Abstract
The small GTP-binding protein Ran is involved in the regulation of essential cellular processes in interphase but also in mitotic cells: Ran controls the nucleocytoplasmic transport of proteins and RNA, it regulates mitotic spindle formation and nuclear envelope assembly. Deregulations in Ran dependent processes were implicated in the development of severe diseases such as cancer and neurodegenerative disorders. To understand how Ran-function is regulated is therefore of highest importance. Recently, several lysine-acetylation sites in Ran were identified by quantitative mass-spectrometry, some being located in highly important regions such as the P-loop, switch I, switch II and the G5/SAK motif. We recently reported that lysine-acetylation regulates nearly all aspects of Ran-function such as RCC1 catalyzed nucleotide exchange, intrinsic nucleotide hydrolysis, its interaction with NTF2 and the formation of import- and export-complexes. As a hint for its biological importance, we identified Ran-specific lysine-deacetylases (KDACs) and -acetyltransferases (KATs). Also for other small GTPases such as Ras, Rho, Cdc42, and for many effectors and regulators thereof, lysine-acetylation sites were discovered. However, the functional impact of lysine-acetylation as a regulator of protein function has only been marginally investigated so far. We will discuss recent findings of lysine-acetylation as a novel modification to regulate Ras-protein signaling.
Keywords: GNBP, KDAC, KAT, lysine-acylation, lysine-acetylation, Ran, Ras
The Ran-Protein is Structurally and Functionally Distinct From Other Ras-Proteins: Lysine-Acetylation as a Specific Regulator of Ran-Function?
The progress in quantitative proteomics showed that lysine-acetylation is conserved from bacteria to man and enabled the identification of thousands of acetylation sites in all cellular compartments covering all essential cellular functions.1-7 Many acetylation sites were identified in Ras-related proteins as well as regulators and effectors thereof.
Ras-related proteins have a common structural motif, known as the G-domain.8 The Arf/Arl-related proteins have an N-terminal extension that can be myristoylated for membrane attachment, the Rho-proteins have an additional α-helix between β5 and α4, known as the insert region/helix.9 Ran carries an additional C-terminal helix, the C-terminal switch, followed by an acidic tail (211-DEDDDL-216). In the active state, it becomes detached from the G-domain enabling the interaction to effector proteins such as RanBP1, RanBP2 and importin β. In contrast to other Ras-proteins, upon GTP-hydrolysis, the switch I in Ran (and Arf) forms an additional highly ordered intramolecular β-sheet with the interswitch composed of β2 and β3.10-12
Many Ras-proteins are targeted to subcellular membranes by a C-terminally localized poly-basic patch (classical Rho-proteins, K-Ras4A) and/or by post-translational lipidation.13 This lipidation includes the formation of stable thioethers by prenylation (geranylgeranyl: classical Rho proteins, Rab, Rap; farnesyl: Ras, Rnd1-3, TC10, TCL, RhoD, Rif) and/or by reversible fatty acid esterification such as palmitoylation (H-Ras, N-Ras, K-Ras4B, RhoB, Cdc42 isoform1, TC10) or myristoylation (Arf/Arl).14-16 Comparing the primary sequences of Ras-related proteins shows that the C-terminus is the most divergent part. Ran contains neither a C-terminally poly-basic patch nor motifs for lipidation. Although for some Ras-proteins a nuclear localization has been reported, they are predominantly cytosolic either attached to subcellular membranes or solubilized in the cytosol by binding to solubilizing factors such as RhoGDI, RabGDI or PDEδ.17-20 In contrast, Ran was found to be more than 90% nuclear in interphase cells.21
Notably, all mammalian species have multiple Ras-related, Rho-related, Arf-like and Rab-proteins.22,23 However, they contain only a single Ran gene, encoding for a single Ran-protein, being the most abundant protein of the Ras-superfamily in the cell. Notably, the existence of a variety of import- and export-receptors is a specificity determinant for the transport of different cargoes, at least to some extent.24
In interphase cells, Ran regulates the directed exchange of proteins and RNA between nucleus and cytosol.25,26 Furthermore, Ran is an essential regulator during mitosis controlling the assembly of the nuclear envelope.26,27 Considering that the cell has just one single Ran variant a defect would have drastical consequences on cellular function.25,27 However, the cell can thereby ensure that those processes are tightly controlled since no other protein can complement Ran-function. Deregulations in Ran dependent processes were implicated in the development of severe diseases, such as cancer and neurodegenerative disorders.28-32 To understand how Ran is regulated is therefore of highest importance. One way cells can modulate and specify protein-functionalities are post-translational modifications (PTMs). However, while for nearly all Ras-related proteins, PTMs such as phosphorylation, lipidation, and ubiquitylation are reported, none of these regulatory mechanisms were functionally described for Ran so far. However, recent progress in quantitative mass-spectrometry revealed that Ran is extensively targeted by lysine-acetylation. Lysine-acetylation is tightly connected to metabolism in that acetyl-CoA serves as the acetyl-donor molecule for lysine-acetyltransferases (KATs) and NAD+ is an obligatory co-factor of sirtuin (Sirt; silent information regulator)-deacetylases.
