Abstract
Bisphosphonate drugs such as zoledronic acid (ZOL), used for the treatment of common bone disorders, target the skeleton and inhibit bone resorption by preventing the prenylation of small GTPases in bone-destroying osteoclasts. Increasing evidence indicates that bisphosphonates also have pleiotropic effects outside the skeleton, most likely via cells of the monocyte/macrophage lineage exposed to nanomolar circulating drug concentrations. However, no effects of such low concentrations of ZOL have been reported using existing approaches. We have optimized a highly sensitive in vitro prenylation assay utilizing recombinant geranylgeranyltransferases to enable the detection of subtle effects of ZOL on the prenylation of Rab- and Rho-family GTPases. Using this assay, we found for the first time that concentrations of ZOL as low as 10nM caused inhibition of Rab prenylation in J774 macrophages following prolonged cell culture. By combining the assay with quantitative mass spectrometry we identified an accumulation of 18 different unprenylated Rab proteins in J774 cells after nanomolar ZOL treatment, with a >7-fold increase in the unprenylated form of Rab proteins associated with the endophagosome pathway (Rab1, Rab5, Rab6, Rab7, Rab11, Rab14 and Rab21). Finally, we also detected a clear effect of subcutaneous ZOL administration in vivo on the prenylation of Rab1A, Rab5B, Rab7A and Rab14 in mouse peritoneal macrophages, confirming that systemic treatment with bisphosphonate drug can inhibit prenylation in myeloid cells in vivo outside the skeleton. These observations begin a new era in defining the precise pharmacological actions of bisphosphonate drugs on the prenylation of small GTPases in vivo.
Keywords: bisphosphonate, geranylgeranyl transferase, macrophage, prenylation, Rab
Introduction
For more than 3 decades, bisphosphonate (BP) drugs have been the frontline treatment for disorders of excessive bone destruction, such as post-menopausal osteoporosis and tumor-associated bone disease. Nitrogen-containing bisphosphonates (N-BP) such as zoledronic acid (ZOL) and alendronate decrease bone resorption by inhibiting farnesyl diphosphate (FPP) synthase in osteoclasts.1-3 Inhibition of this enzyme, at a critical branch point in the mevalonate pathway, prevents the biosynthesis of the isoprenoid lipids farnesyl diphosphate (FPP) and geranylgeranyldiphosphate (GGPP) that are necessary for post-translational protein prenylation.4,5 Most prenylated proteins belong to the superfamily of small GTPase signaling proteins, the majority of which are geranylgeranylated by either GGTase I (which prenylates Rho family GTPases) or Rab GGTase (which prenylates Rab GTPases).6 The anti-resorptive effect of N-BPs on osteoclasts is the result of inhibition of FPP synthase and loss of geranylgeranylated proteins.7 In the absence of prenylation, these proteins accumulate in the cytosol and disrupt downstream signaling pathways.8,9
In addition to affecting osteoclasts, there is increasing evidence from clinical trials and meta-analyses10-12 that in humans N-BPs may exert pleiotropic effects outside the skeleton. N-BP treatment is associated with prolonged disease-free survival in patients with multiple myeloma and in women with early-stage breast cancer,13-16 and decreased risk of mortality in post-menopausal women.17,18 The cell types responsible for these additional effects of N-BPs are unknown, but monocytes and macrophages are likely candidates since (like osteoclasts and unlike most cell types) they internalize N-BPs rapidly and efficiently by endocytosis.19-23 However, there is little knowledge regarding effects of N-BPs on cells other than osteoclasts, largely because of the difficulty of detecting subtle inhibitory effects of N-BPs on protein prenylation. Inhibition of prenylation is most conveniently studied by western blotting to detect the accumulation of the unprenylated form of the small GTPase Rap1A. This approach has shown that concentrations ≥1 μM N-BP inhibit the prenylation of small GTPases in a wide variety of cultured cell types including macrophages20,24 and osteoclasts25 and that N-BPs inhibit Rap1A prenylation in osteoclasts in vivo.22,26,27
A limitation of the western blot assay for unprenylated Rap1A is that it detects a single protein, whereas at least 300 proteins in the human proteome are estimated to be prenylated28 and the effect of N-BPs on this “prenylome” is largely unknown. Furthermore, the maximum circulating concentration of a standard dose of 4 mg iv ZOL or 70 mg oral alendronate in humans is approximately 1 μM and 120 nM respectively,29,30 decreasing rapidly to <1% of this 24 hours after infusion. Hence most cells outside the skeleton are likely to be exposed to far lower concentrations. To our knowledge, no effect of N-BPs on protein prenylation has ever been described in cultured cells at concentrations lower than 1 μM. Using J774 macrophages, we optimized an assay in which recombinant prenyl transferase enzymes (either GGTase I or GGTase II/Rab GGTase) utilize unprenylated proteins in cell lysates as substrates for in vitro prenylation with a biotinylated isoprenoid lipid31 (Fig. 1). This approach would be expected to be more sensitive than western blot detection of unprenylated Rap1A and, by combining this in vitro prenylation (IVP) assay with Stable Isotope with Amino acids in Cell Culture (SILAC)-mass spectrometry (MS), it allows the simultaneous identification and quantitation of multiple prenylated proteins that are affected by N-BPs. We applied this assay to study, for the first time, the effect of nanomolar concentrations ZOL on the geranylgeranylated prenylome of cultured macrophages, to determine the effect ZOL in vivo on the prenylome of macrophages outside the skeleton in mice, and to identify the specific prenylated proteins affected by ZOL treatment.
Figure 1.
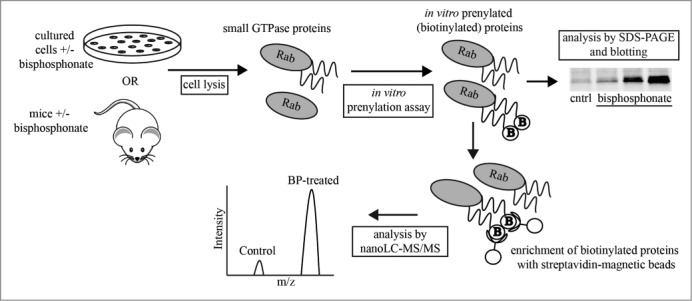
Schematic diagram of the in vitro prenylation assay. Unprenylated proteins (present in cells after bisphosphonate treatment) are prenylated in vitro in cell lysates by recombinant Rab GGTase or GGTase I, utilizing biotinylated-geranyl substrate (“B”). Hence, only unprenylated proteins become biotinylated, allowing detection on protein blots or enrichment for liquid chromatography-mass spectrometric analysis. The assay was used to analyze lysates of cultured cells and macrophages purified from ZOL-treated mice.
