ABSTRACT
The Rho-associated coiled-coil containing kinases (ROCK) were first identified as effectors of the small GTPase RhoA, hence their nomenclature. Since their discovery, two decades ago, scientists have sought to unravel the structure, regulation, and function of these essential kinases. During that time, a consensus model has formed, in which ROCK activity is regulated via both Rho-dependent and independent mechanisms. However, recent findings have raised significant questions regarding this model. In their recent publication in Nature Communications, Truebestein and colleagues present the structure of a full-length Rho kinase for the first time. In contrast to previous reports, the authors could find no evidence for autoinhibition, RhoA binding, or regulation of kinase activity by phosphorylation. Instead, they propose that ROCK functions as a molecular ruler, in which the central coiled-coil bridges the membrane-binding regulatory domains to the kinase domains at a fixed distance from the plasma membrane. Here, we explore the consequences of the new findings, re-examine old data in the context of this model, and emphasize outstanding questions in the field.
KEYWORDS: coiled-coil, cytoskeleton, GTPase, kinase, membrane anchor, molecular ruler, RhoA, ROCK, stress fibers
Introduction
There are two mammalian Rho kinases, ROCK1 and ROCK2, first identified in screens for interacting proteins of the small GTPase, RhoA.1-3 Genetic studies have shown both genes to be essential; knockouts of ROCK1 or ROCK2 are often either embryonic or post-embryonic lethal,4-6 while a recent study revealed the requirement for at least ROCK1 or ROCK2 in permitting cell division and tumor progression in mice.7 In cells, over-expression of kinase-dead ROCK1 or ROCK2 leads to cytoskeletal defects including stress fiber loss,8,9 while application of the ROCK inhibitor Y-27632 or depletion of ROCK1 or ROCK2 leads to a reduction in regulatory myosin light chain (RMLC) phosphorylation.10,11
ROCK1 and ROCK2 comprise N-terminal AGC kinase domains, a central coiled-coil of approximately 700 amino acids, and a C-terminal split-PH domain, into which is spliced a zinc-finger cysteine-rich domain (CRD), also referred to as a C1 domain (Fig. 1). Structural analyses of the kinase domain, and those of the closely related myotonic dystrophy-related Cdc42-binding kinases (MRCK) and dystrophia myotonica protein kinase (DMPK), have revealed that the kinase domain forms a head-to-head dimer mediated by the N-terminal 70 amino acid capped helix bundle (CHB) domain in conjunction with the C-terminal tail of the kinase.12-15 The activation loop, often the site of a regulatory phosphorylation event in eukaryotic protein kinases, exhibits an ordered conformation in the absence of phosphorylation; robust in vitro activity suggests that these kinases may function independently of activation loop phosphorylation.12-15 Biochemical analyses have confirmed that the C-terminal tail and the CHB are both essential for activity.8,12,13 Structural analysis of the regulatory domains reveals them to be classical PH and C1 domains, together forming a unit that binds preferentially to membranes containing acidic phospholipids.16
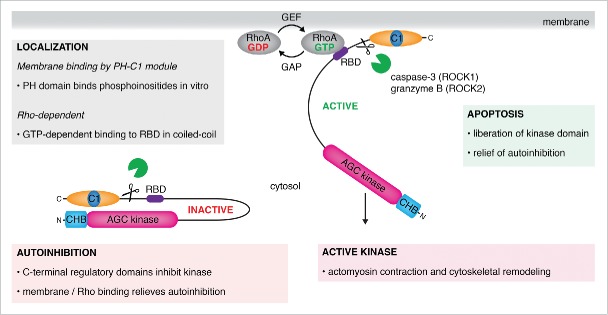
Figure 1.
A historical model of ROCK regulation. ROCK is maintained in an inactive state in the cytosol by autoinhibitory interactions between the carboxy terminal regulatory Rho-binding (RBD), PH and C1 domains. A combination of membrane binding by the PH and C1 domains and the binding of activated, GTP-bound RhoA to the RBD relieves autoinhibiton of the kinase domain and promotes downstream substrate phosphorylation. A second mechanism of activation occurs during apoptosis, in which the kinase domains of ROCK are liberated by proteolytic cleavage of the regulatory PH and C1 domains. The split PH-C1 module has been shown to bind phosphoinositide-containing membranes in vitro.
The PH and C1 domains have been implicated in the regulation of kinase activity, primarily on the basis of three observations: the regulatory portion of ROCK2 was observed to directly inhibit kinase activity in vitro;17 overexpression of the regulatory domains in cells phenocopied over-expression of kinase-dead ROCK, resulting in the loss of stress fibers;8,9 proteolytic cleavage between the coiled-coil and regulatory domains of ROCK1 and ROCK2 by caspase-3 and granzyme B respectively, was observed to enhance ROCK activity and, concomitantly, RMLC phosphorylation in vivo and in vitro.18-20
Early studies revealed that RhoA binds to a portion of the coiled-coil, defined as the Rho-binding domain (RBD), of ROCK1 in a GTP-dependent manner.1,8,21 A subsequent crystal structure of RhoA bound to a fragment of the coiled-coil of ROCK1 showed a heterotetrameric arrangement in which two molecules of RhoA bound in a symmetric fashion to either side of the coiled-coil.22 The RBD was shown to inhibit activity of the catalytic domain of ROCK in vitro and interfere with Rho signaling in vivo.23
These observations and others have led to a model for ROCK activation in cells in which the regulatory domains are displaced from inhibitory interactions with the kinase domain by a combination of membrane binding and RhoA binding (Fig. 1). However, structural and biochemical studies of a full-length ROCK have, until now, not been able to confirm or refute this model. Many of the early studies relied on a bottom-up approach, attempting to understand ROCK function by breaking the protein down into its parts and examining the properties of each part. As is often the case with such studies, the pieces of the jigsaw do not always correspond to the real picture, and should be treated with caution. Significantly, the molecular basis for autoinhibition has, to date, not been demonstrated; the reported influence of RhoA on ROCK activity is very modest (1-2 fold);2,3,24-26 and the structure of the ROCK-RhoA complex raises questions over how it interacts with membranes. Here we take a closer look at these issues in the context of recent structural, biochemical, and cell biological data obtained on full-length human ROCK2.