However, until recently, it was not clear how lysine-acetylation of Ran affects its function or how it is regulated.
Ran-Function is Controlled by Lysine-Acetylation
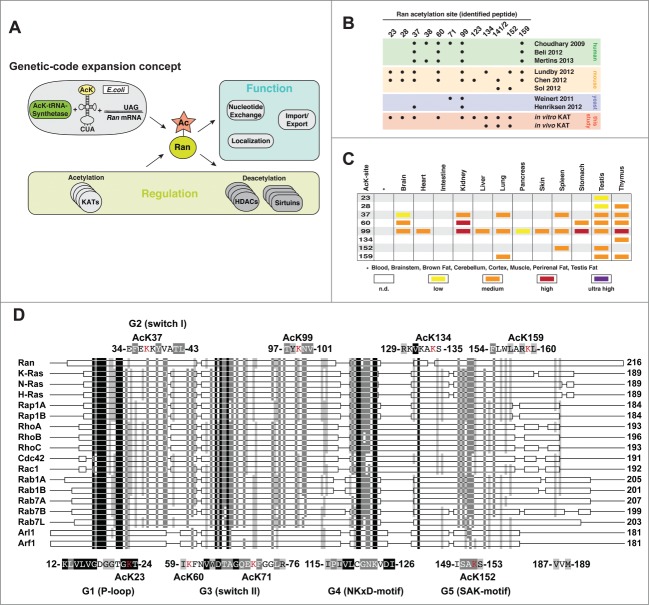
Several mass-spectrometry based screens enabled the identification of a total of 11 lysine-acetylation sites in Ran.5,33 Some of those sites are located in functionally highly important regions such as the P-loop, switch I and switch II as well as the G5/SAK-motif needed for nucleotide binding (K23R, K37R, K71R, K152R; superscript R: Ran) (Fig. 1). We used a synthetic biological approach to site-specifically introduce acetyl-L-lysine at the 5 sites in the Ran-protein originally identified by Choudhary and co-workers in 2009 (K37, K60, K71, K99, K152).34 Using a synthetically evolved acetyl-L-lysyl-tRNA-synthetase/tRNACUA-pair from Methanosarcina barkeri we functionally investigated how lysine-acetylation controls Ran-function (Fig. 1A). We discovered that the acetylations at K37R, K99R and K159R increase the affinity toward import- and export-receptors supporting import- cargo release and export-complex formation in the nucleus. Ran AcK99 has a loss-of-function phenotype lowering RCC1 affinity and the RCC1 catalyzed nucleotide exchange. Furthermore, Ran K99R shows a more cytosolic distribution in HeLa cells by an NTF2 independent mechanism as AcK99R and K99RR both showed no influence on NTF2 binding.34 The acetylation of K71R has a dominant-negative effect by increasing the affinity toward RCC1 and lowering the the RCC1 catalyzed nucleotide exchange activity. Furthermore, it abolishes NTF2 binding leading to a cytosolic accumulation of the Ran•GDP-protein, which, as a consequence, would result in a block of nucleocytoplasmic transport. Mutation of Lys to Arg is often used to preserve a non-acetylatable state in cells to phenotypically characterize an acetylation event in comparison to a Lys to Gln mutation mimicking the acetylated state. We observed a nearly complete cytosolic Ran localization expressing K71QR, mimicking the AcK71R. Notably, K71RR showed a markedly reduced NTF2-binding compared to RanWT. These results show, that these mutational approaches are sometimes misleading as Gln is not a perfect molecular mimic for an acetylated-lysine and Arg sometimes mimics an acetyl-lysine even better if exerting steric rather than electrostatic effects.34,35 Using the genetic-code expansion concept allows to study the real impact of lysine-acetylation on protein-function.
Figure 1.
