ABSTRACT
This review discusses our current understanding of the small ubiquitin-like modifier (SUMO) pathway and how it functionally intersects with Ras signaling in cancer. The Ras family of small GTPases are frequently mutated in cancer. The role of the SUMO pathway in cancer and in Ras signaling is currently not well understood. Recent studies have shown that the SUMO pathway can both regulate Ras/MAPK pathway activity directly and support Ras-driven oncogenesis through the regulation of proteins that are not direct Ras effectors. We recently discovered that in Ras mutant cancer cells, the SUMOylation status of a subset of proteins is altered and one such protein, KAP1, is required for Ras-driven transformation. A better understanding of the functional interaction between the SUMO and Ras pathways could lead to new insights into the mechanism of Ras-driven oncogenesis.
KEYWORDS: colorectal cancer, KAP1, KRAS, non-oncogene addiction, SUMO
Ras genes are among the first oncogenes identified in human cancer and they are also among the most frequently mutated oncogenes.1 In mammals, the Ras family of small GTPases include 3 conserved members KRAS, NRAS and HRAS. Ras are membrane proteins and their activity is regulated through a GTP/GDP exchange cycle. Activation of receptor tyrosine kinases (RTKs) recruits Ras Guanine nucleotide exchange factors (GEFs) to the membrane which in turn promote Ras GTP loading. In addition to RTKs, Ras can also be activated by other signaling molecules including G protein-coupled receptors (GPCR), calcium influx and the T-cell receptor (TCR) complex.2-4 GTP binding leads to a conformational change in Ras and enables it to interact with a number of downstream effector proteins (Fig. 1). The effectors of Ras comprise of the MAP kinase (MAPK) pathway, the PI 3-kinase (PI3K) pathway, the small GTPases Rho, Rac, RalA and RalB, and the lipid enzyme phospholipase-Cε.1,5 These pathways coordinately regulate cell proliferation, survival, growth and motility. The role of Ras signaling in cancer has been recently reviewed.1,5,6 KRAS mutation occurs at a higher frequency than NRAS or HRAS, particularly in solid tumors. This might be due to different isoform-specific functions among Ras proteins as well as the ability of mutant KRAS to confer stem-like properties in cancer cells.7,8 Cancer cells that harbor Ras mutations often exhibit oncogene addiction to Ras, thus the Ras pathway represents a promising drug target in these cells.9,10 Direct inhibition of mutant Ras proteins, particularly KRAS, has proved difficult pharmacologically. However, currently there is a major effort underway to develop novel KRAS inhibitors based on new biochemical and structural insights of its function.1 Downstream of Ras, the MAPK pathway represents an attractive target as it is essential for Ras-dependent cell proliferation.11 However, inhibitors targeting the MAPK pathway, such as RAF and MEK kinase inhibitors, have not been particularly effective at shrinking Ras mutant tumors in patients.12 RAF inhibitors are unable to block MAPK signaling in Ras mutant cells due to their ability to paradoxically activate CRAF in this context.13-15 MEK inhibitors failed demonstrate substantial benefits in phase II trials among patients whose tumors were not genotyped for Ras mutation13-15 In a recent study among lung cancer patients whose tumors harbor KRAS mutation, MEK inhibitor in combination with chemotherapy led to improved progression free survival but not overall survival.16 Thus, Ras mutant cancer remains a major therapeutic challenge. In addition to directly targeting Ras and its effector kinases, synthetic lethal and co-dependency studies have been employed to explore additional genetic vulnerabilities in Ras mutant cancer cells beyond its canonical effectors.17 This approach is based on the idea that cancer cells driven by mutant Ras exhibit non-oncogene addiction to stress-response pathways for the maintenance of cell viability.18 Using this approach, we have recently identified the protein SUMO pathway as being required for Ras-driven transformation.19
Figure 1.
