads to a strongly reduced nucleotide affinity (which is more strongly pronounced for GTP than for GDP),57 while the D136N mutation leads to a dramatic reduction of nucleotide affinity because of the loss of an important specific interaction with the nucleobase. The S34N mutant should be regarded as a hybrid nucleotide-free and GDP-bound mimic. Consistently, a specific interaction has previously been observed in Y2H experiments for Rabin8 (the human homolog of Sec2) and the constitutively inactive mutant Rab8T22N, but not with Rab8WT or the constitutively active Rab8Q67L mutant.23 In addition to the aforementioned effects, reported relative affinities of small GTPases toward GTP and GDP usually show a slightly (approximately 2- to 4-fold) higher nucleotide-affinity of the GTPases toward GTP compared to GDP,57,58 thus making the GDP-bound state a thermodynamically slightly better binding partner of GEFs than the GTP-bound state.
The general applicability of the Y2H approach was shown in this publication using different GEFs together with their known Rab substrates and different mutants thereof. These experiments indicate that comparative Y2H experiments using mutants of small GTPases mimicking the different activity states (i.e. active, inactive or nucleotide-free) can give valuable insights regarding the function of interacting proteins. However, it has to be borne in mind that many protein-protein interactions are of transient nature with KD or KM-values generally in the µM range and the mutations used are only mimics of the different activity states. In some cases they might even hinder GEF binding.59 Therefore only the combination of specific enrichment of potential GEFs combined with different validation steps will give conclusive answers as to whether a certain protein is a GEF or not.
Materials and methods
Pull-down procedure
Purified full length Sec4 (purification as described60) was biotinylated using EZ-Link™ Maleimide-PEG2-Biotin (Life Technologies) as described by the manufacturer and 1.5 nmole biotinylated Sec4 was immobilized on streptavidin magnetic beads (SMBs, New England Biolabs, 250 µl of the bead solution was thoroughly washed with phosphate buffered saline (PBS; 137 mmol/l NaCl, 2.7 mmol/l KCl, 10 mmol/l Na2HPO4, 1.8 mmol/l KH2PO4) pH 7.0 prior to usage).
Yeast cell lysate was prepared by growing yeast cells in YPDA medium to OD600nm=1.7–3.0, harvesting the cells, and resuspending them in PBS pH 7.0 supplemented with protease inhibitors (1 mM phenylmethanesulfonylfluoride and each 5 µg/ml chymostatin, leupeptin, antipain and pepstatinA). Subsequently the cells were lysed using a French press and the supernatant was cleared by centrifugation (75600 g, 10 °C, 1 h).
The yeast lysate (total protein concentration 1 mg/ml) was spiked with 25 units of alkaline phosphatase per ml cell lysate and stored on ice 1 h before usage. A volume of 1 ml lysate was added to the immobilized Sec4 and incubated for 45 min on ice before removing the supernatant and washing three times with 500 µl PBS pH 7.0. Addition of cell lysate and washing with PBS was repeated twice before eluting the bound proteins with 100 µl PBS pH 7.0 containing 500 µM GTP and subsequently 100 µl PBS containing 10 mM GTP. All experiments were performed in triplicate with two biological replicates using a different batch of Sec4 and yeast cell lysate. Additionally similar experiments without immobilized Sec4 were performed as negative controls.
Mass spectrometry
For mass spectrometry, samples from the pull-down were digested in solution using proteomics grade porcine trypsin protease (Sigma Aldrich). A volume of 100 µl of digestion buffer (50 mM Tris pH 7.5, 1 mM DTT, 13.3 % w/v carbamide, 5 µg/ml trypsin) were added to the sample and the mixture was incubated for 1 h at room temperature in a tube rotator at 40 rpm. A volume of 100 µl of alkylation buffer (50 mM Tris pH 7.5, 13.3 % w/v carbamide, 55 mM iodoacetamide) was added and the sample was further incubated overnight at 37°C and 350 rpm in a thermomixer. The reaction was stopped by addition of 2 µl Trifluoroacetic acid.
Digested samples were purified using StageTips made of dual layer blank-outs from Empore™ 2215-C18 extraction discs (3M) as described previously.61 Shortly, StageTips were activated with 100µl methanol and equilibrated by two further centrifugation cycles with 100 µl 0.1 % formic acid in H2O. Samples were applied to the tips twice; after a first cycle of centrifugation, the flow-through was loaded and centrifuged again. StageTips were washed with 100 µl 0.1 % formic acid and subsequently bound peptides were eluted into a clean tube with 2 × 20 µl elution buffer (80 % acetonitrile, 0.1 % formic acid). Eluted samples were dried using a centrifugal evaporator and stored at −20 °C until measurement.