Ran is regulated by lysine-acetylation. (A) We used the genetic-code expansion concept to site-specifically introduce acetyl-L-lysine into Ran at diverse positions (K37, K60, K71, K99, K134, K159) using a synthetically evolved acetyl-lysyl-tRNA-synthetase/tRNACUA pair from Methanosarcina barkeri. We found that the Ran acetylation is regulated by KATs and KDACs and that Ran-function is strongly affected by lysine-acetylation. (B) Lysine-acetylation sites identified in SILAC-based proteomic screens performed in diverse human cells and mouse/rat tissues. These sites were compared to the sites we identified in in vitro and in vivo KAT assays. (C) Ran shows specific distribution of lysine-acetylation in different Rat tissues as shown by Lundby et al.. Some acetylation sites in Ran were found in nearly all tissues such as AcK99, whereas others are specifically acetylated only in some tissues as AcK23, 28, 134 and AcK159 suggesting a tissue specific regulation.5 (D) Lysine acetylation in Ran occurs in or nearby functionally highly important regions such as the P-loop (K23), switch I (K37), switch II (K71) and the SAK-motif (K152). The sequence alignment with Ras-representatives shows that many Ran acetylation sites are not conserved (except from P-loop K23R and SAK K152R).
Regulation of Ran Lysine-Acetylation and Therapeutic Implications of Ran-Acetylation to Design Sirtuin Inhibitors
Lysine-acetylation is tightly connected to the cellular metabolic and energetic state. The writers, lysine acetyltransferases (KATs), using acetyl-CoA as acetyl donor molecule and the erasers, sirtuin deacetylases, using NAD+ as essential cofactors for catalysis, directly connect lysine-acetylation to the cellular metabolism. We identified the KATs CBP, p300, Tip60 and αTAT as potential Ran-specific acetyl-transferases in in vitro and in vivo assays. Importantly, in contrast to published proteomic screens, we identified K37R, K134R, K142R and K152R as being lysine-acetylated using this combined in vitro and in vivo approach. This does of course not exclude that under specific cellular conditions also other Ran-lysines are acetylated in an enzymatic fashion or that Ran is acetylated by transferases not tested in our study. However, another possibility is that other acetylation sites occur non-enzymatically or at very low stoichiometries. In an in vitro screen for Ran lysine-deacetylases, we found that only 2 sites are deacetylated (AcK37 by Sirt1,-2- and -3 and AcK71 only by Sirt2). Again, this does not exclude the existence of other decetylases catalyzing the deacetylation of acetylated Ran also on the other sites. It is known that KATs and KDACs are often active in multi-protein complexes, that their activities are regulated by post-translational modifications and even by binding to regulators such as fatty acids.36 Notably, we observed that the deacetylation at Ran AcK71 is faster in its active state, suggesting that structural features are important for the deacetylation by Sirt2.34 Based on these results, we are investigating the potency of Ran AcK71-derived peptides as Sirt2 inhibitors, which we hope can ultimately be further developed into therapeutics for the treatment of neurodegenerative diseases.37 In fact, peptide-based molecules are among the most potent and specific Sirt2 inhibitors identified so far.38
Physiological Relevance of Ran-Lysine-Acetylation
Although our data demonstrate the strong impact lysine-acetylation has on Ran protein-function, the question remains under which conditions it is physiologically most significant. Lysine-acetylation might either occur enzymatically by KATs or, under certain conditions, non-enzymatically.39 Although it was shown that, in contrast to the mitochondrial matrix, the nucleus and in the cytosol do not allow for non-enzymatic acetylation to occur to a large extent, high acetyl-CoA levels or events increasing the nucleophilicity at the lysine's-epsilon amino group could favor non-enzymatic lysine-acetylation.40 The latter includes factors that lower the lysine's pKA value such as a more basic pH or the presence of basic residues in vicinity.40 A lysine-acetylation site can only be of biological significance if it accumulates to sufficiently high stoichiometries unless it creates a gain-of-function for which a subpopulation of protein might be enough to mediate an effect. Alternatively, acetylation could also have additive effects if it occurs substoichiometrically in a pathway of multiple consecutive steps or if it influences the cooperativity of multi-subunit proteins. For Ran, the only acetylation site that could substoichiometrically exert a biological function is Ran AcK71 since it has a dominant-negative effect on RCC1 catalyzed nucleotide exchange.34 For the other effects we observed (of Ran AcK71 on nuclear localization of Ran•GDP or of AcK37, 99 and 159 on import and export complex formation) the acetylation needs to reach high stoichiometries to fullfil a biological role. Some Ran-acetylation sites such as AcK71 and AcK99 are downregulating Ran-function, whereas others such as AcK37,99,159 upregulate Ran-function in processes such as import cargo release in the nucleus and export-complex formation. Importantly, several additional lysine-acetylation sites were found in proteomic screens. Acetylation of K23R (K16 in Ras) in the G1/P-loop and K152R (K147 in Ras) in the G5/SAK-motif most likely interferes with nucleotide binding. K134R in Ran makes interactions to the export receptor Crm1 and to the nucleotide release factor Mog1.34 For the basic-patch encompassing K141R and K142R, it was shown by a mutational approach that it affects Crm1, importin β and RanBP1 binding.41 How lysine-acetylation at these sites interferes with those interactions and how they are regulated by KATs and KDACs needs further investigation.34 Except from K23R (K16 in Ras) in the P-loop and K152R (K147 in Ras) in the G5/SAK motif, all the other acetylation sites show a weak conservation between members of the Ras-superfamily, albeit being located in functionally highly important regions. This suggests that this global regulation of Ran-function by lysine-acetylation is Ran-specific (Fig. 1D; Table 1). Interestingly, for many sites where a lysine is found in Ran there are negatively charged residues Glu or Asp at the homologous positions in other Ras-proteins, maybe indicating that the charge is of functional importance (Table 1). The exact physiological conditions under which Ran-acetylation is functionally important might very much depend on the cellular metabolic state. Notably, lysine-acetylation might play important roles in precisely defined physiological or cell cycle states not only in interphase but also in mitotic cells. For Ran-function lysine-acetylation is an important regulator.