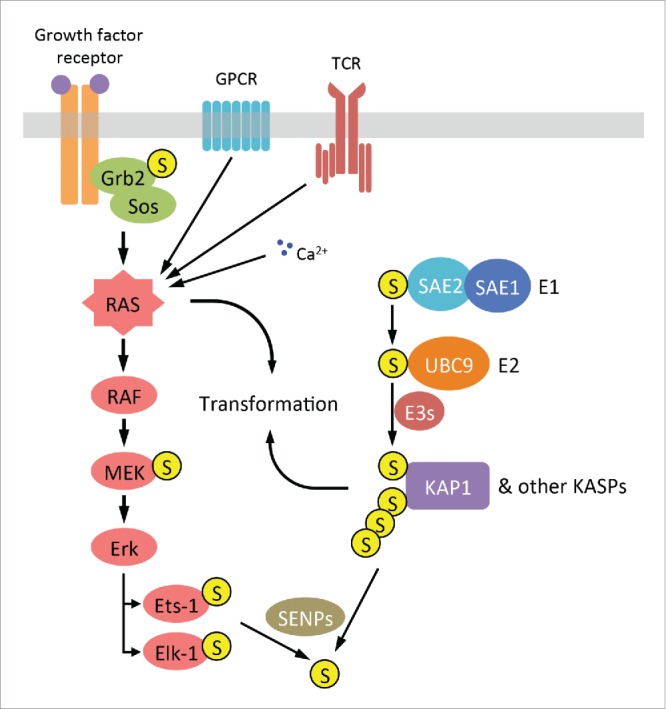
Intersection between the Ras and the SUMO pathway in cancer. Ras can be activated by multiple upstream signaling inputs including receptor tyrosine kinases (RTKs), G protein-coupled receptors (GPCRs), calcium influx (Ca2+) and the T-cell receptor (TCR) complex. Downstream of Ras, several members of MAPK pathway, including MEK kinases and the transcription factors Ets-1 and Elk-1, are SUMOylated. The SUMO pathway consists of a single E1 ligase (SAE1/SAE2 heterodimer), a single E2 ligase (Ubc9) and several E3 proteins. Through the regulation of KRAS-associated SUMOylated proteins (KASPs) such as KAP1, the SUMO pathway plays a supportive role in KRAS-driven transformation. SUMO modification is reversible by sentrin-specific proteases (SENPs).
The SUMO pathway conjugates SUMO proteins onto lysine residues of target proteins (Fig. 1). Current knowledge of this pathway's role in normal cellular function and in stress response has been reviewed recently.20-22 In mammals there are 3 SUMO proteins (SUMO1, 2 and 3). All three isoforms can form mono-conjugates, whereas SUMO2 and SUMO3 are highly homologous and can also form poly-SUMO chains. Thousands of cellular proteins have been shown as substrates for SUMOylation, thus this pathway could regulate a wide array of cellular processes. SUMOylation often occurs on lysine residues in the sequence context ΨKxE/D, where Ψ represents hydrophobic amino acids with large side chains and x represents any amino acids.20 In some instances SUMOylation can also occur on lysine residues not residing in the consensus motif.20 Analogous to protein ubiquitination, SUMOylation occurs in several distinct steps. First, the newly synthesized SUMO precursor protein is cleaved by sentrin-specific proteases (SENPs). SUMO is then activated by the E1 activating enzyme and transferred to the E2 enzyme. In the last step, E3 proteins promote the conjugation of SUMO to either a substrate protein or an existing poly-SUMO chain. SUMOylation is reversible via cleavage by SENPs. In mammalian cells there is a single E1 enzyme formed by the SAE1/SAE2 heterodimer, a single E2 enzyme Ubc9 and several E3s and SENPs. In contrast to the ubiquitin pathway where hundreds of E3s exist to serve different target proteins, the number of functionally validated SUMO E3s is small (less than 20). Thus, how substrate specificity is determined in the SUMO pathway remains an enigma.
The identification of SUMO substrates has been greatly facilitated by protein mass-spectrometry.23,24 For many proteins, SUMOylation serves to regulate their activity and/or subcellular localization rather than their stability. However, poly-SUMOylation can act as a recognition signal for SUMO-targeted ubiquitin ligases (STUBLs) that degrade poly-SUMOylated proteins.25 The majority of SUMOylation appears to occur in the nucleus and many transcription factors/co-factors, DNA binding proteins and chromatin-remodeling proteins are subject to SUMOylation.23,24 SUMOylation can either positively or negatively regulate the function of these proteins through mechanisms including changes in DNA-binding affinity, alterations in subcellular localization, induction or inhibition of protein-protein interaction, and modification of chromatin structure. SUMOylation of individual proteins can be regulated through proximity to the ligase and through phosphorylation (see below). With the exception of a few proteins including PML and RanGAP that show near stoichiometric levels of SUMOylation, most cellular proteins appear to be SUMOylated at low levels. There is evidence that the SUMOylated form of a protein could represent its active form, thus even low levels of SUMOylation could critically influence protein function. Indeed, the SUMO pathway is essential for cell viability: deletion of Ubc9 in mouse is lethal26 and cells without Ubc9 failed to complete mitosis.27
How the SUMO pathway is itself regulated is not well understood. It is known that this pathway plays a critical role in cellular stress response. For example, transient heat shock induces a global increase in SUMOylation that serves to preserve cell viability.23 In the DNA damage response, SUMOylation is required for the proper localization of DNA repair proteins to sites of DNA damage and for their subsequent activation.22 SUMOylation affects protein complex formation in 2 ways. The addition of a bulky SUMO moiety could occlude a protein-binding domain and thus disrupt protein-protein interaction. On the other hand, many proteins possess short, hydrophobic SUMO-interaction motifs (SIMs) that bind to SUMO proteins with moderate affinity and specificity.20 It has been proposed that SUMOylation can serve as a “molecular glue” to hold protein complex together through extensive SUMO-SIM interactions.28