Results
An in vitro prenylation assay with Rab GGTase is more sensitive than detection of unprenylated Rap1A by protein gel blotting
Figure 1 illustrates the approach to label unprenylated Rab proteins present in cell lysates, using recombinant Rab GGTase/REP1 and biotinylated-geranyl substrate (B-GPP). Only unprenylated Rab proteins become prenylated (and hence biotinylated) in vitro, allowing detection on protein blots using streptavidin. Unprenylated Rap1A can also be detected on western blots using an antibody that specifically binds to the unprenylated form of the protein.24
Acute treatment of J774 macrophages for 24 h with 5 μM or 10 μM ZOL caused the accumulation of unprenylated Rap1A, detected by protein gel blotting (Fig. 2A). No effect could be detected after treatment with ≤1 μM ZOL (Fig. 2A), consistent with our previous studies showing that ≥1 μM N-BP is required to inhibit prenylation within 24 hours.4,7,20,24,25,32,33 Similarly, after performing the in vitro prenylation (IVP) assay with GGTase I on the same cell lysates, the presence of unprenylated 21 kDa small GTPases was detectable after treatment with 5 μM or 10 μM ZOL, but ≤1 μM ZOL had no detectable effect (Fig. 2B). After IVP of the same cell lysates with Rab GGTase, an intense cluster of protein bands of molecular mass 22-26 kDa was detected following treatment with 5 μM or 10 μM ZOL. However, these bands of in vitro prenylated Rab proteins were also detectable after treatment with 1 μM or 500 nM ZOL (Fig. 2C). An approximately 75 kDa band of endogenous biotinylated protein (most likely methylcrotonoyl-CoA carboxylase α subunit, and/or propionyl-CoA carboxylase α subunit) present in the cell lysates was used as a loading control on the blots (Fig. 2B–C). These studies demonstrate that the IVP assay with Rab GGTase is at least 10 times more sensitive at detecting effects of acute ZOL treatment on protein prenylation than standard western blotting for unprenylated Rap1A.
Figure 2.
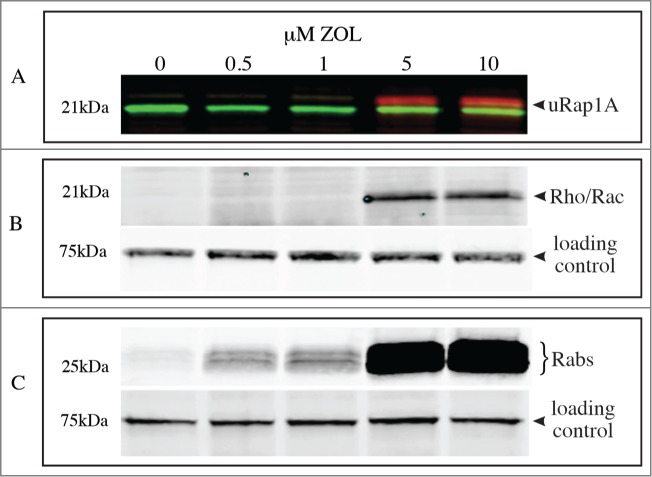
Only the in vitro prenylation assay with Rab GGTase detects acute effects of ≤1 μM ZOL on protein prenylation. J774 macrophages were treated for 48 h with 0.5-10 μM ZOL then cell lysates were analyzed for (A) the presence of unprenylated Rap1A (red, arrowhead) and total Rap1 (green); (B) 21 kDa (eg Rho, Rac) proteins prenylated in vitro by GGTase I (arrowhead); (C) Rab proteins prenylated in vitro by Rab GGTase. The 75 kD band of endogenous biotinylated protein was used as a loading control in (B) and (C).
Prolonged exposure of cultured macrophages to nanomolar concentrations of ZOL inhibits protein prenylation
We then applied the IVP assay, in comparison with protein gel blotting to detect unprenylated Rap1A, to determine whether prolonged exposure to low, nanomolar concentrations of ZOL could inhibit protein prenylation. After 7 days' culture with ≥250 nM ZOL, the IVP assay with Rab GGTase demonstrated an abundance of unprenylated Rab proteins, with an effect also being detectable after treatment with 50 nM and 100nM ZOL (Fig. 3A). By contrast, the IVP assay with GGTase I (Fig. 3B) and the western blot detection of unprenylated Rap1A (Fig. 3C) only showed a very minor effect of 500 nM ZOL after 7 days. After 18 days' culture, ≥50 nM ZOL had a clear effect on the prenylation of Rab proteins in the IVP assay with Rab GGTase (Fig. 3A). However, only 500 nM ZOL had a clear effect (and a minor effect with 250 nM ZOL) on unprenylated Rap1A (Fig. 3C), similar to the IVP assay with GGTase I (Fig. 3B). After 30 days' culture there was a clear effect of 10 nM ZOL on Rab prenylation (and dramatic effects with ≥50 nM ZOL) using the IVP assay with Rab GGTase (Fig. 3A). Concentrations of ZOL ≥50 nM also had a clear effect on prenylation in the IVP assay with GGTase I (Fig. 3B) and on unprenylated Rap1A (Fig. 3C) after 30 days' treatment. The 75 kDa band of endogenous biotinylated protein, used as a loading control on the blots, is shown in Supplementary Figure. 1.
Figure 3.
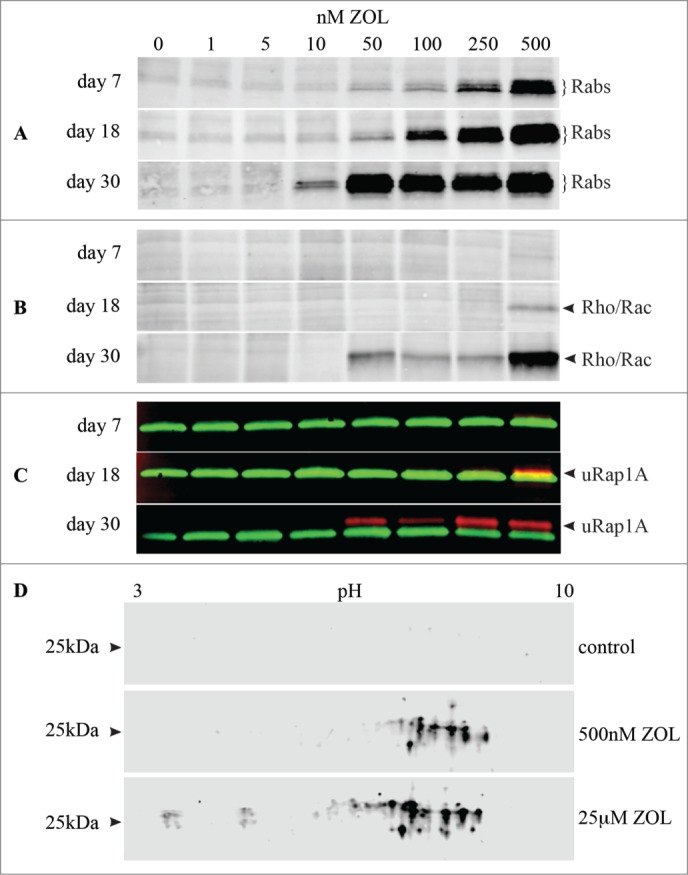
Prolonged exposure to ZOL is required to detect effects of low nanomolar concentrations of ZOL on protein prenylation. J774 macrophages were treated with 1-500 nM ZOL for 7, 18 or 30 days. Cell lysates were then analyzed for (A) Rab proteins prenylated in vitro by Rab GGTase; (B) proteins prenylated in vitro by GGTase I (eg Rho, Rac); (C) the presence of unprenylated Rap1A (red, arrowhead) and total Rap1 (green). The loading control (75 kD band of endogenous biotinylated protein) for (A) and (B) is shown in Fig. S1. (D) 2-dimensional electrophoretic separation of in vitro prenylated Rab proteins following treatment with 500 nM ZOL for 7 days, or with 25 μM ZOL for 30 hr.
