Abstract
Lung cancer is a disease with dismal outcome. We recently reported a detailed intratumor heterogeneity analysis in 7 non-small cell lung cancer samples, revealing spatially separated driver events as well as the temporal dynamics of mutational processes and demonstrating an important role for APOBEC-mediated heterogeneity later in disease evolution.
Keywords: APOBEC, intratumor heterogeneity, lung cancer, multi-region sequencing
Abbreviations
- APOBEC
apolipoprotein-B mRNA editing catalytic polypeptide-like
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- NSCLC
non-small cell lung cancer
- TRACERx
TRAcking Cancer Evolution through therapy (Rx)
Lung cancer, of which non-small cell lung cancer (NSCLC) is the most prevalent subtype, is the leading cause of cancer-related mortality. To better understand NSCLC pathogenesis, several sequencing efforts have been undertaken. These studies demonstrated that NSCLC exhibits a complex genomic landscape and identified several NSCLC driver genes.1-3 Few studies have investigated the clonal architecture of NSCLC. Whole-genome sequencing analysis of 17 NSCLC samples identified biclonal tumors, some of which had potentially targetable mutations in one subclone next to a clonal targetable mutation.3 Intratumor heterogeneity in NSCLC could thus have significant consequences in terms of therapeutic efficacy. We recently showed in renal cancer that single region analyses significantly underestimates the heterogeneity.4 Therefore, our understanding of the clonal architecture of NSCLC and the biological processes driving this disease remain far from complete.
To gain a greater insight into the level of intratumor heterogeneity in NSCLC and improve our understanding of its evolution, we set out to investigate in detail the spatial and temporal heterogeneity of NSCLC.5 We performed multiregion exome and/or whole-genome sequencing on 7 primary NSCLCs, including adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC) samples.
We found spatial heterogeneity of mutations, copy number alterations, and translocations. On average, two-thirds of all mutations identified in a tumor were present in all regions of that tumor, whereas one-third of the mutations were present in only one or a few regions. Importantly, known lung cancer driver genes, including therapeutically targetable drivers, were significantly more often present in all tumor regions. Nevertheless, all tumors revealed candidate driver mutations and/or copy number aberrations present in only one or two regions. Sequence analysis of only that region would have given the illusion that these subclonal driver mutations were fully clonal events (Fig. 1).
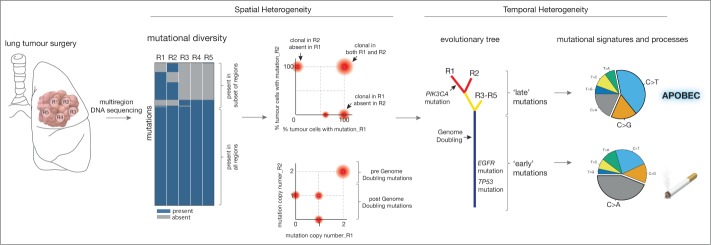
Figure 1.
Multiregion DNA sequencing allows analysis of genetic diversity within a tumor. Certain mutations are present in all tumor regions, whereas others are only present in certain tumor regions, as presented in a heatmap. 2D-Dirichlet analyses of mutations corrected for copy number shows the clonality of each mutation within each region (upper graph). The presence of 2 copies of multiple mutations can indicate a genome-doubling event, in which the entire tumor genome has been duplicated. Mutations present in 2 copies were present prior to the doubling event whereas those present at one copy occurred after the doubling (lower graph). The mutational diversity can also reveal a tumor's life history. Exploring ‘early’ versus ‘late’ mutational signatures sheds light on how mutational processes change over time. In NSCLC adenocarcinomas, ‘early’ mutations are mainly caused by smoking whereas ‘late’ mutations are mainly caused by apolipoprotein-B mRNA editing catalytic polypeptide-like (APOBEC) cytidine deaminase activity.
To investigate the temporal heterogeneity in mutations, we separated ‘early’ mutations (present in all tumor regions) from ‘late’ mutations (present in at least one, but not all regions) and explored the mutational spectra over time. We found that all tumors from former and current smokers showed a decrease in smoking-associated C>A mutations over time, accompanied by an increase in C>T and C>G mutations at TpC sites in the majority of tumors, indicative of apolipoprotein-B mRNA editing catalytic polypeptide-like (APOBEC) cytidine deaminase-mediated mutagenesis.6 On average, 31% of the ‘late’ non-silent mutations occurred in an APOBEC context compared to 11% of the ‘early’ non-silent mutations, indicating a functional impact of APOBEC activity later in NSCLC evolution.