The SUMO pathway's role in cancer is rather complex. Genes in this pathway (E1, E2, E3 and SENPs) are not significantly mutated, amplified or deleted in cancer, thus they are unlikely to be oncogenes or tumor suppressors themselves. Expression of the SUMO E2 ligase Ubc9 is up-regulated in colorectal cancer and multiple myelomas, but down-regulated in advanced breast and lung adenocarcinomas.29,30 In prostate cancer, the expression of Ubc9 appears to be stage-dependent.30 Because SUMOylation is critical for stress protection, cell cycle and DNA repair,20,22,27,31 this pathway is likely to play a supportive role in tumorigenesis. This is consistent with the notion that this pathway is required for KRAS-driven transformation and for the viability of Myc-driven cancer cells.19,32 However, since SUMOylation of both oncoproteins and tumor suppressors has been described, the role of this pathway in cancer is likely to be determined in a context-dependent manner. Among SUMO substrates, several transcription factors with known roles in cancer are repressed by SUMOylation. These include Ets-1, c-Myb, androgen receptor, and MITF1.33-36 In some cases, SUMOylation can promote transcription activation. For example, SUMOylation of the transcription co-factors TBL1 and TBLR1 release them from co-repressor complex and enable them to bind β-catenin and promote the assembly of transcription activator complex.37
A growing body of evidence indicates that the SUMO pathway functionally interacts with the Ras/MAPK pathway. Although human Ras proteins are only found to be ubiquitinated but not SUMOylated,38,39 the Drosophila Ras protein has been identified as a SUMO substrate.40 Downstream in the Ras/MAPK pathway, both MEK1 and MEK2 kinases are direct SUMO targets.41 SUMOylation of a conserved lysine residue on MEK1 and MEK2 leads to inhibition of their kinase activity. Interestingly, Ras oncoprotein, but not BRAF oncoprotein, can down regulate MEK SUMOylation due to Ras' ability to inhibit the binding between MEK and its SUMO E3 ligase MEKK1.41 Further downstream of the Ras/MAPK pathway, the activity of the Ets-family transcription factor Elk-1 is inhibited by SUMOylation.42 Within the PI3K/Akt pathway, Akt1 has been reported as a direct target of SUMOylation, and this modification enhances its kinase activity.43,44 Whether other Ras effectors are subjected to regulation by SUMO is currently unknown.
Genetic studies also support a functional interaction between the SUMO pathway and Ras signaling. In C. elegans, mutations in SUMO pathway genes can modulate RTK-mediated Ras signaling during vulva development.45 The worm Elk-1 ortholog, Lin-1, is inhibited by SUMOylation.46 In Drosophila, SUMO knockdown inhibits ERK activation downstream of wild type (WT) Ras protein but not mutant Ras protein, indicating that SUMO modulates this pathway at or above the level of Ras.40 Through an RNAi screen aimed at identify synthetic lethal partners of the KRAS oncogene in human colorectal cancer cell lines, we found that KRAS mutant cells are more sensitive to the depletion of the E1 ligase SAE1 and the E2 ligase Ubc9, particularly under anchorage-independent conditions.18,19 RNAi-mediated Ubc9 knockdown strongly inhibits the colony growth of cancer cells and KRAS-mediated transformation of immortalized normal epithelial cells. In colorectal cancer cells, inhibition of the SUMO pathway does not appear to affect MAPK signaling, thus the mechanism is likely to be indirect. Using mass-spectrometry to identify global changes in protein SUMOylation that is associated with KRAS mutation, we found that KRAS mutation does not affects global SUMOylation levels but instead alters the SUMOylation of only a small subset of proteins we termed KRAS-associated SUMOylated proteins (KASPs). We further showed that SUMOylation of one such KASP, the KRAB-associated protein 1 (KAP1/TRIM28), functionally contributed to KRAS-driven transformation. KAP1 is a transcriptional co-repressor protein with multiple functions.47 It associates with KRAB-domain zinc-finger proteins through its N-terminus,48 with HP1 through its HP1 binding domain,49-51 and with a number of chromatin-remodeling complexes including N-CoR, NuRD, and SETDB1 through its C-terminus.52-54 In addition to its transcriptional co-repressor activity, KAP1 serves several transcription-independent roles. KAP1 is a scaffold protein for DNA damage repair,55,56 a SUMO E3 ligase,57 and an ubiquitin E3 ligase.58,59 KAP1 is SUMOylated on several lysine residues near its C-terminus, and SUMO modification is required for its transcriptional co-repressor activity.57,60,61 We found that KAP1 is hyper-SUMOylated in KRAS mutant cells, particularly under anchorage-independent conditions, and the expression of a SUMO-KAP1 fusion protein could partially rescue Ubc9 depletion. Furthermore, we found that KAP1 knockdown inhibits the anchorage-independent growth of KRAS mutant cells, and this defect can be rescued by the expression of WT KAP1 but not a SUMO-deficient KAP1.19 KAP1 is not the only KASP that is required for KRAS-driven transformation since KAP1 over-expression can only partially rescue Ubc9 knockdown. It is likely that additional KASPs are also co-opted by the KRAS oncogene to support the growth and survival of cancer cells. The fact that only a small number of KASPs were identified in our mass-spectrometry analysis suggest that KRAS-dependent regulation of their SUMOylation is likely to occur through a specific, yet unknown mechanism that warrants further investigation. In addition, how KAP1 contributes to KRAS-driven transformation, and which of the many functions of KAP1 is relevant in this context remains to be elucidated.
