Abstract
Chromatin compaction represents a barrier for the repair of DNA double-strand breaks (DSBs). However, heterochromatin components are also required for DSB repair by homologous recombination. The BARD1/HP1 interaction, required for the retention of BRCA1, CTIP, and RAD51 at DSB sites, may play a critical role in the crosstalk between chromatin compaction and DSB repair.
Keywords: BARD1, BRCA1, H3K9me2, heterochromatin, homologous recombination, HP1
DNA double-strand breaks (DSBs) are repaired by several different mechanisms, including error-free homologous recombination repair (HRR) and error-prone non-homologous end-joining (NHEJ) mechanisms. The breast and ovarian tumor suppressor BRCA1 plays a pivotal role in HRR, and failure of this function of BRCA1 is important in the etiology of these cancers and also serves as a target of cancer therapy. The antagonistic effects of 53BP1 (TP53BP1, best known as 53BP1)/RIF1, the effectors of NHEJ, and BRCA1-CTIP (also known as RBBP8) on DNA end resection and HRR have recently been a topic of discussion within the field with respect to their importance in clinical applications.
BRCA1 forms 3 distinct complexes through its C-terminal BRCT repeats, which associate with different adaptor proteins: ABRAXAS (FAM175A, also known as ABRA1 or CCDC98), BACH1 (also known as FANCJ or BRIP1) and CTIP. BRCA1 also forms a complex with BRCA2 via PALB2, which interacts with the coiled-coil domain of BRCA1. The best-characterized pathway for recruitment of BRCA1 involves the BRCA1/ABRAXAS complex and is mediated by focal polyubiquitin products at DSB sites (Fig. 1). In response to a DSB, ATM-dependent phosphorylation triggers the subsequent recruitment of the E3 ligases RNF8 and RNF168, and Lys63-linked polyubiquitin chains catalyzed by RNF168 interact with the BRCA1/ABRAXAS/RAP80 complex via the ubiquitin-interacting motif of RAP80. However, although ATM and BRCA1 are both required for HRR, the role of the BRCA1/ABRAXAS complex is not to execute HRR but rather to suppress excess DNA end resection.1,2 Because 53BP1 is also recruited through ATM/RNF8/RNF168-mediated ubiquitination, this pathway is likely the one that reinforces NHEJ. The other BRCA1 complexes, BRCA1/BACH1, BRCA1/CTIP, and BRCA1/PALB2/BRCA2/RAD51, are thought to execute HRR through helicase activity, DNA end resection, and strand invasion, respectively. However, how these BRCA1 complexes are retained at DSB sites remains largely unknown.
Figure 1.
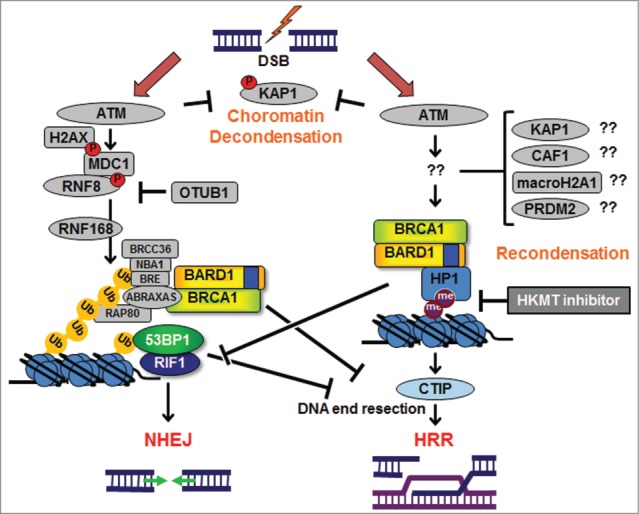
Mechanism of retention of BRCA1/BARD1 complexes at DNA double-strand break sites. In response to DNA double-strand breaks (DSBs), ATM mediates chromatin decondensation in part through KAP1 phosphorylation and activates RNF8/RNF168 pathway that recruit BRCA1/ABRAXAS complex as well as 53BP1/RIF1, the effectors of non-homologous end-joining (NHEJ). BARD1/HP1/H3K9me2 interaction is also mediated by ATM, but not by RNF168, and is required for homologous recombination repair (HRR) and inhibition of RIF1. KAP1, CAF1, macroH2A1, and RPDM2 may be involved in the process of chromatin recondensation in HRR together with the BARD1/HP1 interaction. HKMT, histone lysine methyltransferase; me, methylation, P, phosphorylation; Ub, ubiquitin, blue boxes, BRCT domain.