Nanomolar ZOL treatment inhibits the prenylation of mutiple Rab GTPases
2-dimensional gel electrophoresis of J774 macrophage cell lysates, following the IVP assay with Rab GGTase, confirmed that 500 nM ZOL treatment for 7 days inhibited the prenylation of multiple Rab GTPases (Fig. 3D). This effect was even more pronounced after acute treatment (30 hr) with a high concentration of ZOL (25 μM) (Fig 3D). After SILAC labeling we analyzed the total proteome of J774 cells that had been treated with 500 nM ZOL for 7 days and unambiguously identified and quantified 20 Rab proteins with a false discovery rate <1% (Table S1). The total abundance of the majority of these Rab proteins was not altered following ZOL treatment (<1.2-fold change; Table 1 and Table S1), demonstrating that ZOL treatment did not alter Rab expression.
Table 1.
Proteomic analysis of unprenylated small GTPases in J774 cells following treatment with 50 nM or 500nM ZOL (or saline control) for 7 days. Proteins were identified and quantitated by nano-LC-MS/MS using SILAC. The fold change in abundance of each protein after ZOL treatment was calculated as a ratio of protein abundance in whole cell lysates of 500 nM ZOL-treated/control cells in a single experiment (see Table S1 for additional proteins and details). After in vitro prenylation of cell lysates and enrichment of biotinylated proteins, the relative abundance of each protein was calculated as a percentage of all of the enriched Rab GGTase substrates or all of the GGTase 1 substrates identified. The average fold change in unprenylated small GTPases was calculated as a ratio of abundance of in vitro-prenylated protein in ZOL-treated/control cells from 2 biological replicates (ie inversely labeled by SILAC), +/− SD where the protein was detected in both replicates (see Table S2 for details). NR = No Ratio could be calculated (ie undetectable in either the treated or the control cells)
| Name | Relative % abundance | Fold Change with 500 nM ZOL ± SD | Fold Change with 50 nM ZOL ± SD | Fold Change in Abundance |
|---|---|---|---|---|
| Rab GGTase substrates | ||||
| Rab1B | 14.32 | 9.5±6.7 | 1.6±0.03 | NR |
| Rab14 | 13.41 | 7.0±3.5 | 1.2±0.04 | −1.1 |
| Rab7A | 11.67 | 7.7±2.6 | 1.5±<0.01 | −1.1 |
| Rab21 | 7.32 | 8.8±4.2 | 1.5±0.05 | −1.1 |
| Rab6A/Rab6B | 7.27 | 7.0±3.5 | 1.1±0.04 | 1.0 |
| Rab18 | 5.75 | 4.9±0.4 | 1.3±0.2 | −1.1 |
| Rab5C | 5.20 | 6.7±1.8 | 1.3±0.04 | −1.1 |
| Rab11B/Rab11A | 5.16 | 8.3±4.5 | 1.3±0.2 | −1.1 |
| Rab2A | 4.95 | 5.3±0.3 | 1.2±0.07 | 1.3 |
| Rab1A | 3.95 | 7.9±3.8 | 1.5±0.02 | 1.0 |
| Rab10 | 3.17 | 4.9±0.8 | 1.1±0.3 | 1.0 |
| Rab39A | 2.36 | 2.4±2.3 | NR | No ID |
| Rab3A | 1.91 | 6.2±0.5 | NR | No ID |
| Rab5B | 1.88 | 5.9 | −1.1 | 1.0 |
| Rab5A | 1.87 | 7.0±2.2 | NR | −1.3 |
| Rab31 | 1.51 | 4.4±2.9 | −1.1±0.1 | −1.3 |
| Rab9A | 1.19 | 4.3 | NR | 1.0 |
| Rab43/Rab19 | 1.08 | 3.0 | NR | No ID |
| GGTase I substrates | ||||
| Rac1/Rac3 | 58.25 | 2.6±1.2 | −1.3±0.01 | NR |
| Rac2 | 21.58 | 1.8±0.6 | 1.0±0.2 | 1.0 |
| RhoA | 3.94 | 2.1 | NR | 1.0 |
To quantify the effects of ZOL on protein prenylation, we used magnetic streptavidin beads to specifically enrich biotinylated proteins from cell lysates following the IVP assay with GGTase I and Rab GGTase. Enriched, biotinylated proteins were then identified by mass spectrometry (MS) and the abundance measured as a ratio between ZOL-treated and untreated cells (Table 1 and Table S2). Using the IVP assay with Rab GGTase, 18 Rab proteins (most of which were also identified from the total proteome analysis of whole cell lysates; Table S2) were identified with a false discovery rate <1% (Table 1). Following treatment with 500 nM ZOL, all 18 Rab proteins exhibited an increase in the unprenylated form, with the majority increasing >4-fold (Table 1 and Table S2). The Rab proteins that showed the largest fold increase were also the most abundant eg Rab1B, Rab14, Rab7A, Rab21 and Rab6A/B, which showed a 7- to 9-fold increase in the unprenylated form after treatment with 500 nM ZOL (Table 1). Other Rab proteins that were markedly affected included Rab11A/B (8.3-fold increase), Rab1A (7.9-fold increase) and Rab5 isoforms (5.9-6.7-fold increase). Even after 50 nM ZOL treatment, 7 of the 18 Rab proteins (including Rab1A, Rab1B, Rab5C, Rab14, Rab7A and Rab21) still demonstrated a >1.2 fold increase in the unprenylated form (Table 1).
Using the IVP assay with GGTase I, 6 small GTPase proteins were identified by MS analysis with a false discovery rate <1% (Table 1 and Table S2). Following 500 nM ZOL treatment for 7 days, the fold increases in the unprenylated form of these proteins were smaller than those of Rab proteins: Rac1/Rac3 (the most abundant GGTase I substrate, from whole proteome analysis) showed the largest change (2.6-fold increase), followed by RhoA (2.1-fold increase) and Rac2 (1.8-fold increase). As with the Rab GTPases, the total abundance of these GTPases in whole cell lysates was not altered by ZOL treatment (Table 1 and Table S1). Consistent with the immunoblotting analyses (Fig. 3B), 50 nM ZOL treatment for 7 days had no discernible effect (fold increases <1.2) on the prenylation of these GGTase I substrates that was detectable by MS analysis (Table 1), while the prenylation of Ran, R-Ras and Rap2 isoforms was not inhibited even by 500 nM ZOL treatment (Table S2).
ZOL inhibits Rab prenylation in vivo in macrophages outside the skeleton
CD11b+ macrophages were isolated from the peritoneal lavage fluid of mice, 24 hours after administration of 100 μg/kg ZOL by either intraperitoneal (i.p.), subcutaneous (s.c.) or intravenous (i.v.) injection (3 mice per group). Compared to macrophages from untreated control mice, a single i.p. injection of ZOL caused a dramatic and consistent inhibitory effect on Rab prenylation, detectable using the IVP assay with Rab GGTase (Fig. 4A). By contrast, a single s.c. or i.v. injection of ZOL had no discernible effect (Fig. 4A) .
Figure 4.