For analysis, these tryptic peptides were dissolved in 20 μl 0.1 % TFA in water and analyzed by nano-HPLC/MS/MS. Briefly, the tryptic digests were separated and analyzed on a UltiMate™ 3000 RSLCnano system coupled on-line to a Q-Exactive™ Hybrid Quadrupole-Orbitrap Mass Spectrometer equipped with a nano-spray source (Nanospray Flex Ion Source, all Thermo Scientific, Germany). All solvents were LC-MS grade. 3 µl of the peptide solution were injected onto a pre-column cartridge (5 µm, 100 Å, 300 µm ID × 5 mm, Thermo Scientific, Germany) using 0.1 % TFA in water as eluent with a flow rate of 30 µl/min. Desalting was performed for 5 min with eluent flow to waste followed by back-flushing of the sample during the whole analysis from the pre-column to the PepMap100 RSLC C18 nano-HPLC column (2 μm, 100 Å, 75 μm ID × 25 cm, nanoViper, Thermo Scientific, Germany) using a linear gradient starting with 95 % solvent A (0.1 % formic acid in water) / 5 % solvent B (0.1 % formic acid in acetonitrile) and increasing to 30 % solvent B after 95 min using a flow rate of 300 nl/min.
For coupling of the nano-HPLC to the Quadrupole-Orbitrap Mass Spectrometer, a standard coated SilicaTip (ID 20 μm, Tip-ID 10 μM, New Objective, Woburn, MA, USA) was used. Mass spectra were acquired using a so called TOP10 method, i.e., full scan spectra were acquired using a mass range of m/z 300 to 1650 with a resolution of 70000, followed by up to ten high energy collision dissociation (HCD) MS/MS scans at a resolution of 17500 of the most intense at least doubly charged ions. The dynamic exclusion was set to 20 s.
Data evaluation was performed using MaxQuant software62 (v.1.4.1.2) including the Andromeda search algorithm and searching the yeast reference proteome of the uniprot database. Briefly, the search was performed for full enzymatic trypsin cleavages allowing two miscleavages. For protein modifications carbamidomethylation was chosen as fixed and oxidation of methionine and acetylation of the N-terminus as variable modifications. The mass accuracy for full mass spectra was set to 20 ppm for the first and 4.5 ppm for the second search and to 20 ppm for MS/MS spectra. The false discovery rates for peptide and protein identification were set to 1 %. Only proteins for which at least two peptides were quantified were chosen for further validation. Relative quantification of proteins was carried out using the label free quantification algorithm implemented in MaxQuant. Briefly, samples resulting from affinity enrichments with SEC4 to the solid support and those resulting from similar enrichment using SMBs without Sec4 were grouped. Label free quanitification intensities (LFQ) were logarithmized (log2) and proteins which were not quantified at least two times in at least one of the groups were filtered out. Missing values were imputed using small normal distributed values and a t-test (FDR 0.05) was performed. Proteins which were statistically significant outliers in the t-test were considered as hits if at least two unique peptides were identified in each technical replicate of the Sec4 group and non in the controls or if at least 4 unique peptides were identified in each technical replicate in the Sec4 group.
Cell-free expression and GEF activity assay
All putative targets were cloned into the pDONR201 entry vector (Invitrogen) for Gateway compatible cloning. For proteins bigger than 120 kDa, smaller fragments were designed using the predict protein website (https://www.predictprotein.org/) to avoid fragments containing only parts of a folded domain. In vitro translation was performed using Lexsy cell extract (Jena Bioscience) as described by the manufacturer after transferring the target genes into pCellFree_G01 expression plasmids.41 To verify successful translation, in vitro translated crude protein lysates were used for immunoblotting. Equal amount of lysate was mixed with 2x SDS loading buffer and boiled for 5 min at 95 °C. Samples were run on either 10 % or 15 % SDS-PAGE and transferred to the Polyvinylidenfluorid (PVDF) membrane. Membranes were blocked with TBS/0.05 % Tween 20 containing 5 % powdered skim milk, followed by incubation with monoclonal mouse anti-His antibody (Sigma). After extensive washing with Tris-buffered saline (TBS; 50 mM Tris, 150 mM NaCl, pH 7.6) / 0.05 % Tween 20, a horseradish peroxidase-conjugated secondary antibody (Cayman chemical company) was used for detection. Images were taken with a GelDoc system (Bio Rad) using the SuperSignal West Dura Substrate (Thermo Scientific).