Table 1.
Comparison of the amino-acids present at analogous positions in selected Ras-proteins representing the 5 major Ras-subfamilies to Ran lysine-residues identified as being lysine-acetylated. In cases where no residue is shown, at those positions there is the same residue as shown above
| Residues in selected Ras-proteins at acetylaed lysines in Ran |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GNBP | 23 | 37 | 38 | 60 | 71 | 99 | 134 | 141 | 142 | 152 | 159 |
| Ran | K | K | K | K | K | K | K | K | K | K | K |
| K-Ras | D | E | L | E | E | Y | G | D | |||
| N-Ras | A | ||||||||||
| H-Ras | E | R | |||||||||
| Rap1A | E | K | M | Q | N | E | W | C | E | ||
| Rap1B | N | ||||||||||
| RhoA | V | E | D | E | I | G | |||||
| RhoB | D | Q | |||||||||
| RhoC | E | S | |||||||||
| Cdc42 | S | E | T | L | K | L | N | ||||
| Rac1 | G | N | P | I | G | L | T | ||||
| Rab1A | E | S | K | R | N | Y | S | L | K | Q | |
| Rab1B | A | N | |||||||||
| Rab7A | N | Q | T | K | K | K | N | ||||
| Rab7B | E | E | K | E | E | D | |||||
| Rab7L | K | H | R | S | D | N | G | E | |||
| Arl1 | T | T | K | S | G | S | R | K | T | ||
| Arf1 | S | K | N | K | N | ||||||
Lysine-Acetylation as a Regulator in Ras-Signaling
Although many acetylation sites were identified in Ras-proteins, effectors and regulators thereof, by quantitative proteomics, the functional consequences of these modifications are mostly unknown. Comparing acetylation sites in Ras-proteins identified in human cells, in mice and rat tissues reveals some overlap but also differences (Table 2). In total, the Ran-acetylation sites identified in tissues resemble more the range of sites we identified in the in vitro KAT assays (Fig. 1B, upper panel). Therefore, results derived from tissue samples compared to cultured cells might better reflect the physiological situation (Fig. 1B, Table 2). Notably, Ran-acetylation can be tissue-specific for some sites, whereas others show a broader tissue distribution (Fig. 1B, lower panel). Interestingly, other Ras-proteins were not found to be lysine-acetylated to the same extent as Ran. Even the totally conserved K23R (K16 in Ras, Cdc42, Rac1; K18 in RhoA/B/C) in the P-loop has not been identified as being lysine-acetylated in any other Ras-protein so far (Fig. 1C). This could be due to the subcellular localization of the Ras-proteins. Ran as the only Ras-protein being predominantly nuclear is always in close spatial proximity to KATs, which are, like Ran, often tightly connected to chromatin. However, also for other Ras-proteins and regulators a regulatory function of lysine-acetylation has been reported. A recent study shows that K104-acetylation on K-Ras 4B G12V impairs its oncogenicity, reducing the SOS catalyzed nucleotide exchange. However, those studies were only performed with the Gln and Arg mutants, used as a mimetic for lysine-acetylation or to conserve the non-acetylated state, respectively. This Ras acetylation was shown to be counterbalanced by the cytosolic deacetylases Sirt2 and HDAC6.42,43 If this occurs directly or indirectly is not known. Future studies will show if acetylation has the same impact on SOS-catalyzed nucleotide exchange on K-Ras 4B and if K-Ras 4B is a direct Sirt2/HDAC6 target. For the Rho-nucleotide exchange factor Net1A, a recent study shows that the treatment of HeLa cells with deacetylase inhibitors results in cytosolic accumulation of Net1A. Mechanistic studies showed that this is mediated by lysine-acetylation within Net1A NLS-sequences. Also for Cdc42 and RhoA acetylation sites have been found in the insert-helix. In RhoGDIα, lysine-acetylation sites were found in its immunoglobulin domain as well as its N-terminal domain suggesting to interfere with the Rho-protein's GDP/GTP and/or membrane/cytosol cycle. In the Rho-effector protein mDia1, lysine-acetylation sites have been identified in functionally highly important regions such as the Formin-homology 2 domain, essential for its actin nucleation capability. To get a deeper molecular understanding of how signaling of Ras-related proteins is affected by lysine-acetylation, the genetic-code expansion concept in