It is currently unclear what activates APOBEC enzymes in NSCLCs, or other tumor types.6-8 A striking observation from our study is the more pronounced enrichment of APOBEC-associated ‘late’ mutations in LUAD compared to LUSC, suggesting a different regulatory route for APOBEC activity between histological subtypes. We furthermore noticed that chromosomal instability, including whole-genome doubling events, often preceded APOBEC activity. We could not, however, find an association with chromosomal breakpoints, nor did we find any evidence for clusters of APOBEC mutations.
A number of other questions arise from our findings, such as: What drives spatial heterogeneity? Does it result from random genetic drift with spatially different selective pressures, or is there a spatial barrier between the subclones preventing subclonal intermixing? Having determined the tumor cell fraction of each mutation within each region, we found very few subclonal mutations shared between regions (Fig. 1), indicating that the regions may evolve through a process similar to allopatric speciation, with geographically distinct separation of subclones. The striking regional differences in APOBEC activity in some tumors provide evidence for spatial heterogeneity in mutational processes, leading to increased mutational intratumor heterogeneity.
Multiregion sequencing of 10 renal cancer samples revealed that many known driver mutations were always subclonally present.4 It would be very interesting to determine whether certain driver mutations are predominantly subclonal or always clonal using larger NSCLC cohorts. Furthermore, this approach can be used to increase the statistical power to identify novel drivers of subclonal expansions.
Intriguingly, by combining smoking cessation information with the relative timing of clonal genome doubling events, we found evidence for a prolonged latency period of these tumors. In these cases, all ‘early’ driver mutations had occurred more than 2 decades prior to surgery, indicating a long period during which these tumors have been shaped, most likely involving many processes prior to clinical presentation.
Another important question for NSCLC is whether the observed intratumor heterogeneity has clinical consequences. Importantly, we found that the mutations present in metastasized tumor cells of patients with lymph node involvement closely correlated with one particular region within the primary tumor, indicating that a minor subclones present in a distinct region in the primary tumor can determine clinical outcome. Intriguingly, the level of intratumor heterogeneity varied from 4% to 63% across the samples. Such variation indicates that tumors could potentially be classified into discrete categories based on their heterogeneity, and these categories may hold clinical relevance. Interestingly, an accompanying study of 11 early-stage NSCLCs by Zhang et al. showed that primary tumors from the 3 patients with relapsed disease had significantly larger subclonal fractions than tumors from the other patients.9
In conclusion, our study showed varying levels of intratumor heterogeneity in NSCLC, and revealed insights into the processes that shape the NSCLC evolution, with APOBEC enzyme activity predominantly later in disease evolution. Our study justifies larger longitudinal studies, such as the lung TRACERx study,10 to assess the clinical impact of this heterogeneity on patient outcome.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
EB is funded by Rosetrees Trust; CS is a senior Cancer Research UK clinical research fellow and is funded by Cancer Research UK, the Rosetrees Trust, EU FP7 (projects PREDICT and RESPONSIFY, ID:259303), the Prostate Cancer Foundation, the European Research Council and the Breast Cancer Research Foundation. This research is supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
References
- 1.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008; 455:1069-75; PMID:18948947; http://dx.doi.org/ 10.1038/nature07423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012; 150:1107-20; PMID:22980975; http://dx.doi.org/ 10.1016/j.cell.2012.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L, Wallis J, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012; 150:1121-34; PMID:22980976; http://dx.doi.org/ 10.1016/j.cell.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014; 46:225-33; PMID:24487277; http://dx.doi.org/ 10.1038/ng.2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014; 346:251-56; PMID:25301630; http://dx.doi.org/ 10.1126/science.1253462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, et al. Signatures of mutational processes in human cancer. Nature 2013; 500:415-21; PMID:23945592; http://dx.doi.org/ 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet 2013; 45:977-83; PMID:23852168; http://dx.doi.org/ 10.1038/ng.2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 2013; 45:970-6; PMID:23852170; http://dx.doi.org/ 10.1038/ng.2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, Seth S, Chow CW, Cao Y, Gumbs C, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014; 346:256-9; PMID:25301631; http://dx.doi.org/ 10.1126/science.1256930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jamal-Hanjani M, Hackshaw A, Ngai Y, Shaw J, Dive C, Quezada S, Middleton G, de Bruin E, Le Quesne J, Shafi S, et al. Tracking genomic cancer evolution for precision medicine: the lung TRACERx study. PLoS Biol 2014; 12:e1001906; PMID:25003521 [DOI] [PMC free article] [PubMed] [Google Scholar]