Recently, Lee et al. demonstrated that heterochromatin protein 1 (HP1) is required for the retention of BRCA1 at DSB sites, providing a hint to this critical issue.3 As another clue, we have recently revealed a mechanism by which the non-BRCA1/ABRAXAS BRCA1 complex is retained at DSB sites.4 In addition to the C-terminal BRCT domains, BRCA1 contains an N-terminal RING domain that binds BARD1 to form a RING heterodimer core complex. In response to DNA damage, BARD1 interacts with Lys9-dimethylated histone H3 (H3K9me2). This interaction is mediated by HP1 through direct binding of the chromoshadow domain of HP1 to a conserved PxVxL motif in the BRCT domain of BARD1. Interestingly, the interaction is ATM-dependent but RNF168-independent. The replacement of endogenous BARD1 with an exogenous BARD1 mutant that abrogates the BARD1/HP1 interaction abolishes retention of BRCA1/BARD1 at DSB sites. Importantly, inhibition of the BARD1/HP1 interaction severely disturbs CTIP and RAD51 retention and allows ectopic RIF1 accumulation at DSB sites in S-phase cells.4 These results illustrate an alternative ATM-dependent mechanism for BRCA1 retention that is required for HRR but is distinct from the ATM/RNF8/RNF168 pathway (Fig. 1).
The mechanism that mediates ATM activity and the BARD1/HP1 interaction is currently unknown (Fig. 1). Excluding the proteins in the RNF8/RNF168 pathway, KAP1 (TRIM28, best known as KAP1), a PxVxL motif-containing transcriptional repressor, is an important candidate ATM substrate that may play a role in the BARD1/HP1 interaction. KAP1 promotes chromatin condensation, and KAP1 phosphorylation by ATM at DSB sites is crucial for chromatin decondensation, which is a prerequisite for NHEJ in heterochromatic regions. KAP1 functions with another PxVxL motif-containing protein, p150/CAF-1 (CHAF1A), to recruit HP1 to DSB sites via the interaction between their PxVxL motifs and the chromoshadow domain.5 Consistent with the idea that KAP1 may regulate the interaction of BARD1 with HP1, KAP1 depletion strongly impairs RAD51 recruitment to DSB sites without affecting H2AX phosphorylation.5 Furthermore, it has been reported that the release of KAP1 from DSB sites is critical for local chromatin relaxation during NHEJ as well as in the initial step of HRR; however, KAP1 re-accumulation is also required for further HRR progress, suggesting that the restoration of heterochromatic structure contributes to HRR completion.6 The BARD1/HP1 interaction may be involved in this process. Whether Lys9-methylated histone H3 is required to anchor BARD1/HP1 at DSB sites or is free from the nucleosome is currently unknown. However, the first interpretation is strongly supported by a recent report of the dynamic depletion and re-accumulation of H3K9me2 at DSB sites, accompanied by dynamic changes in repressive chromatin components, including macroH2A1 (H2AFY) and the H3K9 methyltransferase PRDM2.7 The inhibition of BRCA1/BARD1 retention at DSB sites by the histone methyltransferase inhibitor UNC0638, which specifically inhibits methylation at Lys9 of histone H3, also supports the first hypothesis.4
The HP1 family contains 3 members, HP1α (CBX5), HP1β (CBX1), and HP1γ (CBX3). Although HP1γ was predominantly co-immunoprecipitated with BARD1 after DNA damage, its specificity among the other members of the HP1 family is currently unclear. Lee et al. demonstrated that other HP1 family members are also important for BRCA1 functions.3 Simultaneous inhibition of multiple HP1 family members suppresses BRCA1 retention much more effectively than the inhibition of a single member, suggesting that the family members have redundant functions.4 However, several lines of evidence have indicated the specificity of HP1γ for the interaction with BRCA1. HP1γ interacts with BRCA1 and represses BRCA1-mediated Gadd45a transcription.8 The loss of Wip1 induces ATM activation and subsequently the BRCA1/HP1γ interaction, which is important for the heterochromatic silencing of repetitive DNA elements.9 Furthermore, a recent systematic screen for molecules that interact with BRCA1/BARD1 using a yeast 2-hybrid system combined with tandem affinity purification followed by mass spectrometric analysis identified HP1γ, but not HP1α or HP1β.10 HP1α and HP1β play a critical role in maintaining constitutive heterochromatin, whereas HP1γ and H3K9me2 play roles in maintaining facultative heterochromatin or silencing euchromatin. Therefore, the preference for HP1γ in the BARD1 complex after DSB damage could indicate that euchromatic regions with more abundant damage may require heterochromatic structural changes in order to be repaired by HRR.