One mechanism by which the Ras and SUMO pathways functionally interact is through the co-regulation of substrate proteins. It has been shown that the phosphorylation status of a protein can both positively and negatively influence its SUMOylation. For example, the ETS family transcription factor Elk-1 is SUMOylated under basal condition and this serves to represse its activity. Upon activation of the MAPK pathway, ERK phosphorylates Elk-1 and this inhibits Elk-1 SUMOylation and activates Elk-1.42 On the other hand, ERK-dependent phosphorylation of the nuclear body protein PML enhanced its SUMOylation in response to arsenic oxide treatment.62 These prior studies thus suggest the possibility that some of the KASPs uncovered in our study could be regulated by ERK phosphorylation in an analogous fahsion. In some cases, regulation of SUMOylation by phosphorylation occurs through a phosphorylation-dependent SUMOylation motif (PDSM) within the amino acid sequence context ΨKxExxS/T. In this scenario, SUMOylation on the lysine is directly controlled by the phosphorylation status of the adjacent serine/threonine residue.63,64 A PDSM of the sequence ΨKxExxS/TP has the potential to be regulated by ERK. Bioinformatics analyses have discovered a number of transcription factors containing this PDSMs.63,65,66 To date, however, few of these ERK-regulated PDSMs have been demonstrated experimentally. Nevertheless, this represents an attractive mechanism by which the Ras and SUMO pathways could functionally intersect. It would be interesting to determine if some of the KASPs we uncovered can be regulated through an ERK-directed PDSM.
The SUMO pathway presents a potential targeting opportunity in cancer. Significant efforts have been devoted toward targeting the ubiquitin pathway, and inhibitors of ubiquitin E1, E2, E3s and de-ubiquitnases have been developed.67 This prior experience could guide the development of SUMO pathway inhibitors. Several inhibitors of this pathway have been reported68-75 and the development of highly specific SUMO ligase inhibitors will be critical for evaluating the druggability of this pathway in cancer. Given the essentiality of this pathway, SUMO inhibitors could lead to significant on-target toxicity. However, the clinical success of proteasome inhibitors and the development of neddylation inhibitors67 suggest that inhibitors of the SUMO pathway could also hold potentials as anti-cancer agents.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. John Schneekloth for suggestions and comments. Due to space limitation we apologize for omitting the citations of some of the primary literatures.
Funding
The work is supported by an NCI Intramural Program Award to JL (ZIA BC 011303).