combination with cell-biological studies is a powerful approach to unravel functional consequences of this post-translational modification. Recently, it was shown that KATs and KDACs can not only catalyze the transfer and the removal of acetyl-groups to/from lysine-side chains but can also transfer and remove longer acyl-groups such as butyryl-, propionyl-, crotonyl-, succinyl- myristoyl and even palmitoyl-groups to/from lysine-side chains.44,45 For Ran, it was shown that K37 and K99 (the sites for which we observed an effect on import- and export-complex formation upon acetylation) are also targeted by succinylation.46 Future investigations are needed to show to which extent acylations occur on lysines, if they occur simultaneously on distinct subpools and how the acylations, particularly of negatively charged acylations such as succinylation, exert mechanistically different functions. Particularly, poly-basic regions of Ras-proteins, due to their increased reactivity, might likely be targeted by these post-translational modifications.
Table 2.
Lysine-residues in Ras-proteins representing the 5 major Ras-subfamilies identified to be lysine-acetylated by quantitative proteomics as found in PhosphoSitePlus.49 The sites in human were found in A549 (pulmonary), HeLa (cervical), Jurkat (T lymphocyte), K562 (erythroid) and/or MV4-11 (macrophage) cells.33,46,50 The sites in mouse were identified in liver tissue and in the ones in rat were found in 11 different tissues (brain, heart, kidney, liver, lung, pancreas, skin, spleen, stomach, testis, thymus)5
| Lysine-acetylation sites found by MS |
|||
|---|---|---|---|
| GNBP | Human | Mouse | Rat |
| Ran | K37,60,71,99, 152,159 | K60,71,159 | K23,28,37,60,99, 134,152,159 |
| K-Ras | K104 | — | — |
| N-Ras | — | — | — |
| H-Ras | — | — | — |
| Rap1A | — | — | K151 |
| Rap1B | — | — | K151 |
| RhoA | — | — | K133,135 |
| RhoB | — | — | — |
| RhoC | — | — | — |
| Cdc42 | K135,144,153 | — | K128,133,135, 144,150,153,166 |
| Rac1 | — | K132, K133 | K147,153 |
| Rab1A | K61 | — | K61,132,140 |
| Rab1B | K58 | — | — |
| Rab7A | — | — | K32 |
| Rab7B | — | — | — |
| Rab7L | — | — | — |
| Arl1 | — | — | K152 |
| Arf1 | K36,142 | K142 | K36,142 |
Conclusions and Future Directions
Lysine-acetylation is tightly connected to the cellular metabolism. For Ran, this represents a regulatory system to adapt processes such as nucleocytoplasmic protein transport or the mitotic spindle formation to the cellular energetic state. Future studies will show to what extent signaling of Ras-related proteins is regulated by lysine-acylation. One of the major challenges in the acetylation/acylation research field is to distinguish between biologically relevant and low-stoichiometric biologically irrelevant sites. Most proteomics studies performed use SILAC-based approaches to quantify lysine-acetylation in a relative rather than an absolute manner, the latter being essential to judge the stoichiometry of a specific lysine-acetylation site at defined cellular conditions. Although there is some progress in quantitative high-resolution mass-spectrometry to determine absolute quantities, these methods need further technical improvement.2,47 Moreover, to show which of the sites are enzymatically acetylated, future studies should address which KATs have a robust lysine-acetyltransferase activity. Due to its low sequence homology, the identification of KATs is challenging. A recent report suggested that, apart from the classical KATs, there are additional orphan-KATs with putative lysine-acetyltransferase activity.48 Future studies are needed to show how KATs are regulated, how they are subcellularly localized and what their substrates are. As a summary, lysine-acetylation can represent a dynamic and powerful post-translational modification to control protein-function far beyond just neutralizing a positive-charge. The identification of differently charged or uncharged acylations of different acyl-chain lengths adds another level of complexity on this post-translational modification and it will be interesting to see how these acylations exert mechanistically different regulatory functions. Tackling the lysine-acetylation machinery might allow the development of novel therapeutic strategies in Ras-signaling.