A variety of therapeutic agents that target histone modifications, including histone methyltransferase inhibitors (as described above) or histone deacetylase inhibitors, have been developed. The importance of heterochromatin-related factors in the BRCA1/BARD1-mediated DNA damage response suggests that these inhibitors may well affect chemosensitivity by exploiting HRR failure.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev 2011; 25:685-700; PMID:21406551; http://dx.doi.org/ 10.1101/gad.2011011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coleman KA, Greenberg RA. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem 2011; 286:13669-80; PMID:21335604; http://dx.doi.org/ 10.1074/jbc.M110.213728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y-H, Kuo C-Y, Stark JM, Shih H-M, Ann DK. HP1 promotes tumor suppressor BRCA1 functions during the DNA damage response. Nucleic Acids Res 2013; 8111:1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu W, Nishikawa H, Fukuda T, Vittal V, Asano M, Miyoshi Y, Klevit RE, Ohta T. Interaction of BARD1 and HP1 is required for BRCA1 retention at sites of DNA damage. Cancer Res 2015; PMID:25634209; http://dx.doi.org/ 10.1158/0008-5472.CAN-14-2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldeyron C, Soria G, Roche D, Cook AJL, Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol 2011; 193:81-95; PMID:21464229; http://dx.doi.org/ 10.1083/jcb.201101030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geuting V, Reul C, Löbrich M. ATM release at resected double-strand breaks provides heterochromatin reconstitution to facilitate homologous recombination. PLoS Genet 2013; 9:e1003667; PMID:23935532; http://dx.doi.org/ 10.1371/journal.pgen.1003667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khurana S, Kruhlak MJ, Kim J, Tran AD, Liu J, Nyswaner K, Shi L, Jailwala P, Sung MH, Hakim O, et al.. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep 2014; 8:1049-62; PMID:25131201; http://dx.doi.org/ 10.1016/j.celrep.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi JD, Park MA, Lee J-S. Suppression and recovery of BRCA1-mediated transcription by HP1γ via modulation of promoter occupancy. Nucleic Acids Res 2012; 40:11321-38; PMID:23074186; http://dx.doi.org/ 10.1093/nar/gks947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filipponi D, Muller J, Emelyanov A, Bulavin D V. Wip1 controls global heterochromatin silencing via ATM/BRCA1-dependent DNA methylation. Cancer Cell 2013; 24:528-41; PMID:24135283; http://dx.doi.org/ 10.1016/j.ccr.2013.08.022 [DOI] [PubMed] [Google Scholar]
- 10.Hill SJ, Rolland T, Adelmant G, Xia X, Owen MS, Dricot A, Zack TI, Sahni N, Jacob Y, Hao T, et al.. Systematic screening reveals a role for BRCA1 in the response to transcription-associated DNA damage. Genes Dev 2014; 28:1957-75; PMID:25184681; http://dx.doi.org/ 10.1101/gad.241620.114 [DOI] [PMC free article] [PubMed] [Google Scholar]