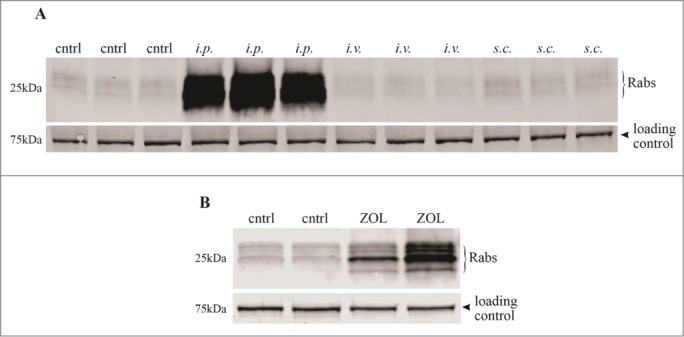
ZOL treatment inhibits Rab prenylation in vivo in mouse peritoneal macrophages. Mice (3 per group) were administered either (A) a single i.p., s.c. or i.v. injection of 100 μg/kg ZOL or left untreated (cntrl), or (B) administered s.c. ZOL (n = 2 mice) or saline control (2 mice per group) once-weekly for 8 weeks. 24 hours after the final injection, cell lysates were prepared from CD11b+ macrophages isolated from peritoneal lavage fluid. After in vitro prenylation with Rab GGTase, samples were transferred onto blots and analyzed for the presence of biotinylated Rab proteins. The 75 kD band of endogenous biotinylated protein was used as a loading control.
To better reflect clinical treatment in humans, with repeated exposure to low circulating doses of N-BP, mice were administered one s.c. injection of ZOL (2 mice) or saline (2 mice) once per week for 8 weeks. The IVP assay with Rab GGTase then revealed a clear effect of s.c. ZOL on the prenylation of multiple Rab proteins in CD11b+ peritoneal macrophages from the 2 ZOL-treated mice (although the magnitude of the effect varied between the 2 mice), compared to the peritoneal macrophages from the 2 control mice (Fig. 4B). Enrichment of the in vitro-prenylated Rab proteins, followed by label-free quantitative MS analysis (Table 2), identified increases in unprenylated Rab5b (estimated >20-fold increase in both mice), Rab7a (2.9-fold and 14.6-fold increase in the 2 mice), Rab1a (4.4-fold and 11.8-fold increase) and Rab14 (1.1-fold and 4.7-fold increase) in the 2 ZOL-treated mice compared to the control mice.
Table 2.
Identification and quantification of in vitro-prenylated Rab GTPase proteins in peritoneal macrophages from ZOL-treated mice, using label-free, quantitative-MS. The fold change in in vitro-prenylated Rab protein in cells from each ZOL-treated mouse was calculated as a ratio with the average level of in vitro-prenylated Rab protein in cells from the 2 control mice. PEP = Posterior Error Probability
| Name | Accession Number | Number of peptides | Fold Increase Mouse 1 | Fold Increase Mouse 2 | PEP |
|---|---|---|---|---|---|
| Rab7A | P51150 | 3 | 2.9 | 14.6 | 1.22E-09 |
| Rab5B | P61021 | 3 | >20(a) | >20(a) | 7.02E-09 |
| Rab14 | Q91V41 | 3 | 1.1 | 4.7 | 1.32E-07 |
| Rab1A | P62821 | 7 | 4.4 | 11.8 | 1.59E-18 |
Estimated fold change, since the in vitro-prenylated protein was not detected in lysates from control mice.
Discussion
In this study we describe a highly sensitive new assay to detect and profile the pharmacologic effect of N-BP drugs such as ZOL on the prenylation of specific small GTPase proteins. The in vitro prenylation (IVP) assay (Fig. 1), originally described by Nguyen and colleagues,31 was optimized in order to detect effects of ZOL on the prenylation of small GTPases in cultured J774 macrophages, using short-term treatment conditions that we have previously shown to inhibit the prenylation of Rap1A.20,24,25 Once optimized, the IVP assay using Rab GGTase was at least 10-fold more sensitive at detecting unprenylated Rab GTPases than detection of unprenylated Rap1A by protein gel blotting and, for the first time to our knowledge, demonstrated an effect of acute (24 hour) treatment with a nanomolar concentration of ZOL on protein prenylation in cultured cells. Interestingly, the IVP assay with GGTase I was less sensitive than with Rab GGTase and comparable to western blotting for unprenylated Rap1A, perhaps reflecting the higher abundance of Rab proteins.
We then applied the IVP assay to determine whether protein prenylation is inhibited in cultured cells by prolonged exposure to the low, nanomolar concentrations of N-BP that may be present in the circulation after iv or oral administration. Studies in rats demonstrated that circulating levels of 14C-labeled ZOL in the nanomolar range are detectable up to 8 months after a single iv injection,34 while single administration of 70 mg alendronate (the standard weekly dose) in humans gave rise to maximum plasma concentrations of about 120 nM.30 This raises the possibility that some cells types outside the skeleton, particularly those of the myeloid lineage, such as monocytes and macrophages,23 may be exposed to nanomolar concentrations of N-BP for prolonged periods of time even after a single iv dose of ZOL, or exposed repeatedly with weekly oral administration of N-BPs such as risedronate or alendronate. In keeping with this, we have previously shown that even a short (2 hour) exposure of monocytes to N-BP is sufficient to inhibit the mevalonate pathway.21 In the present study, using the IVP assay with Rab GGTase we detected a clear effect of 50 nM ZOL on Rab prenylation in J774 macrophages after 1 week of treatment. Remarkably, ZOL had a detectable effect at a concentration of just 10nM ZOL after 30 days' treatment. To our knowledge this concentration is at least 100 times lower than the previous limit of detection for an effect of ZOL on prenylation.
In contrast to the high sensitivity of the IVP assay with RabGGTase, the sensitivity of the IVP assay with GGTase I was comparable to the sensitivity of detection of unprenylated Rap1A by western blotting. However, even the western blot assay could detect the effect of 30 days' exposure of J774 macrophages to ZOL at concentrations ≥50 nM. These observations demonstrate that the inhibitory effect of N-BP on protein prenylation is cumulative over time, and that longer (or repeated) exposure of cells to low concentrations of N-BP is required to have a detectable effect. This is consistent with our previous finding that higher concentrations of N-BP have a more rapid inhibitory effect on the prenylation of small GTPases even when cells are treated acutely ie for 24 hours or less.24
As well as the high sensitivity of the IVP assay with Rab GGTase, an additional benefit is the ability to identify the individual small GTPases that are affected using nano-liquid chromatography mass spectrometry (LC-MS). J774 cells were metabolically labeled using SILAC in order to quantitatively compare individual proteins enriched from cell lysates of untreated and ZOL-treated cells using the IVP assay. For the first time, we demonstrate the effect of a pharmacologically relevant concentration of ZOL (50 nM and 500 nM) on a profile of proteins that are normally prenylated by Rab GGTase and GGTase I in macrophages. Of the 60 or more Rab GTPases known to exist in mammalian cells, 18 Rabs showed an increase >2-fold in the unprenylated form after ZOL treatment. In particular, Rab 1B, Rab14, Rab7A, Rab21 and Rab6A/B exhibited >7-fold increases in their unprenylated form after 500nM ZOL treatment. These Rabs were also the 5 most abundant Rab proteins in macrophage whole cell lysates, by whole proteome MS analysis. Together with Rab5C (>6-fold increase after 500 nM ZOL treatment) and Rab11A/B (>8-fold increase after 500 nM ZOL treatment), these are among the family of most commonly identified, endosome and phagosome-associated Rab proteins.35 Rabs that were less abundant generally showed a less marked increase in the unprenylated form. Previous studies have suggested a hierarchy of Rab prenylation dependent on the efficiency of prenylation by Rab GGTase/REP1 and therefore that some Rab proteins may be more susceptible to inhibition of prenylation.36 However, our observations indicate that ZOL, by depleting GGPP (the substrate required for prenylation of Rab GTPases by Rab GGTase) probably inhibits the prenylation of all Rab proteins but the effect is most pronounced in those that are most highly expressed (since prenylation occurs on newly-synthesized proteins, higher rates of transcription/translation of certain Rabs would lead to higher levels of unprenylated protein).