References
- [1].Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell 2014; 25:272-81; PMID:24651010; http://dx.doi.org/ 10.1016/j.ccr.2014.02.017 [DOI] [PubMed] [Google Scholar]
- [2].Kranenburg O, Moolenaar WH. Ras-MAP kinase signaling by lysophosphatidic acid and other G protein-coupled receptor agonists. Oncogene 2001; 20:1540-6; PMID:11313900; http://dx.doi.org/ 10.1038/sj.onc.1204187 [DOI] [PubMed] [Google Scholar]
- [3].Farnsworth CL, Freshney NW, Rosen LB, Ghosh A, Greenberg ME, Feig LA. Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature 1995; 376:524-7; PMID:7637786; http://dx.doi.org/ 10.1038/376524a0 [DOI] [PubMed] [Google Scholar]
- [4].Finco TS, Kadlecek T, Zhang W, Samelson LE, Weiss A. LAT is required for TCR-mediated activation of PLCgamma1 and the Ras pathway. Immunity 1998; 9:617-26; PMID:9846483; http://dx.doi.org/ 10.1016/S1074-7613(00)80659-7 [DOI] [PubMed] [Google Scholar]
- [5].Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 2014; 13:828-51; PMID:25323927; http://dx.doi.org/ 10.1038/nrd4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 2011; 11:761-74; PMID:21993244; http://dx.doi.org/ 10.1038/nrc3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wang MT, Holderfield M, Galeas J, Delrosario R, To MD, Balmain A, McCormick F. K-Ras Promotes Tumorigenicity through Suppression of Non-canonical Wnt Signaling. Cell 2015; 163:1237-51; PMID:26590425; http://dx.doi.org/ 10.1016/j.cell.2015.10.041 [DOI] [PubMed] [Google Scholar]
- [8].Quinlan MP, Settleman J. Isoform-specific ras functions in development and cancer. Future Oncol 2009; 5:105-16; PMID:19243303; http://dx.doi.org/ 10.2217/14796694.5.1.105 [DOI] [PubMed] [Google Scholar]
- [9].Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer, 2003:11-22; PMID:12509763; http://dx.doi.org/ 10.1038/nrc969 [DOI] [PubMed] [Google Scholar]
- [10].Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 2009; 15:489-500; PMID:19477428; http://dx.doi.org/ 10.1016/j.ccr.2009.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Drosten M, Dhawahir A, Sum EYM, Urosevic J, Lechuga CG, Esteban LM, Castellano E, Guerra C, Santos E, Barbacid M. Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J 2010:1091-104; PMID:NOT_FOUND; http://dx.doi.org/ 10.1038/emboj.2010.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Samatar AA, Poulikakos PI. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov 2014; 13:928-42; PMID:25435214; http://dx.doi.org/ 10.1038/nrd4281 [DOI] [PubMed] [Google Scholar]
- [13].Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, et al.. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 2010; 464:431-5; PMID:20130576; http://dx.doi.org/ 10.1038/nature08833 [DOI] [PubMed] [Google Scholar]
- [14].Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 2010; 464:427-30; PMID:20179705; http://dx.doi.org/ 10.1038/nature08902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, et al.. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 2010; 140:209-21; PMID:20141835; http://dx.doi.org/ 10.1016/j.cell.2009.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Janne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, Franke FA, Grinsted L, Zazulina V, Smith P, et al.. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol 2013; 14:38-47; PMID:23200175; http://dx.doi.org/ 10.1016/S1470-2045(12)70489-8 [DOI] [PubMed] [Google Scholar]
- [17].Yu B, Luo J. Synthetic lethal genetic screens in Ras mutant cancers. Enzymes 2013; 34 Pt. B:201-19; PMID:25034106; http://dx.doi.org/ 10.1016/B978-0-12-420146-0.00009-3 [DOI] [PubMed] [Google Scholar]
- [18].Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell, 2009:823-37; PMID:19269363; http://dx.doi.org/ 10.1016/j.cell.2009.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yu B, Swatkoski S, Holly A, Lee LC, Giroux V, Lee CS, Hsu D, Smith JL, Yuen G, Yue J, et al.. Oncogenesis driven by the Ras/Raf pathway requires the SUMO E2 ligase Ubc9. Proc Natl Acad Sci U S A 2015; 112:E1724-33; PMID:25805818; http://dx.doi.org/ 10.1073/pnas.1415569112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 2010; 11:861-71; PMID:21102611; http://dx.doi.org/ 10.1038/nrm3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bettermann K, Benesch M, Weis S, Haybaeck J. SUMOylation in carcinogenesis. Cancer Lett 2012; 316:113-25; PMID:22138131; http://dx.doi.org/ 10.1016/j.canlet.2011.10.036 [DOI] [PubMed] [Google Scholar]