ROCK – a molecular ruler
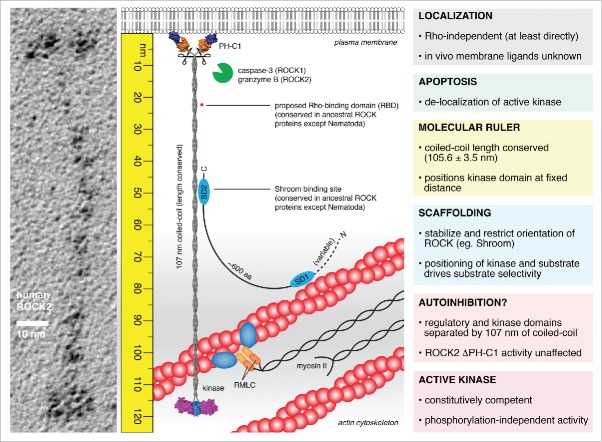
In their recent publication, Truebestein et al. challenge the existing model of ROCK regulation.27 The structure of full-length human ROCK2, which the authors determined by electron microscopy of rotary shadowed specimens, reveals it to be an extended dimer, 120 nm in length. Combining structural, biochemical, and cell biological tools, the authors make the case that ROCK2 is not autoinhibited, is not bound or activated by RhoA, and that its activity is not regulated by canonical mechanisms of AGC kinase phosphorylation. Instead, the authors claim that ROCK exhibits constitutive activity, which is regulated in vivo by the length of the coiled-coil (107 nm) that joins the kinase domain to the regulatory domains. Remarkably, the length, but not the sequence, of this coiled-coil has been conserved throughout 650 million years of evolution. The authors show that truncation of the coiled-coil, while not compromising activity, results in the loss of actin stress fibers in cells. Finally, they propose a new model in which substrate phosphorylation in the cell is controlled by the length of the coiled-coil of ROCK, restricting activity to a fixed distance from the plasma membrane (Fig. 2). The authors propose that it is not strictly necessary to maintain a kinase in an inactive state in the absence of its substrates. By spatially and temporally regulating the positioning of kinase and substrate in the cell, it should be possible to restrict activity to a specific location and, conversely, insulate the rest of the cell from such activity. Put another way, the kinase can only transfer phosphate to a substrate when presented with it. The model has implications for the flux and fidelity of substrate phosphorylation, much akin to the scaffolding of enzymes (and substrates) in the MAPK cascade28 or to the signaling at discrete cellular loci regulated by A-kinase anchoring proteins (AKAPs), a family of scaffold proteins.29
Figure 2.
The molecular ruler model of ROCK function. ROCK2 is an extended homodimer, 120 nm in length in which the kinase and regulatory domains are separated by 107 nm of parallel, semi-rigid coiled-coil. The kinase domains exist in a constitutively competent conformation mediated by the capped helix bundle (CHB) dimerization domain and an ordered, active conformation activation loop in the absence of phosphorylation. The regulatory domains exert no influence on catalytic activity. RhoA does not bind directly to either ROCK1 or ROCK2 in solution. The coiled-coil of ROCK is highly divergent in sequence, but remarkably conserved in length. Truncations in the coiled-coil, while retaining activity, cause the loss of actin stress fibers in cells, indicating the functional significance of its length. As such, the coiled-coil of ROCK bridges the kinase domains to a fixed distance from the plasma membrane. Scaffold proteins, such as Shroom, may bind to the coiled-coil of ROCK, stabilizing the orientation of the coiled-coil and perhaps restricting the positioning of the kinase domains. This leads to a model in which the phosphorylation of substrates by ROCK is governed by the spatial positioning of both kinase and substrate in the actin cytoskeleton. Proteolytic cleavage during apoptosis presumably liberates the active kinase domains, leading to delocalized activity, unregulated actomyosin contraction, and cell fragmentation.
While the model proposed by Truebestein and colleagues suggests that the coiled-coil of ROCK is oriented perpendicular to the plasma membrane, it should be noted that the region joining the coiled-coil to the regulatory domains is intrinsically flexible and could therefore allow the pivoting of the coiled-coil. This hinge motion may in fact be necessary to accommodate conformational changes in the cytoskeleton that arise from actomyosin contraction. As such, the possibility that ROCK could phosphorylate substrates in an arc defined by the length of its tether should be considered.
Constitutive competency
The structure of ROCK2 has a number of important consequences. With a 107 nm coiled-coil separating kinase and regulatory domains, it is difficult to imagine how the regulatory domains could exert an influence on kinase activity. Indeed, Truebestein et al. show that truncation of the coiled-coil or deletion of the coiled-coil and regulatory domains has no effect on activity in vitro. In the context of these findings, it is also worth pointing out that the evidence for a direct influence of the regulatory domains on kinase activity is inconsistent. While Amano et al. showed that titrating small amounts of a construct containing the RBD into a kinase reaction could specifically inhibit native Rho kinase in the presence of RhoA, it had no effect on the isolated, recombinant catalytic domain.23 In a later study, only a single regulatory domain construct of six tested showed a capacity to inhibit catalytic activity of the isolated kinase domain. In the single construct reported to specifically inhibit activity, the authors also introduced a double point mutation into the RBD.17 The inhibition of the kinase domain by a regulatory domain construct mutated in its RBD is counter to the proposed involvement of the RBD in autoinhibition. On the basis of the proposed autoinhibition by the regulatory domains, a structural study of a fragment of the coiled-coil sought to rationalize their interaction with the kinase domain by proposing that a discontinuity in the coiled-coil could act as a molecular hinge, thereby allowing the regulatory domains to approach the kinase domain.30 However, while this is an attractive hypothesis, the authors did not present any biochemical evidence to substantiate this claim.
RhoA – effect(or) not?
The publication of the crystal structure of RhoA in complex with a fragment of the coiled-coil of ROCK1 appeared to provide a neat explanation for the direct interaction of these two proteins. The previously delineated RBD,1,8,21 conserved in sequence, bound to RhoA in a stoichiometric manner, with switch I and II regions contacted by the two helices of the ROCK1 coiled-coil. However, the authors did not provide evidence for the existence of the complex in solution or validate the interface by site directed mutagenesis. While the authors claim to have isolated the complex by size exclusion chromatography, which would imply a sub-micromolar binding constant, Truebestein et al. found that the two proteins co-elute at the size of their respective masses. More significantly, neither the RBD of ROCK1 nor ROCK2 could be observed to interact with RhoA in solution, even when presented at high concentrations in a fluorescence polarization assay. These observations question whether the RhoA-ROCK1 crystal structure is actually an artifact of high protein concentrations and the global free energy minimum of lattice formation.31 Indeed, computational analysis of the RhoA:ROCK1 interface indicates that, while the coiled-coil is predicted to be a stable assembly, the interaction with RhoA is especially weak (complexation significance scores (CSS) of 0.08 and 0.10 for the interaction of RhoA with the two helices of the coiled-coil respectively; as a reference, the CSS score for the intermolecular coiled-coil interaction is 1.00).32
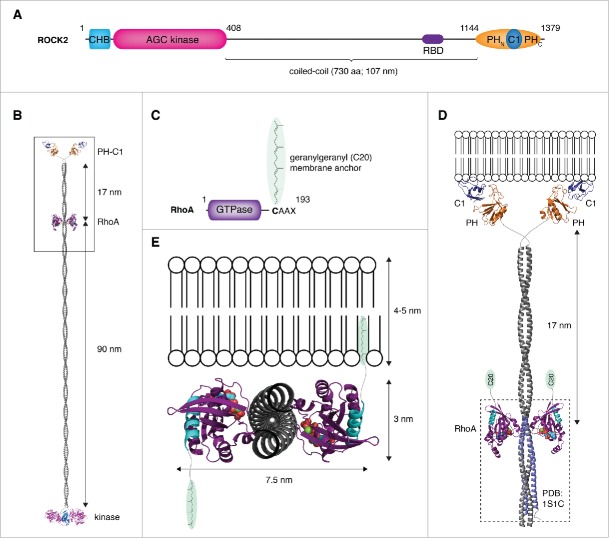
Notwithstanding the question marks surrounding the structure of the ROCK-RhoA complex, a second issue arises from the structure of full-length ROCK2. A notable consequence of the structure is the location of the RBD. The RBD is positioned 90 nm distal to the kinase domains but, more significantly, 17 nm proximal to the membrane-binding regulatory domains. By docking the crystal structure of the complex between RhoA and a fragment of the coiled-coil22 to the high-resolution model of full-length ROCK2 presented by Truebestein et al.27 the putative RhoA-ROCK complex can be visualized (Fig. 3A–B). The problem is immediately evident: how to reconcile the binding of ROCK to a membrane via its regulatory domains with the simultaneous anchoring in the membrane of geranylgeranylated RhoA? If ROCK is arranged such that its coiled-coil is perpendicular to the plane of the membrane, RhoA is located too far from the membrane for its C-terminal lipid anchor to reach (Fig. 3C–D); if ROCK is arranged such that its coiled-coil is parallel to the plane of the membrane, the C-termini of RhoA project in opposite directions (Fig. 3E). While, in theory, it would be possible to anchor one molecule of RhoA in the membrane in this topology, the lipid anchor of the second molecule would be presented to the cytosol. The solution to this problem is therefore also apparent: ROCK must surely undergo a conformational change upon RhoA binding. And herein lies the problem: neither ROCK1 nor ROCK2 could be shown to bind directly to RhoA.27 Not surprisingly, RhoA also exerts no activating effect on ROCK activity in vitro.27
Figure 3.