Consistent with our previous studies that showed the accumulation of unprenylated, GTP-bound Rho and Rac following acute treatment of J774 macrophages with N-BPs,8 we also detected an increase in the unprenylated form of the GGTase I substrates Rac1/3, Rac2, and RhoA after treatment with 500 nM ZOL (although the fold increases in unprenylated protein were not as high as we observed for Rab proteins). These observations, together with the greater sensitivity of the IVP assay with Rab GGTase to detect effects of low nanomolar concentrations of ZOL, suggest that prenylation by Rab GGTase is more sensitive to depletion of GGPP than prenylation by GGTase I, or that the IVP reaction with recombinant GGTase I is less efficient than with recombinant Rab GGTase.
Although we unequivocally identified 20 Rab proteins in J774 macrophages by whole proteome MS analysis, some Rabs were not identified, perhaps because of low abundance or the similarity in sequence between Rab proteins. Surprisingly, we also did not detect in vitro prenylated Rap1A by MS, although unprenylated Rap1A was detectable by western blotting.
Finally, we applied the highly sensitive IVP assay with Rab GGTase to determine the effect of ZOL on protein prenylation in vivo in cells outside the skeleton in mice. Perhaps not surprisingly, direct injection of a single dose of ZOL into the peritoneal cavity caused a dramatic effect on Rab prenylation in peritoneal macrophages, whereas a single dose of ZOL by s.c. or i.v. injection had no discernible effect. However, repeated systemic administration of s.c. ZOL over the course of 8 weeks had a clear inhibitory effect on Rab prenylation in peritoneal macrophages, consistent with the cell culture studies showing that prolonged exposure to low concentrations of ZOL had a cumulative effect. Using cells from the mice treated repeatedly with s.c. ZOL, the IVP assay and MS analysis revealed that ZOL caused the accumulation of unprenylated Rab1A, Rab5B, Rab7A and Rab14 in peritoneal macrophages. Crucially, these endophagosome-associated Rabs35 were among those that we also found (using quantitative SILAC analysis) to be among those most highly affected by prolonged treatment of cultured J774 macrophages with nanomolar concentrations of ZOL. To our knowledge, this is the first study to identify the in vivo effect of an N-BP drug on specific Rab proteins in macrophage cells outside the skeleton. This challenges the long-held view that BP drugs act solely on osteoclasts in bone in vivo. Our finding that the same Rab proteins were affected by ZOL in peritoneal macrophages in vivo and in cultured J774 macrophages suggests that prolonged exposure of cells in culture to nanomolar concentrations of N-BPs such as ZOL is an appropriate model to study the in vivo effect of low, circulating concentrations of N-BPs on cells outside bone.
Further studies are now needed to examine how inhibition of prenylation of small GTPases such as phagosome-associated Rab1, Rab5, Rab7 or Rab14 by ZOL influences the function of peritoneal macrophages and other myeloid cell populations in vivo, since effects on these myeloid cells may underscore the beneficial, pleiotropic actions of N-BPs. For example, we recently showed that the anti-cancer effects of N-BPs on the growth and metastasis of soft tissue tumors such as mammary carcinomas outside bone are likely mediated by effects on tumor-associated macrophages.19 Effects of N-BPs on macrophages may also account for adverse effects such as bisphosphonate-related osteonecrosis of the jaw.37,38 The highly sensitive IVP assay with Rab GGTase, capable of detecting effects of nanomolar concentrations of N-BP drugs, now provides a much-needed approach for examining effects on these and other cell types outside the skeleton that may be affected in vivo by N-BPs, as well as for further defining the cell types affected in vivo by other drugs that inhibit small GTPase prenylation such as statins and GGTase I inhibitors.
Materials and Methods
Reagents
Recombinant Rab escort protein-1 (REP1), GGTase I, Rab GGTase and biotinylated GPP (B-GPP) were prepared as described previously.31 Zoledronic acid (ZOL), a kind gift from Novartis, was dissolved in PBS, final pH 7.4. The murine J774.2 macrophage cell line (J774) was obtained from the European Collection of Cell Cultures and was maintained in DMEM (Life Technologies) with 10% foetal bovine serum (Hyclone, Thermo Fisher Scientific). Cultures were confirmed to be clear of mycoplasma.
Cell culture and bisphosphonate treatment
1 million murine J774 macrophage-like cells were seeded into 150 cm2 plates. ZOL was added to culture medium at a final concentration of 10 nM-10,000 nM and cells were incubated for between 1-30 days. For extended treatment, the medium was changed every 3 days; adherent cells were collected by gentle scraping and pooled with non-adherent cells, then re-seeded into fresh tissue culture plates with fresh medium containing ZOL.
In vitro prenylation (IVP) assay
Following ZOL treatment, J774 cells were collected by gentle scraping, washed twice with PBS and lysed by sonication in prenylation buffer containing 50mM HEPES, 50mM NaCl, 2 mM MgCl2, 100 µM GDP, 1 × Roche complete EDTA-free protease inhibitor cocktail. Protein was quantified using a BCA Protein Assay Kit (Pierce) and, unless state otherwise, 75 ug of total protein lysate was used per reaction. DTT was added to the lysates to a final concentration of 2 mM, and final concentrations of either 1 µM GGTase I plus 20 µM ZnCl2 and 5 µM biotinylated-geranyl substrate (B-GPP) were added, or 2 µM Rab GGTase plus 2 µM recombinant Danio rerio Rab escort protein-1 (REP1) and 0.5 µM B-GPP were added, in a total volume of approximately 40 µl. The IVP reactions were carried out for 4 h at room temperature. These optimal concentrations of prenyltransferase and B-GPP, and the incubation time for the IVP reaction, were determined empirically using lysate from J774 cells that had been treated for 24 h with 5 μM ZOL.
Immunoblotting
IVP reactions, or 50 μg J774 whole cell lysate, were boiled with Laemmli sample buffer then electrophoretically separated using 10% or 10-20% precast criterion TGX gels (Bio-Rad) and transferred to PVDF-FL blots using a Trans-Blot Turbo (Bio-Rad). For 2-dimensional gel electrophoresis, 250 μg of lysate were used for the IVP reaction with Rab GGTase. Samples were then buffer exchanged with 7M urea, 2M thiourea and 4% CHAPS using 5 kDa cut-off membrane concentrators until a conductivity of ≤ 300 µS/cm2 was achieved. 20 mM Tris-base was added and samples were reduced with 5mM TBP (90 min at RT) and 10mM acrylamide (90 min at RT). Samples were then loaded onto 11cm, pH 1-10 linear IPG strips for passive rehydration for 6 h and RT. Rehydrated IPG strips were iso-electrically focused for 2h at 300 V, then linearly ramped to 8000 V over 8 hours and then held constant for at 8000V for a total of 85 KV hours with the current limited to 50 µAmps per IPG strip (Ittan IPGphor, GE Healthcare). Strips were then subjected to second dimension protein separation using 8-16% polyacrylamide gel electrophoresis at 4 mA for 30 min and then 30 mA until the dye was run off the gel. Separated proteins were then transferred onto PVDF-FL membranes using semi-dry electro-blotters at 300 mAmps for 60 min (Towbin buffer system).