GEF activities of in vitro translated crude extracts were measured with Sec4 preparatively loaded with mantGDP as described previously42,60 in a buffer containing 20 mM Hepes pH 7.5, 50 mM NaCl, 2 mM MgCl2 and 5 mM DTE on a Fluromax-3 fluorescence spectrometer (Horiba Jobin Yvon Inc.) at 25°C. The change in mant fluorescence (excitation λex =360 nm; emission λem =440 nm) was fitted to a single exponential equation using Origin 9.0.
Yeast two-hybrid experiments
Gateway compatible yeast two-hybrid vectors were designed based on the commercially available pGBKT7 vector (Clontech Laboratories, Inc.). The Gateway cloning cassette was introduced into the multiple cloning site using the NdeI and NotI restriction sites. Additionally the kanamycine resistance gene was replaced with a gentamicin resistance gene to be compatible with the donor vector pDONR201 (Invitrogen) used in this study (a vector map of the resulting Y2H vector is shown in Fig. S3).
The coding sequences for all putative targets from the pull-down screens were transferred from the corresponding pDONR201 entry vectors (see above) into the Gateway compatible pGBKT7 bait vector, the coding sequences for Sec4WTΔC and the different mutants thereof were cloned into the pGADT7 prey vector (Clontech Laboratories, Inc.). Using the one-step transformation protocol for yeast in stationary phase,63 the generated vectors containing the putative targets or the Sec4 mutants as well as the empty vectors (to test for auto-activation) were transformed into yeast strains AH109 (pGBKT7 bait vectors) or Y187 (pGADT7 prey vector) and transformants were selected on SD plates deficient of tryptophane (SD-Trp) or leucine (SD-Leu), respectively. Subsequently, all different combinations of the AH109 yeast strain containing the bait vectors and the Y187 yeast strain containing the prey vectors were mated and plated on SD plates deficient of tryptophane and leucine (SD-Trp-Leu) and grown for 3 days (30 °C). Protein-protein interactions were subsequently screened by resuspending a single colony from the mating plates in 50 µl 0.9 % NaCl solution and plating 3 µl thereof on SD-Trp-Leu plates containing X-α-Gal with and without histidine and growing the cells for 3–4 days at 30 °C. To further analyze the strength of interactions of known Rab:GEF pairs, the expression of two or three reporter genes have been tested, using varying stringent growth conditions. The strength of interaction was estimated from the comparative growth of yeast cells on minimal media lacking either adenine or histidine or both. Mating of yeast cells (Y2HGold or AH109 strain) containing either the pGBKT7–53 or pGBKT7-Lam vectors with yeast cells (Y187 strain) containing the pGADT7-T vector (all Clontech Laboratories, Inc.) served as positive and negative controls, respectively.
For expression control of the different proteins in yeast, proteins were extracted according to 64 and expression was confirmed via western-blot (primary antibodies rabbit Gal4 DBD antibody sc-577 for bait proteins and mouse anti GAL4-TA sc-1663 for prey proteins, Santa Cruz Biotechnology, Inc.). The BCIP-NBT solution from Santa Cruz Biotechnology Inc. was used for detection using an alkaline phosphatase coupled secondary antibody.
Supplementary Material
Abbreviations
- DBD
DNA binding domain
- GAP
GTPase activating protein
- GEF
guanine nucleotide-exchange factor
- GDP
Guanosine-5′-diphosphat
- GppNHp
Guanosine-5′-[β,γ-imido]-triphosphat
- GTP
Guanosine-5′-triphosphat
- GXP
Guanine nucleotide
- IVTT
in vitro transcription/translation
- LFQ
label free quantification
- MS
mass spectrometry
- PBS
phosphate buffered saline
- SMBs
streptavidin magnetic beads
- TA
transcriptional activation domain
- WT
wild type
- XTP
Xanthosine-5′-triphosphat
- Y2H
yeast two-hybrid
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Prof. Kirill Alexandrov for the generous gift of Gateway-compatible expression vectors for cell-free expression. The DPF (Dortmund Protein Facility) is acknowledged for cloning of different constructs used in this publication.
Funding
The work was funded by DFG/ANR grant GO 284/8-1.