Membrane binding by the ROCK:RhoA complex – a topological problem. (A) Schematic illustrating the primary domain composition and structure of ROCK. (B) Structural model of ROCK2 bound to RhoA. A model of ROCK2 was constructed by combining the high-resolution structures of the kinase and regulatory domains (PDB IDs: 2F2U, 2ROV, 2ROW) together with a modeled parallel coiled-coil, 107 nm in length. The complex of ROCK1:RhoA was docked onto the coiled-coil at a position corresponding to the location of the RBD in the primary sequence using the region of canonical coiled-coil in the structure (PDB ID: 1S1C) to which RhoA was observed to bind. In this model, RhoA binds to the coiled-coil at a position 90 nm distal to the kinase domains and 17 nm proximal to the membrane-binding domains. (C) RhoA is anchored in the membrane by a geranylgeranyl (C20) lipid anchor covalently attached to its C-terminus. (D) Docking the complex to a membrane such that the coiled-coil of ROCK is oriented perpendicular to the plane of the membrane results in the lipid anchors of RhoA being too far from the membrane. (E) Docking the complex to a membrane such that the coiled-coil of ROCK is parallel to the plane of the membrane results in the C-termini of RhoA (cyan) projecting in opposite directions. In this topology, only one molecule of RhoA would be capable of inserting its lipid anchor in the membrane.
What, then, is the evidence for the direct interaction of ROCK with RhoA? Using GST-RhoA bound to GTPγS as a probe, either in protein overlay assays or bound to a column as an affinity matrix, early studies aimed at identifying novel RhoA effector proteins.1-3 In the case of ROCK2, screening of a cDNA expression library in one study purported to identify the RBD,1 which, in a parallel study, was also localized to the coiled-coil.3 Using RhoA loaded with GDP, GTPγS, or an effector domain mutant of RhoA, RhoAT37A, these studies demonstrated that the identified proteins bound RhoA in a GTP-dependent manner and that RhoA modestly (1.5-2 fold) stimulated kinase activity in vitro. Later studies identified a homolog of Rho kinase in Drosophila, named Drok, which also bound a constitutively active form of RhoA, RhoAG14V, as shown by yeast two hybrid analysis33 or GST-RhoA pull-down assay.34 In the latter study, the effector domain mutation, RhoAT37A, abolished the interaction. However, the literature is, again, inconsistent. Identification of the RBD by protein overlay assay and yeast two hybrid analysis yielded a minimal interaction domain between residues 934 and 1015 of ROCK1.21 However, two constructs containing the region of interaction later identified in the crystal structure do not interact in either assay. While Truebestein et al. could obtain a weak interaction with a construct containing the RBD by yeast two hybrid, the interaction was lost upon reversal of the fusion partners. Neither full-length ROCK1 nor ROCK2 showed any interaction with RhoA in the same assay.27 In a separate study, site directed mutagenesis of the RBD identified three mutants that abrogated the interaction with RhoA,8 though two of the three do not correspond to surfaces of interaction with RhoA in the subsequently reported crystal structure.22 In a study of cytoskeletal signaling pathways involving RhoA, Sahai et al. explored the binding properties of RhoA effector loop mutants, again using GST pull-down assays and yeast two hybrid analysis.35 However, the data supporting RhoA interactions with ROCK1, PKN, and the diaphanous homolog mDia2 are also partially inconsistent with the crystal structures of ROCK1-RBD:RhoA,22 PKN:RhoA,36 and mDia1:Cdc4237 respectively. For example, mutation of F39 to alanine appears to abrogate the interaction with both PKN and ROCK1, but it does not affect the interaction with mDia2. While F39 is found in the interface between RhoA and the coiled-coil of ROCK1, it does not participate in any direct interactions in complex with PKN, while its counterpart in Cdc42 binds in a hydrophobic pocket on mDia1.
Remarkably, until now, not a single solution binding experiment between RhoA and either ROCK1 or ROCK2 has been reported in the literature. Using inhibition of nucleotide dissociation as a proxy for effector domain binding, the affinity of RhoA for the RBD of ROCK1 was derived to be 130 nM,38 a value that would imply stable complex formation in solution. If this value is to be believed, it is unclear as to why Truebestein et al. failed to recapitulate the interaction in any of their experiments.
In conclusion, while there appears to be abundant evidence for the interaction of RhoA with both ROCK1 and ROCK2, the lack of quantitation of these interactions is notable. Given recent findings, it is questionable whether a direct interaction of measurable affinity actually exists. Coupled to the topological issues presented by the hypothetical RhoA:ROCK complex, we are faced with the very real question of whether in fact the Rho kinases directly interact with RhoA. On the other hand, it is impossible to ignore the numerous studies that have reported phenotypic similarities between RhoA and ROCK perturbation.39-46 One should, therefore, be cautious in discounting a role for RhoA in ROCK-mediated cytoskeletal regulation.