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics 2009; 8:215-25; http://dx.doi.org/ 10.1074/mcp.M800187-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinert BT, Iesmantavicius V, Moustafa T, Schölz C, Wagner SA, Magnes C, Zechner R, Choudhary C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol 2014; 10:716; PMID:24489116; http://dx.doi.org/ 10.1002/msb.134766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al.. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 2006; 23:607-18; PMID:16916647; http://dx.doi.org/ 10.1016/j.molcel.2006.06.026 [DOI] [PubMed] [Google Scholar]
- 4.Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell 2012; 46:212-25; PMID:22424773; http://dx.doi.org/ 10.1016/j.molcel.2012.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lundby A, Lage K, Weinert BT, Bekker-Jensen DB, Secher A, Skovgaard T, Kelstrup CD, Dmytriyev A, Choudhary C, Lundby C, et al.. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep 2012; 2:419-31; PMID:22902405; http://dx.doi.org/ 10.1016/j.celrep.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, Carr SA. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods 2013; 10, 634-7; http://dx.doi.org/ 10.1038/nmeth.2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sol EM, Wagner SA, Weinert BT, Kumar A, Kim HS, Deng CX, Choudhary C. Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3. PLoS One 2012; 7:e50545; PMID:23236377; http://dx.doi.org/ 10.1371/journal.pone.0050545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science 2001; 294:1299-304; PMID:11701921; http://dx.doi.org/ 10.1126/science.1062023 [DOI] [PubMed] [Google Scholar]
- 9.Randazzo PA, Terui T, Sturch S, Fales HM, Ferrige AG, Kahn RA. The myristoylated amino terminus of ADP-ribosylation factor 1 is a phospholipid- and GTP-sensitive switch. J Biol Chem 1995; 270:14809-15; PMID:7782347; http://dx.doi.org/ 10.1074/jbc.270.24.14809 [DOI] [PubMed] [Google Scholar]
- 10.Monecke T, Güttler T, Neumann P, Dickmanns A, Görlich D, Ficner R. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science 2009; 324:1087-91; PMID:19389996; http://dx.doi.org/ 10.1126/science.1173388 [DOI] [PubMed] [Google Scholar]
- 11.Scheffzek K, Klebe C, Fritz-Wolf K, Kabsch W, Wittinghofer A. Crystal structure of the nuclear Ras-related protein Ran in its GDP-bound form. Nature 1995; 374:378-81; PMID:7885480; http://dx.doi.org/ 10.1038/374378a0 [DOI] [PubMed] [Google Scholar]
- 12.Seewald MJ, Kraemer A, Farkasovsky M, Körner C, Wittinghofer A, Vetter IR. Biochemical characterization of the Ran-RanBP1-RanGAP system: are RanBP proteins and the acidic tail of RanGAP required for the Ran-RanGAP GTPase reaction? Mol Cell Biol 2003; 23:8124-36; PMID:14585972; http://dx.doi.org/ 10.1128/MCB.23.22.8124-8136.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cadwallader KA, Paterson H, Macdonald SG, Hancock JF. N-terminally myristoylated Ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol Cell Biol 1994; 14:4722-30; PMID:8007974; http://dx.doi.org/ 10.1128/MCB.14.7.4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson JH, Cochrane CG, Bourne JR, Solski PA, Buss JE, Der CJ. Farnesol modification of Kirsten-ras exon 4B protein is essential for transformation. Proc Natl Acad Sci U S A 1990; 87:3042-6; PMID:2183224; http://dx.doi.org/ 10.1073/pnas.87.8.3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenberg S, Laude AJ, Beckett AJ, Mageean CJ, Aran V, Hernandez-Valladares M, Henis YI, Prior IA. The role of palmitoylation in regulating Ras localization and function. Biochem Soc Trans 2013; 41:79-83; PMID:23356262; http://dx.doi.org/ 10.1042/BST20120268 [DOI] [PubMed] [Google Scholar]
- 16.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 2005; 307:1746-52; PMID:15705808; http://dx.doi.org/ 10.1126/science.1105654 [DOI] [PubMed] [Google Scholar]