References
- [1].Hurley JB, Simon MI, Teplow DB, Robishaw JD, Gilman AG. Homologies between signal transducing G proteins and ras gene products. Science 1984; 226:860-2; PMID:6436980; http://dx.doi.org/ 10.1126/science.6436980 [DOI] [PubMed] [Google Scholar]
- [2].Wittinghofer A. Signal transduction via Ras. Biol Chem 1998; 379:933-7; PMID:9792425. [PubMed] [Google Scholar]
- [3].Burridge K, Wennerberg K. Rho and Rac take center stage. Cell 2004; 116:167-79; PMID:14744429; http://dx.doi.org/ 10.1016/S0092-8674(04)00003-0 [DOI] [PubMed] [Google Scholar]
- [4].Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J 1996; 15:5584-94; PMID:8896452. [PMC free article] [PubMed] [Google Scholar]
- [5].Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev 2011; 91:119-49; PMID:21248164; http://dx.doi.org/ 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 2009; 10:513-25; PMID:19603039; http://dx.doi.org/ 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- [7].Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev 2001; 81:153-208; PMID:11152757. [DOI] [PubMed] [Google Scholar]
- [8].Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell 2007; 129:865-77; PMID:17540168; http://dx.doi.org/ 10.1016/j.cell.2007.05.018 [DOI] [PubMed] [Google Scholar]
- [9].Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 2013; 93:269-309; PMID:23303910; http://dx.doi.org/ 10.1152/physrev.00003.2012 [DOI] [PubMed] [Google Scholar]
- [10].Broek D, Toda T, Michaeli T, Levin L, Birchmeier C, Zoller M, Powers S, Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell 1987; 48:789-99; PMID:3545497; http://dx.doi.org/ 10.1016/0092-8674(87)90076-6 [DOI] [PubMed] [Google Scholar]
- [11].Eva A, Vecchio G, Rao CD, Tronick SR, Aaronson SA. The predicted DBL oncogene product defines a distinct class of transforming proteins. Proc Natl Acad Sci U S A 1988; 85:2061-5; PMID:3281159; http://dx.doi.org/ 10.1073/pnas.85.7.2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bischoff FR, Maier G, Tilz G, Ponstingl H. A 47-kDa human nuclear protein recognized by antikinetochore autoimmune sera is homologous with the protein encoded by RCC1, a gene implicated in onset of chromosome condensation. Proc Natl Acad Sci U S A 1990; 87:8617-21; PMID:2236072; http://dx.doi.org/ 10.1073/pnas.87.21.8617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Downward J, Riehl R, Wu L, Weinberg RA. Identification of a nucleotide exchange-promoting activity for p21ras. Proc Natl Acad Sci U S A 1990; 87:5998-6002; PMID:2116014; http://dx.doi.org/ 10.1073/pnas.87.15.5998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huang YK, Kung HF, Kamata T. Purification of a factor capable of stimulating the guanine nucleotide exchange reaction of ras proteins and its effect on ras-related small molecular mass G proteins. Proc Natl Acad Sci U S A 1990; 87:8008-12; PMID:2172971; http://dx.doi.org/ 10.1073/pnas.87.20.8008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hart MJ, Eva A, Evans T, Aaronson SA, Cerione RA. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the dbl oncogene product. Nature 1991; 354:311-4; PMID:1956381; http://dx.doi.org/ 10.1038/354311a0 [DOI] [PubMed] [Google Scholar]
- [16].Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 1991; 354:80-2; PMID:1944575; http://dx.doi.org/ 10.1038/354080a0 [DOI] [PubMed] [Google Scholar]