Location, location, location
Truebestein et al.27 propose that the coiled-coil of ROCK functions as a molecular ruler, restricting kinase activity to a fixed distance from the plasma membrane. The model raises a number of questions, not least those related to how ROCK is localized or positioned in the cell. Early studies on ROCK2 reported a predominantly cytosolic distribution.1,3 Overexpression of a constitutively active RhoA, locked in its GTP-bound state, resulted in the localization of ROCK2 to the plasma membrane, coincident with the distribution of activated RhoA,1 though a separate study found that activated RhoA did not promote the translocation of ROCK2 to specific sites.3 More recent studies have revealed ROCK2 accumulation at the cleavage furrow, implying a role in formation of the contractile ring during cytokinesis.47 However, the lack of RhoA binding in solution challenges those studies that have proposed that activated RhoA translocates ROCK to the membrane by binding to its coiled-coil. If RhoA is not responsible for ROCK localization, what is?
A comparative study of ROCK1 and ROCK2 activity against myosin II indicated a different specificity of the PH domains for phosphoinositides and reported a modest enhancement of ROCK2 kinase activity (1-2 fold) by both PIP2 and PIP3.10 A subsequent structural study of the regulatory PH and C1 domains concluded that the PH-C1 supramodule binds preferentially to membranes containing 3′-phosphoinositides, and therefore may act as a PIP3 sensor. Mutation of positively charged residues on the surface of the domain reduced binding to phosphoinositide-containing liposomes in vitro.16 However, comparison of the structure with canonical PIP3-binding PH domains, such as those of Akt48 or Btk,49 indicates that none of the residues required for phosphoinositide binding are conserved in either ROCK1 or ROCK2 and a global study of PH domains failed to detect any basal or stimulated translocation to the plasma membrane of the PH domain of ROCK1 in NIH3T3 cells.50 It is perhaps worth noting, in the context of the liposome binding data, that the regulatory domains of ROCK2 have an isoelectric point of 9.4 and the protein exhibits a tendency to bind liposomes of increasing negative charge density, suggesting that, at least in these in vitro experiments, the binding may be driven by non-specific electrostatics.
The C1 domain is an atypical C1 domain, in so far as it does not bind the lipid second messenger diacylglycerol, but like many atypical C1 domains, the identity of the ligand to which it binds is still a mystery. Though the PH-C1 module of ROCK can be shown to bind membranes of varying lipid composition in vitro, it remains to be determined to which ligands and in which membranes both the C1 and PH domains bind. Given the spatial restriction of ROCK activity implied by the molecular ruler model, the identification of precisely what these domains bind to in the cell is of paramount importance.
Finally, the structure of ROCK2 has profound implications simply from the length of its coiled-coil. At 120 nm, ROCK is approximately half of the diameter of a mycoplasm (0.15–0.3 μm) in length. What is the significance of its length? Truebestein et al. propose that it corresponds to a distance from the plasma membrane at which substrates of ROCK, such as RMLC and myosin phosphatase-targeting subunit 1 (MYPT1), are expected to be found in the actin cortex. However, the actin cortex is not a uniform layer at 120 nm,51,52 and questions remain about what, precisely, the coiled-coil is bridging. While the sequence of the coiled-coil is divergent, it also remains to be seen whether the mechanical properties of the coiled-coil are important, and whether small segments of surface conservation along the coiled-coil are important for the binding of scaffold proteins.
A prominent scaffold protein, Shroom, essential for formation of the neural tube, gut, and eye in vertebrates,53 localizes ROCK to apical cell junctions by binding to a region of the coiled-coil.54-56 In addition to binding ROCK, Shroom also contains an actin-binding domain, thereby linking ROCK directly to the actin cytoskeleton (Fig. 2). Scaffold proteins, such as Shroom, may serve to orient the coiled-coil of ROCK, thereby positioning the kinase domain more precisely with respect to its substrates. In the same vein, the positioning of kinase and substrate with respect to each other may also be a function of cytoskeletal tension, which has the capacity to regulate the spatial positioning of substrates such as RMLC and MYPT1. The local organization of signaling hubs to drive physiological processes is now widely recognized as a means to guide the flow of molecular information.29,57
Encoding substrate selectivity in the face of constitutive catalytic competence
How can a constitutively competent enzyme ensure adequate substrate selectivity in the cell? In the interests of promoting robust, switch-like signaling behavior, kinase-substrate interactions are intrinsically weak. It therefore follows that local concentration effects will play a significant role in determining kinase signaling. In the case of ROCK, a further avidity effect may arise from the homodimeric arrangement of its kinase domains, which implies the phosphorylation of dimeric substrates. This is likely the consequence of the fixed orientation of the kinase domains with respect to each other,12,13 though we cannot exclude the possibility that they might also act independently. Enzymatic studies with full-length monomeric and dimeric substrates are likely to provide the answer to this question. For dimeric substrates, efficient phosphorylation may not only be regulated by the combined affinities of the monomeric substrates, but by their precise spatial arrangement. It is notable that the most well characterized substrates of ROCK, RMLC and MYPT1, are both dimeric in cells. While RMLC is monomeric by itself in solution, in the cell it forms a constitutive complex with myosin II in which the two copies of RMLC are closely associated and related by a pseudo-2-fold rotation. In a recent cryo-electron microscopy study of myosin II, the phosphorylation sites on each of the RMLC monomers appear on the same face of the complex at an approximate distance of 60 Å,58 while docking of a crystal structure of the RMLC-myosin complex suggests this distance may be as much as 74 Å.59 Given the intermolecular spacing of substrate phospho-acceptor residues of 65 Å implied by the structure of the dimeric ROCK kinase,12,13 it is tempting to speculate that ROCK may recognize a particular arrangement of RMLC in the context of myosin II and that phosphorylation of both chains is necessary for the coordinated regulation of myosin function. However, it is not clear how ROCK could access the two copies of RMLC in the presence of the myosin stalk, which projects outwards from the same face of the complex as the N-termini of RMLC. Furthermore, it remains to be unambiguously demonstrated that RMLC is in fact a direct substrate for ROCK in vivo.
MYPT1 targets protein phosphatase 1 (PP1) to myosin where it acts to dampen actomyosin contractility by removing the activating phosphates from RMLC. In addition to structuring the PP1 catalytic subunit for efficient dephosphorylation of RMLC,60 MYPT1 contains two inhibitory phosphorylation sites at T696 and T866 that have been shown to be ROCK substrates.61,62 Phosphorylation by ROCK, particularly at T696, enhances actomyosin contraction by inhibiting myosin phosphatase.63 A lack of structural information for autoinhibited MYPT1 prohibits a detailed picture of how phosphorylation promotes PP1 inhibition, but the mechanism is thought to be competitive binding to the phosphatase active site.64 MYPT1 contains a leucine zipper in its C-terminus, which mediates its dimerization,65 thereby mirroring the oligomeric states of both ROCK and RMLC. However, the precise arrangement of the phosphorylation sites in MYPT1 is not known, therefore it is not possible to comment on their interaction with ROCK beyond noting that MYPT1 is itself dimeric.