- 17.Dubash AD, Guilluy C, Srougi MC, Boulter E, Burridge K, García-Mata R. The small GTPase RhoA localizes to the nucleus and is activated by Net1 and DNA damage signals. PLoS One 2011; 6:e17380; PMID:21390328; http://dx.doi.org/ 10.1371/journal.pone.0017380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longenecker K, Read P, Derewenda U, Dauter Z, Liu X, Garrard S, Walker L, Somlyo AV, Nakamoto RK, Somlyo AP, et al.. How RhoGDI binds Rho. Acta Crystallogr D Biol Crystallogr 1999; 55:1503-15; PMID:10489445; http://dx.doi.org/ 10.1107/S090744499900801X [DOI] [PubMed] [Google Scholar]
- 19.Ismail SA, Chen YX, Rusinova A, Chandra A, Bierbaum M, Gremer L, Triola G, Waldmann H, Bastiaens PI, Wittinghofer A. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat Chem Biol 2011; 7:942-9; PMID:22002721; http://dx.doi.org/ 10.1038/nchembio.686 [DOI] [PubMed] [Google Scholar]
- 20.Goody RS, Rak A, Alexandrov K. The structural and mechanistic basis for recycling of Rab proteins between membrane compartments. Cell Mol Life Sci 2005; 62:1657-70; PMID:15924270; http://dx.doi.org/ 10.1007/s00018-005-4486-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plafker K, Macara IG. Facilitated nucleocytoplasmic shuttling of the Ran binding protein RanBP1. Mol Cell Biol 2000; 20:3510-21; PMID:10779340; http://dx.doi.org/ 10.1128/MCB.20.10.3510-3521.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rojas AM, Fuentes G, Rausell A, Valencia A. The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J Cell Biol 2012; 196:189-201; PMID:22270915; http://dx.doi.org/ 10.1083/jcb.201103008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leipe DD, Koonin EV, Aravind L. Evolution and classification of P-loop kinases and related proteins. J Mol Biol 2003; 333:781-815; PMID:14568537; http://dx.doi.org/ 10.1016/j.jmb.2003.08.040 [DOI] [PubMed] [Google Scholar]
- 24.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 2005; 6:187-98; PMID:15702987; http://dx.doi.org/ 10.1111/j.1600-0854.2005.00270.x [DOI] [PubMed] [Google Scholar]
- 25.Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev 2001; 65:570-94, table of contents; PMID:11729264; http://dx.doi.org/ 10.1128/MMBR.65.4.570-594.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol 2008; 9:464-77; PMID:18478030; http://dx.doi.org/ 10.1038/nrm2410 [DOI] [PubMed] [Google Scholar]
- 27.Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J Cell Sci 2008; 121:1577-86; PMID:18469014; http://dx.doi.org/ 10.1242/jcs.005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mastroeni D, Chouliaras L, Grover A, Liang WS, Hauns K, Rogers J, Coleman PD. Reduced RAN expression and disrupted transport between cytoplasm and nucleus; a key event in Alzheimer's disease pathophysiology. PLoS One 2013; 8:e53349; PMID:23308199; http://dx.doi.org/ 10.1371/journal.pone.0053349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng L, Lu Y, Zhao X, Sun Y, Shi Y, Fan H, Liu C, Zhou J, Nie Y, Wu K, et al.. Ran GTPase protein promotes human pancreatic cancer proliferation by deregulating the expression of Survivin and cell cycle proteins. Biochem Biophys Res Commun 2013; 440:322-9; PMID:24076388; http://dx.doi.org/ 10.1016/j.bbrc.2013.09.079 [DOI] [PubMed] [Google Scholar]
- 30.Fan H, Lu Y, Qin H, Zhou Y, Gu Y, Zhou J, Wang X, Fan D. High Ran level is correlated with poor prognosis in patients with colorectal cancer. Int J Clin Oncol 2013; 18:856-63; PMID:22956174; http://dx.doi.org/ 10.1007/s10147-012-0465-x [DOI] [PubMed] [Google Scholar]
- 31.Matchett KB, McFarlane S, Hamilton SE, Eltuhamy YS, Davidson MA, Murray JT, Faheem AM, El-Tanani M. Ran GTPase in nuclear envelope formation and cancer Metastasis. Adv Exp Med Biol 2014; 773:323-51; PMID:24563355; http://dx.doi.org/ 10.1007/978-1-4899-8032-8_15 [DOI] [PubMed] [Google Scholar]