- [22].Dou H, Huang C, Van Nguyen T, Lu LS, Yeh ET. SUMOylation and de-SUMOylation in response to DNA damage. FEBS Lett 2011; 585:2891-6; PMID:21486569; http://dx.doi.org/ 10.1016/j.febslet.2011.04.002 [DOI] [PubMed] [Google Scholar]
- [23].Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2009; 2:ra24; PMID:19471022; http://dx.doi.org/ 10.1126/scisignal.2000282 [DOI] [PubMed] [Google Scholar]
- [24].Matic I, Schimmel J, Hendriks IA, van Santen MA, van de Rijke F, van Dam H, Gnad F, Mann M, Vertegaal AC. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell 2010; 39:641-52; PMID:20797634; http://dx.doi.org/ 10.1016/j.molcel.2010.07.026 [DOI] [PubMed] [Google Scholar]
- [25].Sriramachandran AM, Dohmen RJ. SUMO-targeted ubiquitin ligases. Biochim Biophys Acta 2014; 1843:75-85; PMID:24018209; http://dx.doi.org/ 10.1016/j.bbamcr.2013.08.022 [DOI] [PubMed] [Google Scholar]
- [26].Demarque MD, Nacerddine K, Neyret-Kahn H, Andrieux A, Danenberg E, Jouvion G, Bomme P, Hamard G, Romagnolo B, Terris B, et al.. Sumoylation by Ubc9 regulates the stem cell compartment and structure and function of the intestinal epithelium in mice. Gastroenterology 2011; 140:286-96; PMID:20951138; http://dx.doi.org/ 10.1053/j.gastro.2010.10.002 [DOI] [PubMed] [Google Scholar]
- [27].Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell 2005; 9:769-79; PMID:16326389; http://dx.doi.org/ 10.1016/j.devcel.2005.10.007 [DOI] [PubMed] [Google Scholar]
- [28].Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 2012; 151:807-20; PMID:23122649; http://dx.doi.org/ 10.1016/j.cell.2012.10.021 [DOI] [PubMed] [Google Scholar]
- [29].Driscoll JJ, Pelluru D, Lefkimmiatis K, Fulciniti M, Prabhala RH, Greipp PR, Barlogie B, Tai YT, Anderson KC, Shaughnessy JD Jr., et al.. The sumoylation pathway is dysregulated in multiple myeloma and is associated with adverse patient outcome. Blood 2010; 115:2827-34; PMID:19965618; http://dx.doi.org/ 10.1182/blood-2009-03-211045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Moschos SJ, Jukic DM, Athanassiou C, Bhargava R, Dacic S, Wang X, Kuan SF, Fayewicz SL, Galambos C, Acquafondata M, et al.. Expression analysis of Ubc9, the single small ubiquitin-like modifier (SUMO) E2 conjugating enzyme, in normal and malignant tissues. Hum Pathol 2010; 41:1286-98; PMID:20561671; http://dx.doi.org/ 10.1016/j.humpath.2010.02.007 [DOI] [PubMed] [Google Scholar]
- [31].Mukhopadhyay D, Dasso M. The fate of metaphase kinetochores is weighed in the balance of SUMOylation during S phase. Cell Cycle 2010; 9:3194-201; PMID:20724819; http://dx.doi.org/ 10.4161/cc.9.16.12619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al.. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 2012; 335:348-53; PMID:22157079; http://dx.doi.org/ 10.1126/science.1212728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ji Z, Degerny C, Vintonenko N, Deheuninck J, Foveau B, Leroy C, Coll J, Tulasne D, Baert JL, Fafeur V. Regulation of the Ets-1 transcription factor by sumoylation and ubiquitinylation. Oncogene 2007; 26:395-406; PMID:16862185; http://dx.doi.org/ 10.1038/sj.onc.1209789 [DOI] [PubMed] [Google Scholar]
- [34].Sramko M, Markus J, Kabat J, Wolff L, Bies J. Stress-induced inactivation of the c-Myb transcription factor through conjugation of SUMO-2/3 proteins. J Biol Chem 2006; 281:40065-75; PMID:17077080; http://dx.doi.org/ 10.1074/jbc.M609404200 [DOI] [PubMed] [Google Scholar]
- [35].Cheng J, Wang D, Wang Z, Yeh ET. SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol Cell Biol 2004; 24:6021-8; PMID:15199155; http://dx.doi.org/ 10.1128/MCB.24.13.6021-6028.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bertolotto C, Lesueur F, Giuliano S, Strub T, de Lichy M, Bille K, Dessen P, d'Hayer B, Mohamdi H, Remenieras A, et al.. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 2011; 480:94-8; PMID:22012259; http://dx.doi.org/ 10.1038/nature10539 [DOI] [PubMed] [Google Scholar]
- [37].Choi HK, Choi KC, Yoo JY, Song M, Ko SJ, Kim CH, Ahn JH, Chun KH, Yook JI, Yoon HG. Reversible SUMOylation of TBL1-TBLR1 regulates beta-catenin-mediated Wnt signaling. Mol Cell 2011; 43:203-16; PMID:21777810; http://dx.doi.org/ 10.1016/j.molcel.2011.05.027 [DOI] [PubMed] [Google Scholar]
- [38].Sasaki AT, Carracedo A, Locasale JW, Anastasiou D, Takeuchi K, Kahoud ER, Haviv S, Asara JM, Pandolfi PP, Cantley LC. Ubiquitination of K-Ras enhances activation and facilitates binding to select downstream effectors. Sci Signal 2011; 4:ra13; PMID:21386094; http://dx.doi.org/ 10.1126/scisignal.2001518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D. Differential modification of Ras proteins by ubiquitination. Mol Cell 2006; 21:679-87; PMID:16507365; http://dx.doi.org/ 10.1016/j.molcel.2006.02.011 [DOI] [PubMed] [Google Scholar]