While RMLC and MYPT1 are dimeric in nature, a well-characterized monomeric substrate of ROCK1 is Rnd3/RhoE. RhoE is an unconventional small GTPase that inhibits RhoA-induced stress fiber formation through currently unknown mechanisms. RhoE is a direct substrate of ROCK1, but not ROCK2,66 which phosphorylates it on multiple sites in its N- and C-termini, increasing RhoE stability and promoting its cytosolic re-localization.67 While effector domain mutations in RhoE attenuate the capacity of RhoE to disassemble stress fibers in vivo, mutations in the region interacting with ROCK1 have no effect.68 The phosphorylation of RhoE by ROCK1 is therefore a transient event that most likely occurs in the context of freely available, cytosolic RhoE.
Apoptosis – activation or de-regulation?
During apoptosis, the contraction of actomyosin provides the force for cell shrinking and eventual fragmentation. ROCK1 and ROCK2 encode proteolytic cleavage sites for the apoptotic proteases caspase-3 and granzyme B respectively, either side of a short conserved sequence of unknown function in the linker between the coiled-coil and the PH domain. In two independent studies, cleavage of ROCK1 by caspase-3 was shown to promote Rho-independent membrane blebbing and RMLC phosphorylation,18,20 despite the fact that proteolytic cleavage of the regulatory domains would not, in fact, separate the RBD from the kinase domain. Immunoprecipitated ROCK1 truncated at the caspase-3 cleavage site showed significantly enhanced activity against histone H1, presumed to be due to the relief of autoinhibition by removal of the regulatory domains.18 In contrast to wild type ROCK1, cells expressing truncated ROCK1 exhibited thick, and often stellate, actin stress fibers similar to cells expressing the catalytic domain alone (often referred to as constitutively active ROCK).8,9,23 In the second study, RMLC phosphorylation was observed to increase concomitantly with ROCK1 cleavage, and could be blocked with the ROCK inhibitor Y-27632, suggesting that ROCK1 had been activated during apoptosis.20 ROCK2 has since been shown to also participate in membrane blebbing and apoptosis following cleavage by granzyme B. Release of cleaved ROCK2 also enhanced MLC phosphorylation that could be inhibited by Y-27632.19
How can we reconcile these observations with those of Truebestein et al.? In purified, recombinant ROCK2, the regulatory domains are separated from the kinase domains by 107 nm of semi-rigid coiled-coil. Deletion of the regulatory domains has no effect on kinase activity in vitro. Could it be that activation of ROCK during apoptosis is not due to the enhancement of intrinsic catalytic activity, but rather due to the de-localization of the enzyme? In other words, could release of the catalytic domains induce actomyosin contraction and cell shrinkage by permitting unregulated RMLC phosphorylation at other sites in the cell? While the findings of Truebestein et al. would appear to support this hypothesis, the enhanced activity of immunoprecipitated ROCK in vitro rather implies the direct activation of ROCK itself. Future studies will therefore be needed to clarify the situation.
Regulation by phosphorylation
While many eukaryotic protein kinases are regulated by activation loop phosphorylation, the lack of regulation at this site in ROCK is unambiguous. Consistent with an active conformation of the activation loop in the absence of phosphorylation,12,13 Truebestein et al. show that mutation of the canonical phosphoacceptor residue in ROCK2 to either alanine or a glutamate phosphomimetic has no effect on activity in vitro or the formation of stress fibers in vivo.27 Similarly, while many of the growth factor-stimulated AGC kinases are activated by phosphorylation of a hydrophobic motif in their C-terminal tail, ROCK and the related kinases MRCK and DMPK appear not to be.12-15 In fact, replacement of the canonical phospho-acceptor threonine of the hydrophobic motif of ROCK2 with a phosphomimetic residue inhibited dimerization and resulted in a loss of activity,69 consistent with the loss of activity observed when the C-terminal tail is deleted.26
What about other sites of regulation? While early studies of ROCK2 found that it was capable of autophosphorylation in vitro2,3,9 and a recent proteomic study identified phosphorylation sites in the C-terminus,70 no study has found evidence of kinase activation in vitro. In contrast, there is some evidence to suggest that phosphorylation of the C-terminus of ROCK1 may regulate its subcellular localization.71 While future studies may yet identify further sites of phosphorylation, it should be noted that the C-terminus of ancestral ROCK proteins is highly divergent; any regulatory sites are therefore likely to be species- or isoform-specific adaptations rather than fundamental properties of all ROCK proteins.
Conclusions and future directions
Having derived their name from their interaction with RhoA, the Rho-associated coiled-coil kinases have largely been studied within the framework of Rho signaling ever since their discovery 20 years ago. While the absence of evidence is not evidence of absence, the lack of a measurable interaction between these two proteins in vitro is alarming, especially in the context of a crystal structure that purports to describe the intermolecular complex. Further studies will be required to reconcile the identification of ROCK as a direct RhoA effector with the failure of Truebestein et al to recapitulate the interaction with purified proteins in solution. Given the clear role of RhoA in cytoskeletal remodeling, the identification of the missing link between RhoA and ROCK will be essential.
The concept of a molecular ruler in regulating kinase activity at a fixed distance is elegant, but raises significant questions about precisely where and how ROCK is localized in the cell. Our understanding of ROCK function would therefore be aided enormously by the identification of the ligands to which its regulatory domains bind and their distribution within cellular membranes. Enzymatic studies would additionally address the activity of ROCK toward dimeric versus monomeric substrates. While the new work on the ROCK2 holoenzyme still leaves many questions unanswered, it challenges our existing hypotheses and assumptions about ROCK structure, function, and regulation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was financed by the Austrian Science Fund (FWF): P 28135.