- [17].Burd CG, Mustol PA, Schu PV, Emr SD. A yeast protein related to a mammalian Ras-binding protein, Vps9p, is required for localization of vacuolar proteins. Mol Cell Biol 1996; 16:2369-77; PMID:8628304; http://dx.doi.org/ 10.1128/MCB.16.5.2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sato M, Sato K, Liou W, Pant S, Harada A, Grant BD. Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J 2008; 27:1183-96; PMID:18354496; http://dx.doi.org/ 10.1038/emboj.2008.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Allaire PD, Marat AL, Dall'Armi C, Di Paolo G, McPherson PS, Ritter B. The Connecdenn DENN domain: a GEF for Rab35 mediating cargo-specific exit from early endosomes. Mol Cell 2010; 37:370-82; PMID:20159556; http://dx.doi.org/ 10.1016/j.molcel.2009.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hama H, Tall GG, Horazdovsky BF. Vps9p is a guanine nucleotide exchange factor involved in vesicle-mediated vacuolar protein transport. J Biol Chem 1999; 274:15284-91; PMID:10329739; http://dx.doi.org/ 10.1074/jbc.274.21.15284 [DOI] [PubMed] [Google Scholar]
- [21].Berken A, Thomas C, Wittinghofer A. A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 2005; 436:1176-80; PMID:15980860; http://dx.doi.org/ 10.1038/nature03883 [DOI] [PubMed] [Google Scholar]
- [22].Sakaguchi A, Sato M, Sato K, Gengyo-Ando K, Yorimitsu T, Nakai J, Hara T, Sato K, Sato K. REI-1 Is a Guanine Nucleotide Exchange Factor Regulating RAB-11 Localization and Function in C. elegans Embryos. Dev Cell 2015; 35:211-21; PMID:26506309; http://dx.doi.org/ 10.1016/j.devcel.2015.09.013 [DOI] [PubMed] [Google Scholar]
- [23].Hattula K, Furuhjelm J, Arffman A, Peränen JA. Rab8-specific GDP/GTP Exchange Factor Is Involved in Actin Remodeling and Polarized Membrane Transport. Mol Biol Cell 2002; 13:3268-3280; PMID:12221131; http://dx.doi.org/ 10.1091/mbc.E02-03-0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mori Y, Matsui T, Fukuda M. Rabex-5 protein regulates dendritic localization of small GTPase Rab17 and neurite morphogenesis in hippocampal neurons. J Biol Chem 2013; 288:9835-47.; PMID:23430262; http://dx.doi.org/ 10.1074/jbc.M112.427591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tamura K, Ohbayashi N, Maruta Y, Kanno E, Itoh T, Fukuda M. Varp is a novel Rab32/38-binding protein that regulates Tyrp1 trafficking in melanocytes. Mol Biol Cell 2009; 20:2900-8; PMID:19403694; http://dx.doi.org/ 10.1091/mbc.E08-12-1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Guo Z, Ahmadian MR, Goody RS. Guanine nucleotide exchange factors operate by a simple allosteric competitive mechanism. Biochemistry 2005; 44:15423-9; PMID:16300389; http://dx.doi.org/ 10.1021/bi0518601 [DOI] [PubMed] [Google Scholar]
- [27].Klebe C, Prinz H, Wittinghofer A, Goody RS. The kinetic mechanism of Ran–nucleotide exchange catalyzed by RCC1. Biochemistry 1995; 34:12543-52; PMID:7548002; http://dx.doi.org/ 10.1021/bi00039a008 [DOI] [PubMed] [Google Scholar]
- [28].Goody RS, Hofmann-Goody W. Exchange factors, effectors, GAPs and motor proteins: common thermodynamic and kinetic principles for different functions. Eur Biophys J 2002; 31:268-74; PMID:12122473; http://dx.doi.org/ 10.1007/s00249-002-0225-3 [DOI] [PubMed] [Google Scholar]
- [29].Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem 1994; 140:1-22; PMID:7877593; http://dx.doi.org/ 10.1007/BF00928361 [DOI] [PubMed] [Google Scholar]
- [30].Schoebel S, Oesterlin LK, Blankenfeldt W, Goody RS, Itzen A. RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol Cell 2009; 36:1060-72; PMID:20064470; http://dx.doi.org/ 10.1016/j.molcel.2009.11.014 [DOI] [PubMed] [Google Scholar]
- [31].Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature 1998; 394:337-43; PMID:9690470; http://dx.doi.org/ 10.1038/28548 [DOI] [PubMed] [Google Scholar]
- [32].Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1). Cell 2001; 105:245-55; PMID:11336674; http://dx.doi.org/ 10.1016/S0092-8674(01)00315-4 [DOI] [PubMed] [Google Scholar]