- [40].Nie M, Xie Y, Loo JA, Courey AJ. Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PloS One 2009; 4:e5905; PMID:19529778; http://dx.doi.org/ 10.1371/journal.pone.0005905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kubota Y, O'Grady P, Saito H, Takekawa M. Oncogenic Ras abrogates MEK SUMOylation that suppresses the ERK pathway and cell transformation. Nat Cell Biol 2011; 13:282-91; PMID:21336309; http://dx.doi.org/ 10.1038/ncb2169 [DOI] [PubMed] [Google Scholar]
- [42].Yang SH, Jaffray E, Hay RT, Sharrocks AD. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol Cell 2003; 12:63-74; PMID:12887893; http://dx.doi.org/ 10.1016/S1097-2765(03)00265-X [DOI] [PubMed] [Google Scholar]
- [43].Li R, Wei J, Jiang C, Liu D, Deng L, Zhang K, Wang P. Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res 2013; 73:5742-53; PMID:23884910; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-0538 [DOI] [PubMed] [Google Scholar]
- [44].Risso G, Pelisch F, Pozzi B, Mammi P, Blaustein M, Colman-Lerner A, Srebrow A. Modification of Akt by SUMO conjugation regulates alternative splicing and cell cycle. Cell Cycle 2013; 12:3165-74; PMID:24013425; http://dx.doi.org/ 10.4161/cc.26183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Poulin G, Dong Y, Fraser AG, Hopper NA, Ahringer J. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J 2005; 24:2613-23; PMID:15990876; http://dx.doi.org/ 10.1038/sj.emboj.7600726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Leight ER, Murphy JT, Fantz DA, Pepin D, Schneider DL, Ratliff TM, Mohammad DH, Herman MA, Kornfeld K. Conversion of the LIN-1 ETS protein of Caenorhabditis elegans from a SUMOylated transcriptional repressor to a phosphorylated transcriptional activator. Genetics 2015; 199:761-75; PMID:25567989; http://dx.doi.org/ 10.1534/genetics.114.172668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cheng CT, Kuo CY, Ann DK. KAPtain in charge of multiple missions: Emerging roles of KAP1. World J Biol Chem 2014; 5:308-20; PMID:25225599; http://dx.doi.org/ 10.4331/wjbc.v5.i3.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Peng H, Gibson LC, Capili AD, Borden KL, Osborne MJ, Harper SL, Speicher DW, Zhao K, Marmorstein R, Rock TA, et al.. The structurally disordered KRAB repression domain is incorporated into a protease resistant core upon binding to KAP-1-RBCC domain. J Mol Biol 2007; 370:269-89; PMID:17512541; http://dx.doi.org/ 10.1016/j.jmb.2007.03.047 [DOI] [PubMed] [Google Scholar]
- [49].Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J 1999; 18:6385-95; PMID:10562550; http://dx.doi.org/ 10.1093/emboj/18.22.6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ 3rd. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol 1999; 19:4366-78; PMID:10330177; http://dx.doi.org/ 10.1128/MCB.19.6.4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lechner MS, Begg GE, Speicher DW, Rauscher FJ 3rd. Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol Cell Biol 2000; 20:6449-65; PMID:10938122; http://dx.doi.org/ 10.1128/MCB.20.17.6449-6465.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem 2000; 275:40463-70; PMID:11013263; http://dx.doi.org/ 10.1074/jbc.M007864200 [DOI] [PubMed] [Google Scholar]
- [53].Schultz DC, Friedman JR, Rauscher FJ 3rd. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev 2001; 15:428-43; PMID:11230151; http://dx.doi.org/ 10.1101/gad.869501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 3rd. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 2002; 16:919-32; PMID:11959841; http://dx.doi.org/ 10.1101/gad.973302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 2006; 8:870-6; PMID:16862143; http://dx.doi.org/ 10.1038/ncb1446 [DOI] [PubMed] [Google Scholar]
- [56].White DE, Negorev D, Peng H, Ivanov AV, Maul GG, Rauscher FJ 3rd. KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res 2006; 66:11594-9; PMID:17178852; http://dx.doi.org/ 10.1158/0008-5472.CAN-06-4138 [DOI] [PubMed] [Google Scholar]