References
- [1].Leung T, Manser E, Tan L, Lim L. A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J Biol Chem 1995; 270:29051-4; PMID:7493923; http://dx.doi.org/ 10.1074/jbc.270.42.25107 [DOI] [PubMed] [Google Scholar]
- [2].Ishizaki T, Maekawa M, Fujisawa K, Okawa K, Iwamatsu A, Fujita A, Watanabe N, Saito Y, Kakizuka A, Morii N, et al.. The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J 1996; 15:1885-93; PMID:8617235. [PMC free article] [PubMed] [Google Scholar]
- [3].Matsui T, Amano M, Yamamoto T, Chihara K, Nakafuku M, Ito M, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Rho-associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho. EMBO J 1996; 15:2208-16; PMID:8641286. [PMC free article] [PubMed] [Google Scholar]
- [4].Shimizu Y, Thumkeo D, Keel J, Ishizaki T, Oshima H, Oshima M, Noda Y, Matsumura F, Taketo MM, Narumiya S. ROCK-I regulates closure of the eyelids and ventral body wall by inducing assembly of actomyosin bundles. J Cell Biol 2005; 168:941-53; PMID:15753128; http://dx.doi.org/ 10.1083/jcb.200411179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Thumkeo D, Keel J, Ishizaki T, Hirose M, Nonomura K, Oshima H, Oshima M, Taketo MM, Narumiya S. Targeted disruption of the mouse rho-associated kinase 2 gene results in intrauterine growth retardation and fetal death. Mol Cell Biol 2003; 23:5043-55; PMID:12832488; http://dx.doi.org/ 10.1128/MCB.23.14.5043-5055.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Thumkeo D, Shimizu Y, Sakamoto S, Yamada S, Narumiya S. ROCK-I and ROCK-II cooperatively regulate closure of eyelid and ventral body wall in mouse embryo. Genes Cells 2005; 10:825-34; PMID:16098146; http://dx.doi.org/ 10.1111/j.1365-2443.2005.00882.x [DOI] [PubMed] [Google Scholar]
- [7].Kumper S, Mardakheh FK, McCarthy A, Yeo M, Stamp GW, Paul A, Worboys J, Sadok A, Jorgensen C, Guichard S, et al.. Rho-associated kinase (ROCK) function is essential for cell cycle progression, senescence and tumorigenesis. Elife 2016; 5:e12203; PMID:26765561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Leung T, Chen XQ, Manser E, Lim L. The p160 RhoA-binding kinase ROK α is a member of a kinase family and is involved in the reorganization of the cytoskeleton. Mol Cell Biol 1996; 16:5313-27; PMID:8816443; http://dx.doi.org/ 10.1128/MCB.16.10.5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ishizaki T, Naito M, Fujisawa K, Maekawa M, Watanabe N, Saito Y, Narumiya S. p160ROCK, a Rho-associated coiled-coil forming protein kinase, works downstream of Rho and induces focal adhesions. FEBS Lett 1997; 404:118-24; PMID:9119047; http://dx.doi.org/ 10.1016/S0014-5793(97)00107-5 [DOI] [PubMed] [Google Scholar]
- [10].Yoneda A, Multhaupt HA, Couchman JR. The Rho kinases I and II regulate different aspects of myosin II activity. J Cell Biol 2005; 170:443-53; PMID:16043513; http://dx.doi.org/ 10.1083/jcb.200412043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shi J, Wu X, Surma M, Vemula S, Zhang L, Yang Y, Kapur R, Wei L. Distinct roles for ROCK1 and ROCK2 in the regulation of cell detachment. Cell Death Dis 2013; 4:e483; PMID:23392171; http://dx.doi.org/ 10.1038/cddis.2013.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yamaguchi H, Kasa M, Amano M, Kaibuchi K, Hakoshima T. Molecular mechanism for the regulation of rho-kinase by dimerization and its inhibition by fasudil. Structure 2006; 14:589-600; PMID:16531242; http://dx.doi.org/ 10.1016/j.str.2005.11.024 [DOI] [PubMed] [Google Scholar]
- [13].Jacobs M, Hayakawa K, Swenson L, Bellon S, Fleming M, Taslimi P, Doran J. The structure of dimeric ROCK I reveals the mechanism for ligand selectivity. J Biol Chem 2006; 281:260-8; PMID:16249185; http://dx.doi.org/ 10.1074/jbc.M508847200 [DOI] [PubMed] [Google Scholar]
- [14].Heikkila T, Wheatley E, Crighton D, Schroder E, Boakes A, Kaye SJ, Mezna M, Pang L, Rushbrooke M, Turnbull A, et al.. Co-crystal structures of inhibitors with MRCKbeta, a key regulator of tumor cell invasion. PLoS One 2011; 6:e24825; PMID:21949762; http://dx.doi.org/ 10.1371/journal.pone.0024825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Elkins JM, Amos A, Niesen FH, Pike AC, Fedorov O, Knapp S. Structure of dystrophia myotonica protein kinase. Protein Sci 2009; 18:782-91; PMID:19309729; http://dx.doi.org/ 10.1002/pro.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wen W, Liu W, Yan J, Zhang M. Structure basis and unconventional lipid membrane binding properties of the PH-C1 tandem of rho kinases. J Biol Chem 2008; 283:26263-73; PMID:18640982; http://dx.doi.org/ 10.1074/jbc.M803417200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Amano M, Chihara K, Nakamura N, Kaneko T, Matsuura Y, Kaibuchi K. The COOH terminus of Rho-kinase negatively regulates rho-kinase activity. J Biol Chem 1999; 274:32418-24; PMID:10542285; http://dx.doi.org/ 10.1074/jbc.274.45.32418 [DOI] [PubMed] [Google Scholar]
- [18].Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol 2001; 3:339-45; PMID:11283606; http://dx.doi.org/ 10.1038/35070009 [DOI] [PubMed] [Google Scholar]
- [19].Sebbagh M, Hamelin J, Bertoglio J, Solary E, Breard J. Direct cleavage of ROCK II by granzyme B induces target cell membrane blebbing in a caspase-independent manner. J Exp Med 2005; 201:465-71; PMID:15699075; http://dx.doi.org/ 10.1084/jem.20031877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sebbagh M, Renvoize C, Hamelin J, Riche N, Bertoglio J, Breard J. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat Cell Biol 2001; 3:346-52; PMID:11283607; http://dx.doi.org/ 10.1038/35070019 [DOI] [PubMed] [Google Scholar]
- [21].Fujisawa K, Fujita A, Ishizaki T, Saito Y, Narumiya S. Identification of the Rho-binding domain of p160ROCK, a Rho-associated coiled-coil containing protein kinase. J Biol Chem 1996; 271:23022-8; PMID:8798490; http://dx.doi.org/ 10.1074/jbc.271.38.23022 [DOI] [PubMed] [Google Scholar]
- [22].Dvorsky R, Blumenstein L, Vetter IR, Ahmadian MR. Structural insights into the interaction of ROCKI with the switch regions of RhoA. J Biol Chem 2004; 279:7098-104; PMID:14660612; http://dx.doi.org/ 10.1074/jbc.M311911200 [DOI] [PubMed] [Google Scholar]
- [23].Amano M, Chihara K, Kimura K, Fukata Y, Nakamura N, Matsuura Y, Kaibuchi K. Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science 1997; 275:1308-11; PMID:9036856; http://dx.doi.org/ 10.1126/science.275.5304.1308 [DOI] [PubMed] [Google Scholar]