- [33].Dong G, Medkova M, Novick P, Reinisch KM. A catalytic coiled coil: structural insights into the activation of the Rab GTPase Sec4p by Sec2p. Mol Cell 2007; 25:455-62; PMID:17289591; http://dx.doi.org/ 10.1016/j.molcel.2007.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sato Y, Fukai S, Ishitani R, Nureki O. Crystal structure of the Sec4p.Sec2p complex in the nucleotide exchanging intermediate state. Proc Natl Acad Sci U S A 2007; 104:8305-10; PMID:17488829; http://dx.doi.org/ 10.1073/pnas.0701550104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Burton J, Roberts D, Montaldi M, Novick P, De Camilli P. A mammalian guanine-nucleotide-releasing protein enhances function of yeast secretory protein Sec4. Nature 1993; 361:464-7; PMID:8429887; http://dx.doi.org/ 10.1038/361464a0 [DOI] [PubMed] [Google Scholar]
- [36].Walch-Solimena C, Collins RN, Novick PJ. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol 1997; 137:1495-509; PMID:9199166; http://dx.doi.org/ 10.1083/jcb.137.7.1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Numrich J, Péli-Gulli MP, Arlt H, Sardu A, Griffith J, Levine T, Engelbrecht-Vandré S, Reggiori F, De Virgilio C, Ungermann C. The I-BAR protein Ivy1 is an effector of the Rab7 GTPase Ypt7 involved in vacuole membrane homeostasis. J Cell Sci 2015; 128(13):2278-92; PMID:25999476; http://dx.doi.org/ 10.1242/jcs.164905 [DOI] [PubMed] [Google Scholar]
- [38].Bauer BE, Lorenzetti S, Miaczynska M, Bui DM, Schweyen RJ, Ragnini A. Amino- and carboxy-terminal domains of the yeast Rab escort protein are both required for binding of Ypt small G proteins. Mol Biol Cell 1996; 7:1521-33; PMID:8898359; http://dx.doi.org/ 10.1091/mbc.7.10.1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Grosshans BL, Andreeva A, Gangar A, Niessen S, Yates JR 3rd, Brennwald P, Novick P. The yeast lgl family member Sro7p is an effector of the secretory Rab GTPase Sec4p. J Cell Biol 2006; 172:55-66; PMID:16390997; http://dx.doi.org/ 10.1083/jcb.200510016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mureev S, Kovtun O, Nguyen UT, Alexandrov K. Species-independent translational leaders facilitate cell-free expression. Nat Biotechnol 2009; 27:747-52; PMID:19648909; http://dx.doi.org/ 10.1038/nbt.1556 [DOI] [PubMed] [Google Scholar]
- [41].Gagoski D, Mureev S, Giles N, Johnston W, Dahmer-Heath M, Škalamera D, Gonda TJ, Alexandrov K. Gateway-compatible vectors for high-throughput protein expression in pro- and eukaryotic cell-free systems. J Biotechnol 2015; 195:1-7; PMID:25529348; http://dx.doi.org/ 10.1016/j.jbiotec.2014.12.006 [DOI] [PubMed] [Google Scholar]
- [42].Esters H, Alexandrov K, Iakovenko A, Ivanova T, Thomä N, Rybin V, Zerial M, Scheidig AJ, Goody RS. Vps9, Rabex-5 and DSS4: proteins with weak but distinct nucleotide-exchange activities for Rab proteins. J Mol Biol 2001; 310:141-56; http://dx.doi.org/ 10.1006/jmbi.2001.4735 [DOI] [PubMed] [Google Scholar]
- [43].Itzen A, Pylypenko O, Goody RS, Alexandrov K, Rak A. Nucleotide exchange via local protein unfolding–structure of Rab8 in complex with MSS4. EMBO J 2006; 25:1445-55; PMID:16541104; http://dx.doi.org/ 10.1038/sj.emboj.7601044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jones S, Litt RJ, Richardson CJ, Segev N. Requirement of nucleotide exchange factor for Ypt1 GTPase mediated protein transport. J Cell Biol 1995; 130:1051-61; PMID:7657691; http://dx.doi.org/ 10.1083/jcb.130.5.1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Seifert R. Constitutive Activity of β-Adrenoceptors: Analysis in Membrane Systems in G Protein-Coupled Receptors as Drug Targets 121-140 (Wiley-VCH Verlag GmbH & Co. KGaA, 2006) [Google Scholar]
- [46].Schmidt G, Lenzen C, Simon I, Deuter R, Cool RH, Goody RS, Wittinghofer A. Biochemical and biological consequences of changing the specificity of p21ras from guanosine to xanthosine nucleotides. Oncogene 1996; 12:87-96; PMID:8552403. [PubMed] [Google Scholar]