- 32.Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al.. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 2015; 525:56-61; PMID:26308891; http://dx.doi.org/ 10.1038/nature14973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 2009; 325:834-40; PMID:19608861; http://dx.doi.org/ 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- 34.De Boor S Knyphausen P, Kuhlmann N, Wroblowski S, Brenig J, Scislowski L, Baldus L, Nolte H, Krüger M, Lammers M. Small GTP-binding protein Ran is regulated by posttranslational lysine acetylation. Proc Natl Acad Sci U S A 2015; 112:E3679-3688; PMID:26124124; http://dx.doi.org/ 10.1073/pnas.1505995112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lammers M, Neumann H, Chin JW, James LC. Acetylation regulates cyclophilin A catalysis, immunosuppression and HIV isomerization. Nat Chem Biol 2010; 6:331-7; PMID:20364129; http://dx.doi.org/ 10.1038/nchembio.342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 2003; 370:737-49; PMID:12429021; http://dx.doi.org/ 10.1042/bj20021321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Outeiro TF, Kontopoulos E, Altmann SM, Kufareva I, Strathearn KE, Amore AM, Volk CB, Maxwell MM, Rochet JC, McLean PJ, et al.. Sirtuin 2 inhibitors rescue α-synuclein-mediated toxicity in models of Parkinson's disease. Science 2007; 317:516-9; PMID:17588900; http://dx.doi.org/ 10.1126/science.1143780 [DOI] [PubMed] [Google Scholar]
- 38.Yamagata K, Goto Y, Nishimasu H, Morimoto J, Ishitani R, Dohmae N, Takeda N, Nagai R, Komuro I, Suga H, et al.. Structural basis for potent inhibition of SIRT2 deacetylase by a macrocyclic peptide inducing dynamic structural change. Structure 2014; 22:345-52; PMID:24389023; http://dx.doi.org/ 10.1016/j.str.2013.12.001 [DOI] [PubMed] [Google Scholar]
- 39.Baeza J, Smallegan MJ, Denu JM. Site-specific reactivity of nonenzymatic lysine acetylation. ACS Chem Biol 2015; 10:122-8; PMID:25555129; http://dx.doi.org/ 10.1021/cb500848p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner GR, Payne RM. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem 2013; 288:29036-45; PMID:23946487; http://dx.doi.org/ 10.1074/jbc.M113.486753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nilsson J, Askjaer P, Kjems J. A role for the basic patch and the C terminus of RanGTP in regulating the dynamic interactions with importin β, CRM1 and RanBP1. J Mol Biol 2001; 305:231-43; PMID:11124902; http://dx.doi.org/ 10.1006/jmbi.2000.4313 [DOI] [PubMed] [Google Scholar]
- 42.Yang MH. Laurent G, Bause AS, Spang R, German N, Haigis MC, Haigis KM. HDAC6 and SIRT2 regulate the acetylation state and oncogenic activity of mutant K-RAS. Mol Cancer Res 2013; 11:1072-7; PMID:23723075; http://dx.doi.org/ 10.1158/1541-7786.MCR-13-0040-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang MH, Nickerson S, Kim ET, Liot C, Laurent G, Spang R, Philips MR, Shan Y, Shaw DE, Bar-Sagi D, et al.. Regulation of RAS oncogenicity by acetylation. Proc Natl Acad Sci USA 2012; 109:10843-8; PMID:22711838; http://dx.doi.org/ 10.1073/pnas.1201487109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, et al.. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell 2013; 50:919-30; PMID:23806337; http://dx.doi.org/ 10.1016/j.mo-lcel.2013.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leemhuis H, Packman LC, Nightingale KP, Hollfelder F. The human histone acetyltransferase P/CAF is a promiscuous histone propionyltransferase. Chembiochem 2008; 9:499-503; PMID:18247445; http://dx.doi.org/ 10.1002/cbic.200700556 [DOI] [PubMed] [Google Scholar]
- 46.Weinert BT, Schölz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, Choudhary C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep 2013; 4:842-51; PMID:23954790; http://dx.doi.org/ 10.1016/j.celrep.2013.07.024 [DOI] [PubMed] [Google Scholar]
- 47.Baeza J, Dowell JA, Smallegan MJ, Fan J, Amador-Noguez D, Khan Z, Denu JM. Stoichiometry of site-specific lysine acetylation in an entire proteome. J Biol Chem 2014; 289:21326-38; PMID:24917678; http://dx.doi.org/ 10.1074/jbc.M114.581843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montgomery DC, Sorum AW, Meier JL. Chemoproteomic profiling of lysine acetyltransferases highlights an expanded landscape of catalytic acetylation. J Am Chem Soc 2014; 136:8669-76; PMID:24836640; http://dx.doi.org/ 10.1021/ja502372j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 2004; 4:1551-61; PMID:15174125; http://dx.doi.org/ 10.1002/pmic.200300772 [DOI] [PubMed] [Google Scholar]
- 50.9920, C. C. S. Biosample/Treatment: cell line, K-562/untreated; Disease: chronic myelogenous leukemia; SILAC: -; Specificity of Antibody Used to Purify Peptides prior to MS2: anti-AcK Curated Info 2010 [Google Scholar]