- [24].Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J Biol Chem 1996; 271:20246-9; PMID:8702756; http://dx.doi.org/ 10.1074/jbc.271.34.20246 [DOI] [PubMed] [Google Scholar]
- [25].Feng J, Ito M, Kureishi Y, Ichikawa K, Amano M, Isaka N, Okawa K, Iwamatsu A, Kaibuchi K, Hartshorne DJ, et al.. Rho-associated kinase of chicken gizzard smooth muscle. J Biol Chem 1999; 274:3744-52; PMID:9920927; http://dx.doi.org/ 10.1074/jbc.274.6.3744 [DOI] [PubMed] [Google Scholar]
- [26].Doran JD, Liu X, Taslimi P, Saadat A, Fox T. New insights into the structure-function relationships of Rho-associated kinase: a thermodynamic and hydrodynamic study of the dimer-to-monomer transition and its kinetic implications. Biochem J 2004; 384:255-62; PMID:15291762; http://dx.doi.org/ 10.1042/BJ20040344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Truebestein L, Elsner DJ, Fuchs E, Leonard TA. A molecular ruler regulates cytoskeletal remodelling by the Rho kinases. Nature Communications 2015; 6:10029; PMID:26620183; http://dx.doi.org/ 10.1038/ncomms10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol 2005; 6:827-37; PMID:16227978; http://dx.doi.org/ 10.1038/nrm1743 [DOI] [PubMed] [Google Scholar]
- [29].Langeberg LK, Scott JD. Signalling scaffolds and local organization of cellular behaviour. Nat Rev Mol Cell Biol 2015; 16:232-44; PMID:25785716; http://dx.doi.org/ 10.1038/nrm3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tu D, Li Y, Song HK, Toms AV, Gould CJ, Ficarro SB, Marto JA, Goode BL, Eck MJ. Crystal structure of a coiled-coil domain from human ROCK I. PLoS One 2011; 6:e18080; PMID:21445309; http://dx.doi.org/ 10.1371/journal.pone.0018080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Krissinel E. Crystal contacts as nature's docking solutions. J Comput Chem 2010; 31:133-43; PMID:19421996; http://dx.doi.org/ 10.1002/jcc.21303 [DOI] [PubMed] [Google Scholar]
- [32].Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol 2007; 372:774-97; PMID:17681537; http://dx.doi.org/ 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- [33].Mizuno T, Amano M, Kaibuchi K, Nishida Y. Identification and characterization of Drosophila homolog of Rho-kinase. Gene 1999; 238:437-44; PMID:10570971; http://dx.doi.org/ 10.1016/S0378-1119(99)00351-0 [DOI] [PubMed] [Google Scholar]
- [34].Winter CG, Wang B, Ballew A, Royou A, Karess R, Axelrod JD, Luo L. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell 2001; 105:81-91; PMID:11301004; http://dx.doi.org/ 10.1016/S0092-8674(01)00298-7 [DOI] [PubMed] [Google Scholar]
- [35].Sahai E, Alberts AS, Treisman R. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. EMBO J 1998; 17:1350-61; PMID:9482732; http://dx.doi.org/ 10.1093/emboj/17.5.1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Maesaki R, Ihara K, Shimizu T, Kuroda S, Kaibuchi K, Hakoshima T. The structural basis of Rho effector recognition revealed by the crystal structure of human RhoA complexed with the effector domain of PKN/PRK1. Mol Cell 1999; 4:793-803; PMID:10619026; http://dx.doi.org/ 10.1016/S1097-2765(00)80389-5 [DOI] [PubMed] [Google Scholar]
- [37].Lammers M, Meyer S, Kuhlmann D, Wittinghofer A. Specificity of interactions between mDia isoforms and Rho proteins. J Biol Chem 2008; 283:35236-46; PMID:18829452; http://dx.doi.org/ 10.1074/jbc.M805634200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Blumenstein L, Ahmadian MR. Models of the cooperative mechanism for Rho effector recognition: implications for RhoA-mediated effector activation. J Biol Chem 2004; 279:53419-26; PMID:15475352; http://dx.doi.org/ 10.1074/jbc.M409551200 [DOI] [PubMed] [Google Scholar]
- [39].Saito H, Minamiya Y, Saito S, Ogawa J. Endothelial Rho and Rho kinase regulate neutrophil migration via endothelial myosin light chain phosphorylation. J Leukoc Biol 2002; 72:829-36; PMID:12377953. [PubMed] [Google Scholar]
- [40].Essler M, Amano M, Kruse HJ, Kaibuchi K, Weber PC, Aepfelbacher M. Thrombin inactivates myosin light chain phosphatase via Rho and its target Rho kinase in human endothelial cells. J Biol Chem 1998; 273:21867-74; PMID:9705325; http://dx.doi.org/ 10.1074/jbc.273.34.21867 [DOI] [PubMed] [Google Scholar]
- [41].Mikelis CM, Simaan M, Ando K, Fukuhara S, Sakurai A, Amornphimoltham P, Masedunskas A, Weigert R, Chavakis T, Adams RH, et al.. RhoA and ROCK mediate histamine-induced vascular leakage and anaphylactic shock. Nat Communications 2015; 6:6725; http://dx.doi.org/ 10.1038/ncomms7725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ohta Y, Hartwig JH, Stossel TP. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat Cell Biol 2006; 8:803-14; PMID:16862148; http://dx.doi.org/ 10.1038/ncb1437 [DOI] [PubMed] [Google Scholar]
- [43].Valderrama F, Cordeiro JV, Schleich S, Frischknecht F, Way M. Vaccinia virus-induced cell motility requires F11L-mediated inhibition of RhoA signaling. Science 2006; 311:377-81; PMID:16424340; http://dx.doi.org/ 10.1126/science.1122411 [DOI] [PubMed] [Google Scholar]
- [44].Honing H, van den Berg TK, van der Pol SM, Dijkstra CD, van der Kammen RA, Collard JG, de Vries HE. RhoA activation promotes transendothelial migration of monocytes via ROCK. J Leukoc Biol 2004; 75:523-8; PMID:14634067; http://dx.doi.org/ 10.1189/jlb.0203054 [DOI] [PubMed] [Google Scholar]
- [45].Zhou Y, Su Y, Li B, Liu F, Ryder JW, Wu X, Gonzalez-DeWhitt PA, Gelfanova V, Hale JE, May PC, et al.. Nonsteroidal anti-inflammatory drugs can lower amyloidogenic Abeta42 by inhibiting Rho. Science 2003; 302:1215-7; PMID:14615541; http://dx.doi.org/ 10.1126/science.1090154 [DOI] [PubMed] [Google Scholar]
- [46].Sahai E, Ishizaki T, Narumiya S, Treisman R. Transformation mediated by RhoA requires activity of ROCK kinases. Curr Biol 1999; 9:136-45; PMID:10021386; http://dx.doi.org/ 10.1016/S0960-9822(99)80067-0 [DOI] [PubMed] [Google Scholar]
- [47].Kosako H, Goto H, Yanagida M, Matsuzawa K, Fujita M, Tomono Y, Okigaki T, Odai H, Kaibuchi K, Inagaki M. Specific accumulation of Rho-associated kinase at the cleavage furrow during cytokinesis: cleavage furrow-specific phosphorylation of intermediate filaments. Oncogene 1999; 18:2783-8; PMID:10348354; http://dx.doi.org/ 10.1038/sj.onc.1202633 [DOI] [PubMed] [Google Scholar]
- [48].Thomas CC, Deak M, Alessi DR, van Aalten DM. High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr Biol 2002; 12:1256-62; PMID:12176338; http://dx.doi.org/ 10.1016/S0960-9822(02)00972-7 [DOI] [PubMed] [Google Scholar]