- [47].Guo Z, Hou X, Goody RS, Itzen A. Intermediates in the guanine nucleotide exchange reaction of Rab8 protein catalyzed by guanine nucleotide exchange factors Rabin8 and GRAB. J Biol Chem 2013; 288:32466-74; PMID:24072714; http://dx.doi.org/ 10.1074/jbc.M113.498329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Blumer J, Rey J, Dehmelt L, Mazel T, Wu YW, Bastiaens P, Goody RS, Itzen A. RabGEFs are a major determinant for specific Rab membrane targeting. J Cell Biol 2013; 200:287-300; PMID:23382462; http://dx.doi.org/ 10.1083/jcb.201209113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Orchard RC, Alto NM. Mimicking GEFs: a common theme for bacterial pathogens. Cell Microbiol 2012; 14:10-8; PMID:21951829; http://dx.doi.org/ 10.1111/j.1462-5822.2011.01703.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Haas AK, Yoshimura S, Stephens DJ, Preisinger C, Fuchs E, Barr FA. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci 2007; 120:2997-3010; PMID:17684057; http://dx.doi.org/ 10.1242/jcs.014225 [DOI] [PubMed] [Google Scholar]
- [51].Garcia-Mata R, Wennerberg K, Arthur WT, Noren NK, Ellerbroek SM, Burridge K. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol 2006; 406:425-37; PMID:16472675; http://dx.doi.org/ 10.1016/S0076-6879(06)06031-9 [DOI] [PubMed] [Google Scholar]
- [52].Chen SY, Huff SY, Lai CC, Der CJ, Powers S. Ras-15A protein shares highly similar dominant-negative biological properties with Ras-17N and forms a stable, guanine-nucleotide resistant complex with CDC25 exchange factor. Oncogene 1994; 9:2691-8; PMID:8058333. [PubMed] [Google Scholar]
- [53].Nottingham RM, Pfeffer SR. Defining the boundaries: Rab GEFs and GAPs. Proc Natl Acad Sci U S A 2009; 106:14185-6; PMID:19706500; http://dx.doi.org/ 10.1073/pnas.0907725106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol 2006; 7:952-8; PMID:17139335; http://dx.doi.org/ 10.1038/nrm2067 [DOI] [PubMed] [Google Scholar]
- [55].Randazzo PA, Jian X, Chen PW, Zhai P, Soubias O, Northup JK. Quantitative Analysis of Guanine Nucleotide Exchange Factors (GEFs) as Enzymes. Cell Logist 2013; 3:e27609; PMID:25332840; http://dx.doi.org/ 10.4161/cl.27609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].De Toledo M, Colombo K, Nagase T, Ohara O, Fort P, Blangy A. The yeast exchange assay, a new complementary method to screen for Dbl-like protein specificity: identification of a novel RhoA exchange factor. FEBS Lett 2000; 480:287-92; PMID:11034346; http://dx.doi.org/ 10.1016/S0014-5793(00)01953-0 [DOI] [PubMed] [Google Scholar]
- [57].John J, Rensland H, Schlichting I, Vetter I, Borasio GD, Goody RS, Wittinghofer A. Kinetic and structural analysis of the Mg(2+)-binding site of the guanine nucleotide-binding protein p21H-ras. J Biol Chem 1993; 268:923-9; PMID:8419371. [PubMed] [Google Scholar]
- [58].Zhang B, Zhang Y, Wang Z, Zheng Y. The role of Mg2+ cofactor in the guanine nucleotide exchange and GTP hydrolysis reactions of Rho family GTP-binding proteins. J Biol Chem 2000; 275:25299-307; PMID:10843989; http://dx.doi.org/ 10.1074/jbc.M001027200 [DOI] [PubMed] [Google Scholar]
- [59].Langemeyer L, Nunes Bastos R, Cai Y, Itzen A, Reinisch KM, Barr FA. Diversity and plasticity in Rab GTPase nucleotide release mechanism has consequences for Rab activation and inactivation. Elife 2014; 3:e01623; PMID:24520163; http://dx.doi.org/ 10.7554/eLife.01623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Itzen A, Rak A, Goody RS. Sec2 is a highly efficient exchange factor for the Rab protein Sec4. J Mol Biol 2007; 365:1359-67; PMID:17134721; http://dx.doi.org/ 10.1016/j.jmb.2006.10.096 [DOI] [PubMed] [Google Scholar]
- [61].Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2007; 2:1896-906; PMID:17703201; http://dx.doi.org/ 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- [62].Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008; 26:1367-72; PMID:19029910; http://dx.doi.org/ 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- [63].Chen DC, Yang BC, Kuo TT. One-step transformation of yeast in stationary phase. Current Genetics 1992; 21:83-84; PMID:1735128; http://dx.doi.org/ 10.1007/BF00318659 [DOI] [PubMed] [Google Scholar]