Blots were blocked with Odyssey Blocking Buffer (Li-COR) for 1h at room temperature and washed with Tris-buffered saline, 0.1% Tween 20 (TBS-T) for 1h. Blots were incubated with 1/1000 dilution of primary antibodies to unprenylated Rap1A (Santa Cruz, sc1482) and Rap1 (Cell Signaling, 2399S) for 16 hr at 4°C, and LiCOR secondary antibodies (1/20,000 dilution anti-goat 680 RD, anti-rabbit 800 CW respectively) or 1/20,000 dilution of LiCOR streptavidin-680 RD for 1 h at room temperature, then washed 3 times with TBS-T. Blots were scanned using an Odyssey Infrared Imager (LiCOR). We consistently detected an endogenous biotinylated 75 kDa protein, which was used as a sample loading control.
Isolation and analysis of mouse peritoneal macrophages
Studies with mice were approved by the Garvan Institute of Medical Research/St Vincent's Hospital Animal Ethics Committee. Adult Balb/c mice were untreated or injected with a single dose of 100 μg/kg ZOL either s.c. in the scruff of the neck, i.p. or i.v. via the tail vein (3 mice per group). In a separate study, 2 mice were injected s.c. once per week for 8 weeks with ZOL or phosphate-buffered saline (2 mice per group). Mice were euthanased 24 h after the final injection, then peritoneal cells (already approximately 40% F4/80+ macrophages) were harvested by lavage. CD11b+ macrophages were further purified using Miltenyi anti-CD11b magnetic beads according to the manufacturer's protocol. The purified macrophages from each mouse were lysed in IVP buffer, then 50 μg of lysate were used for IVP reactions with Rab GGTase as described above and analyzed for in vitro-prenylated (biotinylated) Rab proteins and for unprenylated Rap1A by hybridizing blots with streptavidin-680 RD, followed by anti-unprenylated Rap1A and anti-goat-800 CW antibodies. For label-free MS analysis, 70 μg of lysate were made up to a total of 80 µl with IVP buffer containing 2 µM RabGGTase, 2 µM REP1, 1 µM B-GPP, then in vitro-prenylated proteins were enriched with magnetic streptavidin beads and analyzed by nano-LC-MS as described below.
SILAC labeling
For quantitative MS analysis, J774 cells were labeled using Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) as previously described.39 Briefly, cells were cultured in either heavy (containing 0.199mM of C13, N15 Arginine and 0.798 mM of C13, N15 Lysine) or light (containing a proportional amount of C12 N14 Arginine and C12, N14 Lysine) medium for at least 8 cell doublings, with label incorporation being confirmed by nano-LC-MS/MS. Heavy and light isotope-labeled J774 cells were treated with either PBS (control), or with 50 nM or 500 nM ZOL for 7 days in biological duplicates with inversed labeling.
Enrichment of in vitro-prenylated Rab proteins for MS analysis
After SILAC labeling, 4 mg of light and heavy isotope-labeled J774 cell lysate were mixed. Lysates were then made up to a total volume of 3 ml with prenylation buffer containing 2 mM DTT and either 2 µM Rab GGTase, 2 µM REP1, 1 µM B-GPP, or 2 µM GGTase I, 20 µM ZnCl2, 1 µM B-GPP were added and the IVP reaction carried out for 4 hr at room temperature. In vitro-prenylated (and hence biotinylated) Rab-proteins were then enriched using magnetic streptavidin beads (NEB).31 Lysates were incubated with 1.2 ml of beads for 1hr at room temperature. Beads were washed 3 times with 1% NP-40, 4 M urea, 4 M guanidine hydrochloride then 3 times with 40mM ammonium bicarbonate, each for 10min at 4°C. Bound proteins were then digested overnight at 37°C with trypsin (modified Porcrine, Promega) and peptides were desalted using C18 stage tips as previously described.40
Mass Spectrometry and Data Analysis
Peptides were separated by nano-LC using an Easy-Nano LC II HPLC and autosampler system (Thermo) and mass spectra were acquired on an Orbitrap Velos Pro mass spectrometer (MS) (Thermo). Peptides were delivered to the MS with a nano-C18 column (75 µm ID × ∼20 cm, 3 µm, 200 Å Magic, Michrom) and the reverse phase nano-eluent was subject to positive nano-flow electrospray analysis in an information-dependent acquisition mode (IDA) (400 nl/min over 60 min). In IDA mode, survey scans in the range m/z 350-1750 were acquired with lockmass at m/z 445 enabled and a resolution of 60,000. Up to the 15 most abundant ions (> 5,000 counts) with charge states > +2 were sequentially isolated and further subjected to MS/MS fragmentation within the linear ion trap using collisionally-induced dissociation (CID). MS/MS spectra were accumulated with an activation time of 30 ms at a target value of 30,000 ions. m/z ratios selected for MS/MS were dynamically excluded for 30 sec. For whole-lysate total proteome analysis an in-gel digest was performed as previously described.41
Protein identification and data analysis
The nanoLC-MS/MS .raw files were processed with MaxQuant software (version 1.2.7.4), which uses the Andromeda algorithm for data processing, database searching and protein identification.42 Extracted peak lists were searched against the UniProtKB/Swiss-Prot Mus Musculus database (Version 02_2012) containing 64323 entries (including common contaminants) and a proportionally sized decoy database for false discovery rate (FDR) generation. The following search parameters were selected; fixed cysteine carbamidomethylation modification; variable methionine oxidation modification, variable protein N-acetylation, variable phosphorylation of serine, threonine and tyrosine, minimum peptide length of 7 amino acids and up to 2 missed cleavages were allowed. The initial first search mass tolerance was 20 ppm for precursor ions and 0.5 Da for fragment ions, match between run was enabled with default settings, labeled amino acids were filtered and re-quantify was enabled. The FDR was limited to 1% for both protein and peptide identifications. For protein quantitation, normalized ratios between ZOL-treated and control groups, as calculated by MaxQuant, were used for the SILAC experiments with J774 cells and raw protein intensity values were used to determine ratios for the analysis of isolated peritoneal macrophages.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The contents of this manuscript are solely the responsibility of the authors and do not reflect the views of the NHMRC.
Funding
This work was supported by NHMRC project grant APP1079522 and Cancer Council NSW project grant RG14-12 to MJR, and ARC DP grant DP1094080, ARC FF FT0991611, and NHMRC project grant APP569652 and NHMRC program grant APP1037320 to KA.