- [57].Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al.. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell 2007; 28:823-37; PMID:18082607; http://dx.doi.org/ 10.1016/j.molcel.2007.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell 2010; 39:963-74; PMID:20864041; http://dx.doi.org/ 10.1016/j.molcel.2010.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yang B, O'Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM, Gravekamp C, Setaluri V, Peters N, Hoffmann FM, et al.. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer Res 2007; 67:9954-62; PMID:17942928; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-1478 [DOI] [PubMed] [Google Scholar]
- [60].Li X, Lee YK, Jeng JC, Yen Y, Schultz DC, Shih HM, Ann DK. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem 2007; 282:36177-89; PMID:17942393; http://dx.doi.org/ 10.1074/jbc.M706912200 [DOI] [PubMed] [Google Scholar]
- [61].Lee YK, Thomas SN, Yang AJ, Ann DK. Doxorubicin down-regulates Kruppel-associated box domain-associated protein 1 sumoylation that relieves its transcription repression on p21WAF1/CIP1 in breast cancer MCF-7 cells. J Biol Chem 2007; 282:1595-606; PMID:17079232; http://dx.doi.org/ 10.1074/jbc.M606306200 [DOI] [PubMed] [Google Scholar]
- [62].Hayakawa F, Privalsky ML. Phosphorylation of PML by mitogen-activated protein kinases plays a key role in arsenic trioxide-mediated apoptosis. Cancer Cell 2004; 5:389-401; PMID:15093545; http://dx.doi.org/ 10.1016/S1535-6108(04)00082-0 [DOI] [PubMed] [Google Scholar]
- [63].Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A 2006; 103:45-50; PMID:16371476; http://dx.doi.org/ 10.1073/pnas.0503698102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ, Pirkkala L, Sistonen L. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol Cell Biol 2003; 23:2953-68; PMID:12665592; http://dx.doi.org/ 10.1128/MCB.23.8.2953-2968.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 2006; 311:1012-7; PMID:16484498; http://dx.doi.org/ 10.1126/science.1122513 [DOI] [PubMed] [Google Scholar]
- [66].Yang XJ, Gregoire S. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol Cell 2006; 23:779-86; PMID:16973431; http://dx.doi.org/ 10.1016/j.molcel.2006.08.009 [DOI] [PubMed] [Google Scholar]
- [67].Skaar JR, Pagan JK, Pagano M. SCF ubiquitin ligase-targeted therapies. Nat Rev Drug Discov 2014; 13:889-903; PMID:25394868; http://dx.doi.org/ 10.1038/nrd4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kim YS, Nagy K, Keyser S, Schneekloth JS Jr. An electrophoretic mobility shift assay identifies a mechanistically unique inhibitor of protein sumoylation. Chemistry & biology 2013; 20:604-13; PMID:23601649; http://dx.doi.org/ 10.1016/j.chembiol.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Takemoto M, Kawamura Y, Hirohama M, Yamaguchi Y, Handa H, Saitoh H, Nakao Y, Kawada M, Khalid K, Koshino H, et al.. Inhibition of protein SUMOylation by davidiin, an ellagitannin from Davidia involucrata. The Journal of antibiotics 2014; 67:335-8; PMID:24424345; http://dx.doi.org/ 10.1038/ja.2013.142 [DOI] [PubMed] [Google Scholar]
- [70].Fukuda I, Ito A, Hirai G, Nishimura S, Kawasaki H, Saitoh H, Kimura K, Sodeoka M, Yoshida M. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol 2009; 16:133-40; PMID:19246003; http://dx.doi.org/ 10.1016/j.chembiol.2009.01.009 [DOI] [PubMed] [Google Scholar]
- [71].Fukuda I, Ito A, Uramoto M, Saitoh H, Kawasaki H, Osada H, Yoshida M. Kerriamycin B inhibits protein SUMOylation. J Antibiot 2009; 62:221-4; PMID:19265871; http://dx.doi.org/ 10.1038/ja.2009.10 [DOI] [PubMed] [Google Scholar]
- [72].Brandt M, Szewczuk LM, Zhang H, Hong X, McCormick PM, Lewis TS, Graham TI, Hung ST, Harper-Jones AD, Kerrigan JJ, et al.. Development of a high-throughput screen to detect inhibitors of TRPS1 sumoylation. Assay Drug Dev Technol 2013; 11:308-25; PMID:23772552; http://dx.doi.org/ 10.1089/adt.2012.501 [DOI] [PubMed] [Google Scholar]
- [73].Hirohama M, Kumar A, Fukuda I, Matsuoka S, Igarashi Y, Saitoh H, Takagi M, Shin-ya K, Honda K, Kondoh Y, et al.. Spectomycin B1 as a novel SUMOylation inhibitor that directly binds to SUMO E2. ACS Chem Biol 2013; 8:2635-42; PMID:24143955; http://dx.doi.org/ 10.1021/cb400630z [DOI] [PubMed] [Google Scholar]
- [74].Kumar A, Ito A, Hirohama M, Yoshida M, Zhang KY. Identification of sumoylation activating enzyme 1 inhibitors by structure-based virtual screening. J Chem Inf Model 2013; 53:809-20; PMID:23544417; http://dx.doi.org/ 10.1021/ci300618e [DOI] [PubMed] [Google Scholar]
- [75].Suzawa M, Miranda DA, Ramos KA, Ang KK, Faivre EJ, Wilson CG, Caboni L, Arkin MR, Kim YS, Fletterick RJ, et al.. A gene-expression screen identifies a non-toxic sumoylation inhibitor that Mimics SUMO-less human LRH-1 in liver. eLife 2015; 4; PMID:26653140. [DOI] [PMC free article] [PubMed] [Google Scholar]