- [64].Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast 2000; 16:857-860; PMID:10861908; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- [65].Diez D, Sanchez-Jimenez F, Ranea JA. Evolutionary expansion of the Ras switch regulatory module in eukaryotes. Nucleic Acids Res 2011; 39:5526-37; PMID:21447561; http://dx.doi.org/ 10.1093/nar/gkr154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Jun JE, Rubio I, Roose JP. Regulation of ras exchange factors and cellular localization of ras activation by lipid messengers in T cells. Front Immunol 2013; 4:239; PMID:24027568; http://dx.doi.org/ 10.3389/fimmu.2013.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci 2005; 118:843-6; PMID:15731001; http://dx.doi.org/ 10.1242/jcs.01660 [DOI] [PubMed] [Google Scholar]
- [68].Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene 2014; 33:4021-35; PMID:24037532; http://dx.doi.org/ 10.1038/onc.2013.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Mucha E, Fricke I, Schaefer A, Wittinghofer A, Berken A. Rho proteins of plants–functional cycle and regulation of cytoskeletal dynamics. Eur J Cell Biol 2011; 90:934-43; PMID:21277045; http://dx.doi.org/ 10.1016/j.ejcb.2010.11.009 [DOI] [PubMed] [Google Scholar]
- [70].Barr F, Lambright DG. Rab GEFs and GAPs. Curr Opin Cell Biol 2010; 22:461-70; PMID:20466531; http://dx.doi.org/ 10.1016/j.ceb.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Carney DS, Davies BA, Horazdovsky BF. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol 2006; 16:27-35; PMID:16330212; http://dx.doi.org/ 10.1016/j.tcb.2005.11.001 [DOI] [PubMed] [Google Scholar]
- [72].Marat AL, Dokainish H, McPherson PS. DENN domain proteins: regulators of Rab GTPases. J Biol Chem 2011; 286:13791-800; PMID:21330364; http://dx.doi.org/ 10.1074/jbc.R110.217067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Barrowman J, Bhandari D, Reinisch K, Ferro-Novick S. TRAPP complexes in membrane traffic: convergence through a common Rab. Nat Rev Mol Cell Biol 2010; 11:759-63; PMID:20966969; http://dx.doi.org/ 10.1038/nrm2999 [DOI] [PubMed] [Google Scholar]
- [74].Nordmann M, Cabrera M, Perz A, Bröcker C, Ostrowicz C, Engelbrecht-Vandré S, Ungermann C. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol 2010; 20:1654-9; PMID:20797862; http://dx.doi.org/ 10.1016/j.cub.2010.08.002 [DOI] [PubMed] [Google Scholar]
- [75].Pusapati GV, Luchetti G, Pfeffer SR. Ric1-Rgp1 complex is a guanine nucleotide exchange factor for the late Golgi Rab6A GTPase and an effector of the medial Golgi Rab33B GTPase. J Biol Chem 2012; 287:42129-37; PMID:23091056; http://dx.doi.org/ 10.1074/jbc.M112.414565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Siniossoglou S, Peak-Chew SY, Pelham HR. Ric1p and Rgp1p form a complex that catalyses nucleotide exchange on Ypt6p. EMBO J 2000; 19:4885-94; PMID:10990452; http://dx.doi.org/ 10.1093/emboj/19.18.4885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Gerondopoulos A, Langemeyer L, Liang JR, Linford A, Barr FA. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol 2012; 22:2135-9; PMID:23084991; http://dx.doi.org/ 10.1016/j.cub.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Lonhienne TG, Forwood JK, Marfori M, Robin G, Kobe B, Carroll BJ. Importin-β is a GDP-to-GTP exchange factor of Ran: implications for the mechanism of nuclear import. J Biol Chem 2009; 284:22549-58; PMID:19549784; http://dx.doi.org/ 10.1074/jbc.M109.019935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Schulze H, Dose M, Korpal M, Meyer I, Italiano JE Jr, Shivdasani RA. RanBP10 is a cytoplasmic guanine nucleotide exchange factor that modulates noncentrosomal microtubules. J Biol Chem 2008; 283:14109-19; PMID:18347012; http://dx.doi.org/ 10.1074/jbc.M709397200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.