Supplemental Material
Supplemental Material may be downloaded here: publisher's website
References
- 1.Dunford JE, Thompson K, Coxon FP, Luckman SP, Hahn FM, Poulter CD, Ebetino FH, Rogers MJ. Structure-activity relationships for inhibition of farnesyl diphosphate synthase in vitro and inhibition of bone resorption in vivo by nitrogen-containing bisphosphonates. J Pharmacol Exp Ther 2001; 296:235-42; PMID:11160603 [PubMed] [Google Scholar]
- 2.Rondeau JM, Bitsch F, Geiser M, Hemmig R, Kroemer M, Lehmann S, Ramage P, Rieffel S, Strauss A, Green JR, et al.. Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs. J Med Chem 2006; 1:267-73 [DOI] [PubMed] [Google Scholar]
- 3.Kavanagh K, Guo K, Dunford JE, Wu X, Knapp S, Ebetino FH, Rogers MJ, Russell RGG, Oppermann U. The molecular mechanism of nitrogen-containing bisphosphonates as anti-osteoporosis drugs: crystal structure and inhibition of farnesyl pyrophosphate synthase. Proc Natl Acad Sci 2006; 103 7829-34; PMID:16684881; http://dx.doi.org/ 10.1073/pnas.0601643103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luckman SP, Hughes DE, Coxon FP, Russell RGG, Rogers MJ. Nitrogen-containing bisphosphonates inhibit the mevalonate pathway and prevent post-translational prenylation of GTP-binding proteins, including Ras. J Bone Miner Res 1998; 13:581-9; PMID:9556058; http://dx.doi.org/ 10.1359/jbmr.1998.13.4.581 [DOI] [PubMed] [Google Scholar]
- 5.Rogers MJ, Crockett JC, Coxon FP, Monkkonen J. Biochemical and molecular mechanisms of action of bisphosphonates. Bone 2011; 49:34-41; PMID:21111853; http://dx.doi.org/ 10.1016/j.bone.2010.11.008 [DOI] [PubMed] [Google Scholar]
- 6.McTaggart SJ. Isoprenylated proteins. Cell Mol Life Sci 2006; 63:255-67; PMID:16378247; http://dx.doi.org/ 10.1007/s00018-005-5298-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coxon FP, Helfrich MH, van 't Hof RJ, Sebti SM, Ralston SH, Hamilton AD, Rogers MJ. Protein geranylgeranylation is required for osteoclast formation, function, and survival: inhibition by bisphosphonates and GGTI-298. J Bone Miner Res 2000; 15:1467-76; PMID:10934645; http://dx.doi.org/ 10.1359/jbmr.2000.15.8.1467 [DOI] [PubMed] [Google Scholar]
- 8.Dunford JE, Rogers MJ, Ebetino FH, Phipps RJ, Coxon FP. Inhibition of protein prenylation by bisphosphonates causes sustained activation of Rac, Cdc42 and Rho GTPases. J Bone Miner Res 2006; 21 684-94; PMID:16734383; http://dx.doi.org/ 10.1359/jbmr.060118 [DOI] [PubMed] [Google Scholar]
- 9.Itzstein C, Coxon FP, Rogers MJ. The regulation of osteoclast function and bone resorption by small GTPases. Small GTPases 2011; 2:117-30; PMID:21776413; http://dx.doi.org/ 10.4161/sgtp.2.3.16453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan T, Yin W, Zhou Q, Zhou L, Jiang Y, Du Y, Shao Z, Lu J. The efficacy of zoledronic acid in breast cancer adjuvant therapy: a meta-analysis of randomised controlled trials. Eur J Cancer 2012; 48:187-95; PMID:22100904; http://dx.doi.org/ 10.1016/j.ejca.2011.10.021 [DOI] [PubMed] [Google Scholar]
- 11.Valachis A, Polyzos NP, Coleman RE, Gnant M, Eidtmann H, Brufsky AM, Aft R, Tevaarwerk AJ, Swenson K, Lind P, et al.. Adjuvant therapy with zoledronic acid in patients with breast cancer: a systematic review and meta-analysis. Oncologist 2013; 18:353-61; PMID:23404816; http://dx.doi.org/ 10.1634/theoncologist.2012-0261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolland MJ, Grey AB, Gamble GD, Reid IR. Effect of osteoporosis treatment on mortality: a meta-analysis. J Clin Endocrinol Metab 2010; 95:1174-81; PMID:20080842; http://dx.doi.org/ 10.1210/jc.2009-0852 [DOI] [PubMed] [Google Scholar]
- 13.Morgan GJ, Davies FE, Gregory WM, Cocks K, Bell SE, Szubert AJ, Navarro-Coy N, Drayson MT, Owen RG, Feyler S, et al.. First-line treatment with zoledronic acid as compared with clodronic acid in multiple myeloma (MRC Myeloma IX): a randomised controlled trial. Lancet 2010; 376:1989-99; PMID:21131037; http://dx.doi.org/ 10.1016/S0140-6736(10)62051-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gnant M, Mlineritsch B, Schippinger W, Luschin-Ebengreuth G, Postlberger S, Menzel C, Jakesz R, Seifert M, Hubalek M, Bjelic-Radisic V, et al.. Endocrine therapy plus zoledronic acid in premenopausal breast cancer. N Engl J Med 2009; 360:679-91; PMID:19213681; http://dx.doi.org/ 10.1056/NEJMoa0806285 [DOI] [PubMed] [Google Scholar]
- 15.Coleman RE, Marshall H, Cameron D, Dodwell D, Burkinshaw R, Keane M, Gil M, Houston SJ, Grieve RJ, Barrett-Lee PJ, et al.. Breast-cancer adjuvant therapy with zoledronic acid. N Engl J Med 2011; 365:1396-405; PMID:21995387; http://dx.doi.org/ 10.1056/NEJMoa1105195 [DOI] [PubMed] [Google Scholar]
- 16.Coleman R, de Boer R, Eidtmann H, Llombart A, Davidson N, Neven P, von Minckwitz G, Sleeboom HP, Forbes J, Barrios C, et al.. Zoledronic acid (zoledronate) for postmenopausal women with early breast cancer receiving adjuvant letrozole (ZO-FAST study): final 60-month results. Annals Oncol 2013; 24:398-405; http://dx.doi.org/ 10.1093/annonc/mds277 [DOI] [PubMed] [Google Scholar]
- 17.Center JR, Bliuc D, Nguyen ND, Nguyen TV, Eisman JA. Osteoporosis medication and reduced mortality risk in elderly women and men. J Clin Endo Metab 2011; 96:1006-14; http://dx.doi.org/ 10.1210/jc.2010-2730 [DOI] [PubMed] [Google Scholar]
- 18.Lyles KW, Colon-Emeric CS, Magaziner JS, Adachi JD, Pieper CF, Mautalen C, Hyldstrup L, Recknor C, Nordsletten L, Moore KA, et al.. Zoledronic Acid in Reducing Clinical Fracture and Mortality after Hip Fracture. N Engl J Med 2007; 357:1799-809; PMID:17878149; http://dx.doi.org/ 10.1056/NEJMoa074941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Junankar S, Shay G, Jurczyluk J, Ali N, Down J, Pocock N, Parker A, Nguyen A, Sun S, Kashemirov B, et al.. Real time intravital imaging establishes tumour-associated macrophages as the extraskeletal target of bisphosphonate action in cancer. Cancer Discovery 2015; 5:35-42; PMID:25312016; http://dx.doi.org/ 10.1158/2159-8290.CD-14-0621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson K, Rogers MJ, Coxon FP, Crockett JC. Cytosolic entry of bisphosphonate drugs requires acidification of vesicles after fluid-phase endocytosis. Mol Pharmacol 2006; 69(5):1624-32; PMID:16501031; http://dx.doi.org/ 10.1124/mol.105.020776 [DOI] [PubMed] [Google Scholar]