- [49].Baraldi E, Djinovic Carugo K, Hyvonen M, Surdo PL, Riley AM, Potter BV, O'Brien R, Ladbury JE, Saraste M. Structure of the PH domain from Bruton's tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate. Structure 1999; 7:449-60; PMID:10196129; http://dx.doi.org/ 10.1016/S0969-2126(99)80057-4 [DOI] [PubMed] [Google Scholar]
- [50].Park WS, Heo WD, Whalen JH, O'Rourke NA, Bryan HM, Meyer T, Teruel MN. Comprehensive identification of PtdIns(3,4,5)P3-regulated PH domains from C. elegans to H. sapiens by model prediction and live imaging. Mol Cell 2008; 30:381-92; PMID:18471983; http://dx.doi.org/ 10.1016/j.molcel.2008.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Clark AG, Dierkes K, Paluch EK. Monitoring actin cortex thickness in live cells. Biophysical J 2013; 105:570-80; PMID:23931305; http://dx.doi.org/ 10.1016/j.bpj.2013.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nanoscale architecture of integrin-based cell adhesions. Nature 2010; 468:580-4; PMID:21107430; http://dx.doi.org/ 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hildebrand JD, Soriano P. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell 1999; 99:485-97; PMID:10589677; http://dx.doi.org/ 10.1016/S0092-8674(00)81537-8 [DOI] [PubMed] [Google Scholar]
- [54].Nishimura T, Takeichi M. Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development 2008; 135:1493-502; PMID:18339671; http://dx.doi.org/ 10.1242/dev.019646 [DOI] [PubMed] [Google Scholar]
- [55].Mohan S, Das D, Bauer RJ, Heroux A, Zalewski JK, Heber S, Dosunmu-Ogunbi AM, Trakselis MA, Hildebrand JD, Vandemark AP. Structure of a highly conserved domain of rock1 required for shroom-mediated regulation of cell morphology. PLoS One 2013; 8:e81075; PMID:24349032; http://dx.doi.org/ 10.1371/journal.pone.0081075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mohan S, Rizaldy R, Das D, Bauer RJ, Heroux A, Trakselis MA, Hildebrand JD, VanDemark AP. Structure of Shroom domain 2 reveals a three-segmented coiled-coil required for dimerization, Rock binding, and apical constriction. Mol Biol Cell 2012; 23:2131-42; PMID:22493320; http://dx.doi.org/ 10.1091/mbc.E11-11-0937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science 2011; 332:680-6; PMID:21551057; http://dx.doi.org/ 10.1126/science.1198701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Baumann BA, Taylor DW, Huang Z, Tama F, Fagnant PM, Trybus KM, Taylor KA. Phosphorylated smooth muscle heavy meromyosin shows an open conformation linked to activation. J Mol Biol 2012; 415:274-87; PMID:22079364; http://dx.doi.org/ 10.1016/j.jmb.2011.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Brown JH, Kumar VS, O'Neall-Hennessey E, Reshetnikova L, Robinson H, Nguyen-McCarty M, Szent-Gyorgyi AG, Cohen C. Visualizing key hinges and a potential major source of compliance in the lever arm of myosin. Proc Natl Acad Sci U S A 2011; 108:114-9; PMID:21149681; http://dx.doi.org/ 10.1073/pnas.1016288107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Terrak M, Kerff F, Langsetmo K, Tao T, Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature 2004; 429:780-4; PMID:15164081; http://dx.doi.org/ 10.1038/nature02582 [DOI] [PubMed] [Google Scholar]
- [61].Feng J, Ito M, Ichikawa K, Isaka N, Nishikawa M, Hartshorne DJ, Nakano T. Inhibitory phosphorylation site for Rho-associated kinase on smooth muscle myosin phosphatase. J Biol Chem 1999; 274:37385-90; PMID:10601309; http://dx.doi.org/ 10.1074/jbc.274.52.37385 [DOI] [PubMed] [Google Scholar]
- [62].Kawano Y, Fukata Y, Oshiro N, Amano M, Nakamura T, Ito M, Matsumura F, Inagaki M, Kaibuchi K. Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J Cell Biol 1999; 147:1023-38; PMID:10579722; http://dx.doi.org/ 10.1083/jcb.147.5.1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, et al.. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 1996; 273:245-8; PMID:8662509; http://dx.doi.org/ 10.1126/science.273.5272.245 [DOI] [PubMed] [Google Scholar]
- [64].Khromov A, Choudhury N, Stevenson AS, Somlyo AV, Eto M. Phosphorylation-dependent autoinhibition of myosin light chain phosphatase accounts for Ca2+ sensitization force of smooth muscle contraction. J Biol Chem 2009; 284:21569-79; PMID:19531490; http://dx.doi.org/ 10.1074/jbc.M109.019729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sharma AK, Zhou GP, Kupferman J, Surks HK, Christensen EN, Chou JJ, Mendelsohn ME, Rigby AC. Probing the interaction between the coiled coil leucine zipper of cGMP-dependent protein kinase Ialpha and the C terminus of the myosin binding subunit of the myosin light chain phosphatase. J Biol Chem 2008; 283:32860-9; PMID:18782776; http://dx.doi.org/ 10.1074/jbc.M804916200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Riento K, Guasch RM, Garg R, Jin B, Ridley AJ. RhoE binds to ROCK I and inhibits downstream signaling. Mol Cell Biol 2003; 23:4219-29; PMID:12773565; http://dx.doi.org/ 10.1128/MCB.23.12.4219-4229.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Riento K, Totty N, Villalonga P, Garg R, Guasch R, Ridley AJ. RhoE function is regulated by ROCK I-mediated phosphorylation. EMBO J 2005; 24:1170-80; PMID:15775972; http://dx.doi.org/ 10.1038/sj.emboj.7600612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Komander D, Garg R, Wan PT, Ridley AJ, Barford D. Mechanism of multi-site phosphorylation from a ROCK-I:RhoE complex structure. EMBO J 2008; 27:3175-85; PMID:18946488; http://dx.doi.org/ 10.1038/emboj.2008.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Couzens AL, Saridakis V, Scheid MP. The hydrophobic motif of ROCK2 requires association with the N-terminal extension for kinase activity. Biochem J 2009; 419:141-8; PMID:19099536; http://dx.doi.org/ 10.1042/BJ20081376 [DOI] [PubMed] [Google Scholar]
- [70].Lowery DM, Clauser KR, Hjerrild M, Lim D, Alexander J, Kishi K, Ong SE, Gammeltoft S, Carr SA, Yaffe MB. Proteomic screen defines the Polo-box domain interactome and identifies Rock2 as a Plk1 substrate. EMBO J 2007; 26:2262-73; PMID:17446864; http://dx.doi.org/ 10.1038/sj.emboj.7601683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ishiuchi T, Takeichi M. Willin and Par3 cooperatively regulate epithelial apical constriction through aPKC-mediated ROCK phosphorylation. Nat Cell Biol 2011; 13:860-6; PMID:21685893; http://dx.doi.org/ 10.1038/ncb2274 [DOI] [PubMed] [Google Scholar]