- 21.Roelofs AJ, Jauhiainen M, Monkkonen H, Rogers MJ, Monkkonen J, Thompson K. Peripheral blood monocytes are responsible for gammadelta T cell activation induced by zoledronic acid through accumulation of IPP/DMAPP. Br J Haematol 2009; 144:245-50; PMID:19016713; http://dx.doi.org/ 10.1111/j.1365-2141.2008.07435.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roelofs AJ, Coxon FP, Ebetino FH, Lundy MW, Henneman ZJ, Nancollas GH, Sun S, Blazewska KM, Bala JL, Kashemirov BA, et al.. Fluorescent risedronate analogues reveal bisphosphonate uptake by bone marrow monocytes and localization around osteocytes in vivo. J Bone Miner Res 2010; 25:606-16; PMID:20422624; http://dx.doi.org/ 10.1359/jbmr.091009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roelofs AJ, Thompson K, Ebetino FH, Rogers MJ, Coxon FP. Bisphosphonates: Molecular Mechanisms of Action and Effects on Bone Cells, Monocytes and Macrophages. Curr Pharm Des 2010; 16:2950-60; PMID:20722616; http://dx.doi.org/ 10.2174/138161210793563635 [DOI] [PubMed] [Google Scholar]
- 24.Roelofs AJ, Thompson K, Gordon S, Rogers MJ. Molecular mechanisms of action of bisphosphonates: current status. Clin Cancer Res 2006; 12:6222s-30s; PMID:17062705; http://dx.doi.org/ 10.1158/1078-0432.CCR-06-0843 [DOI] [PubMed] [Google Scholar]
- 25.Coxon FP, Thompson K, Roelofs AJ, Ebetino FH, Rogers MJ. Visualizing mineral binding and uptake of bisphosphonate by osteoclasts and non-resorbing cells. Bone 2008; 42:848-60; PMID:18325866; http://dx.doi.org/ 10.1016/j.bone.2007.12.225 [DOI] [PubMed] [Google Scholar]
- 26.Frith JC, Monkkonen J, Auriola S, Monkkonen H, Rogers MJ. The molecular mechanism of action of the anti-resorptive and anti-inflammatory drug clodronate: evidence for the formation in vivo of a metabolite that inhibits bone resorption and causes osteoclast and macrophage apoptosis. Arthritis Rheum 2001; 44:2201-10; PMID:11592386; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 27.Coxon FP, Ebetino FH, Mules EH, Seabra MC, McKenna CE, Rogers MJ. Phosphonocarboxylate inhibitors of Rab geranylgeranyl transferase disrupt the prenylation and membrane localization of Rab proteins in osteoclasts in vitro and in vivo. Bone 2005; 37:349-58; PMID:16006204; http://dx.doi.org/ 10.1016/j.bone.2005.04.021 [DOI] [PubMed] [Google Scholar]
- 28.Sebti SM. Protein farnesylation: implications for normal physiology, malignant transformation, and cancer therapy. Cancer Cell 2005; 7:297-300; PMID:15837619; http://dx.doi.org/ 10.1016/j.ccr.2005.04.005 [DOI] [PubMed] [Google Scholar]
- 29.Chen T, Berenson J, Vescio R, Swift R, Gilchick A, Goodin S, LoRusso P, Ma P, Ravera C, Deckert F, et al.. Pharmacokinetics and pharmacodynamics of zoledronic acid in cancer patients with bone metastases. J Clin Pharmacol 2002; 42:1228-36; PMID:12412821; http://dx.doi.org/ 10.1177/009127002762491316 [DOI] [PubMed] [Google Scholar]
- 30.Rhim SY, Park JH, Park YS, Lee MH, Kim DS, Shaw LM, Yang SC, Kang JS. Bioavailability and bioequivalence of two oral formulations of alendronate sodium 70 mg: an open-label, randomized, two-period crossover comparison in healthy Korean adult male volunteers. Clinical Therapeutics 2009; 31:1037-45; PMID:19539104; http://dx.doi.org/ 10.1016/j.clinthera.2009.05.001 [DOI] [PubMed] [Google Scholar]
- 31.Nguyen UT, Guo Z, Delon C, Wu Y, Deraeve C, Franzel B, Bon RS, Blankenfeldt W, Goody RS, Waldmann H, et al.. Analysis of the eukaryotic prenylome by isoprenoid affinity tagging. Nat Chem Biol 2009; 5:227-35; PMID:19219049; http://dx.doi.org/ 10.1038/nchembio.149 [DOI] [PubMed] [Google Scholar]
- 32.Luckman SP, Coxon FP, Ebetino FH, Russell RGG, Rogers MJ. Heterocycle-containing bisphosphonates cause apoptosis and inhibit bone resorption by preventing protein prenylation: evidence from structure- activity relationships in J774 macrophages. J Bone Miner Res 1998; 13:1668-78; PMID:9797474; http://dx.doi.org/ 10.1359/jbmr.1998.13.11.1668 [DOI] [PubMed] [Google Scholar]
- 33.Frith JC, Rogers MJ. Antagonistic effects of different classes of bisphosphonates in osteoclasts and macrophages in vitro. J Bone Miner Res 2003; 18:204-12; PMID:12568397; http://dx.doi.org/ 10.1359/jbmr.2003.18.2.204 [DOI] [PubMed] [Google Scholar]
- 34.Green JR, Rogers MJ. Pharmacologic profile of zoledronic acid: a highly potent inhibitor of bone resorption. Drug Dev Res 2002; 55:210-24; http://dx.doi.org/ 10.1002/ddr.10071 [DOI] [Google Scholar]
- 35.Gutierrez MG. Functional role(s) of phagosomal Rab GTPases. Small GTPases 2013; 4:148-58; PMID:24088602; http://dx.doi.org/ 10.4161/sgtp.25604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohnke M, Delon C, Hastie ML, Nguyen UT, Wu YW, Waldmann H, Goody RS, Gorman JJ, Alexandrov K. Rab GTPase prenylation hierarchy and its potential role in choroideremia disease. PloS One 2013; 8:e81758; PMID:24358126; http://dx.doi.org/ 10.1371/journal.pone.0081758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pazianas M. Osteonecrosis of the jaw and the role of macrophages. J Natl Cancer Inst 2011; 103:232-40; PMID:21189409; http://dx.doi.org/ 10.1093/jnci/djq516 [DOI] [PubMed] [Google Scholar]
- 38.Zhang Q, Atsuta I, Liu S, Chen C, Shi S, Shi S, Le AD. IL-17-mediated M1/M2 macrophage alteration contributes to pathogenesis of bisphosphonate-related osteonecrosis of the jaws. Clin Cancer Res 2013; 19:3176-88; PMID:23616636; http://dx.doi.org/ 10.1158/1078-0432.CCR-13-0042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 2002; 1:376-86; PMID:12118079; http://dx.doi.org/ 10.1074/mcp.M200025-MCP200 [DOI] [PubMed] [Google Scholar]
- 40.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 2003; 75:663-70; PMID:12585499; http://dx.doi.org/ 10.1021/ac026117i [DOI] [PubMed] [Google Scholar]
- 41.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols 2006; 1:2856-60; PMID:17406544; http://dx.doi.org/ 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- 42.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 2011; 10:1794-805; PMID:21254760; http://dx.doi.org/ 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


