Abstract
Parkinson's disease (PD) is a neurodegenerative disorder with poorly understood etiology. Increasing evidence suggests that age-dependent compromise of the maintenance of mitochondrial function is a key risk factor. Several proteins encoded by PD-related genes are associated with mitochondria including PTEN-induced putative kinase 1 (PINK1), which was first identified as a gene that is upregulated by PTEN. Loss-of-function PINK1 mutations induce mitochondrial dysfunction and, ultimately, neuronal cell death. To mitigate the negative effects of altered cellular functions cells possess a degradation mechanism called autophagy for recycling damaged components; selective elimination of dysfunctional mitochondria by autophagy is termed mitophagy. Our study indicates that autophagy and mitophagy are upregulated in PINK1-deficient cells, and is the first report to demonstrate efficient fluxes by one-step analysis. We propose that autophagy is induced to maintain cellular homeostasis under conditions of non-regulated mitochondrial quality control.
Keywords: autophagy, cancer, mitochondria, mitophagy, Parkinson's disease, PINK1
Abbreviations
- AP
autophagosome
- AL
autolysosome
- ALS
amyotrophic lateral sclerosis
- AV
autophagic vacuole
- Baf. A1
bafilomycin A1
- catD
cathepsin D
- GFP
green fluorescent protein
- HD
Huntington's disease
- LC3
microtubule-associated protein 1 light chain 3
- LTR
Lysotracker red
- MEF
mouse embryonic fibroblast
- mTOR
mammalian target of rapamycin
- MTS
mitochondrial targeting sequence
- PD
Parkinson's disease
- PINK1
PTEN-induced putative kinase 1
- ROS
reactive oxygen species
- TM
transmembrane domain
- VDAC
voltage-dependent anion channel
- WT
wild-type.
Introduction
Parkinson's disease (PD) is the second most common neurodegenerative disorder and presents a complex etiology involving genetic and environmental factors. Mitochondria are important organelles for cellular homeostasis and are involved in essential functions including energy generation and xenobiotic detoxification. Development of PD is highly influenced by the mitochondrial status and several PD-related genes are linked to this organelle, including PTEN-induced putative kinase 1 (PINK1), mutations of which cause autosomal recessive forms of early-onset Parkinsonism. The role of PINK1 has been mainly implicated in mitochondrial regulation because of its N-terminal mitochondrial targeting sequence (MTS), which allows selective localization in this organelle. Thus, PINK1 deficiency or PINK1 mutations result in mitochondrial dysfunction, i.e., morphological abnormalities, reduced mitochondrial membrane potential (Δψm), and increased reactive oxygen species (ROS) generation, thus enhancing neurodegeneration.1-6
Although PD and cancer are different human disorders, epidemiological evidence has shown that there is a low risk of cancer in PD patients.7 Interestingly, in 2001, the PINK1 gene was initially discovered in cancer cells as a gene that is upregulated by the major tumor suppressor PTEN.8 Survival kinases are upregulated in many cancer cells, resulting in cellular resistance to therapies. In this regard, a sensitized RNAi screen of the human kinome and phosphatome identified PINK1 as an essential element for survival.9 Moreover, recent reports have demonstrated that PINK1 is directly involved in mechanisms linked to cancer.10,11
Macroautophagy (referred to hereafter as autophagy) is a highly conserved process that allows cells to eliminate damaged organelles, such as mitochondria, or unfolded proteins. Briefly, this degradative pathway starts with the formation of a double-membraned vesicle called autophagosome (AP), where isoform II of microtubule-associated protein 1 light chain 3 (LC3) protein is localized. In parallel, p62 interacts with ubiquitinated proteins, generating aggregates that are sequestered in APs by p62–LC3-II interaction. Finally, APs fuse with lysosomes, allowing degradation of the cargo by lysosomal enzymes, such as cathepsin D (catD). Some of the interest in this topic is generated by the relationship between autophagy and age-related diseases.12 Defects in autophagy could contribute to accumulation of protein aggregates, which have been linked to neurodegenerative disorders such as PD, amyotrophic lateral sclerosis (ALS), and Huntington's disease (HD). Indeed, the number of autophagic vacuoles (AVs) is abnormally elevated in the brains of patients with PD.13 However, in cancer, autophagy presents a dual role, acting as a tumor suppressor by preventing accumulation of damaged cellular constituents and favoring tumor progression once malignant transformation occurs.14
The role of PINK1 in the autophagic process has mainly focused on mitophagy, in which selective degradation of mitochondria occurs through the regulation of Parkin, an E3 ubiquitin ligase linked to PD. Upon dissipation of Δψm, PINK1 is localized to damaged mitochondria,15-18 driven by calcium-dependent gene expression.19 This localization allows phosphorylation of Parkin on Ser65 by the PINK1 kinase activity, leading to its activation and the subsequent ubiquitination of mitochondrial substrates, such as voltage-dependent anion channel (VDAC) and mitofusins.15,20,21 However, the role of PINK1 in autophagy/mitophagy is still unknown.
The aim of this study was to characterize autophagic and mitophagic activities in PINK1-deficient cells and analyze the role of PINK1 in autophagy. We report that PINK1 is critical for autophagy/mitophagy processes; the absence of PINK1 induces activation of these degradative mechanisms in response to a compromised cell situation. We also show that expression of PINK1 (wild-type [WT] or kinase domain mutants) in PINK1 knockout cells restores autophagy flux to basal levels, further highlighting the essential role of this protein.
Results
Loss of PINK1 promotes autophagy and mitophagy
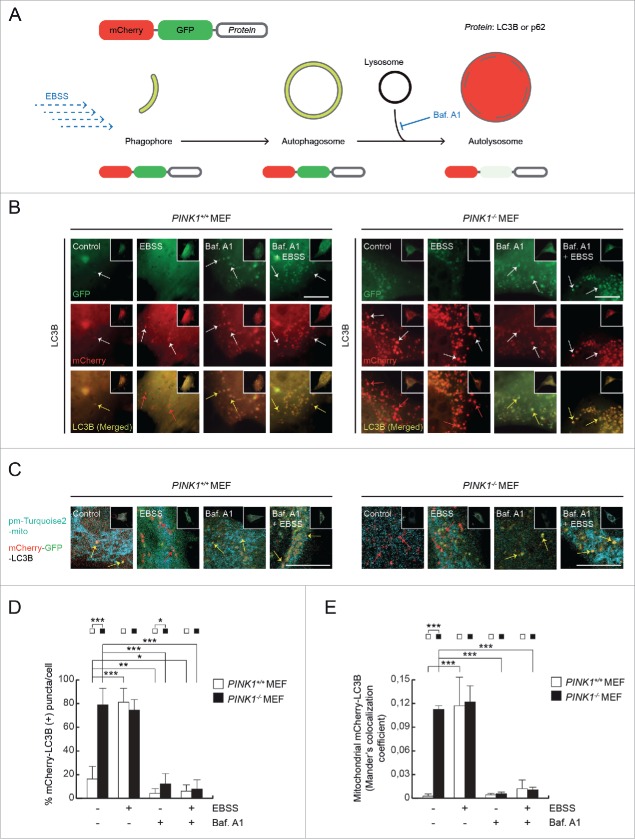
A previous report has shown that the number of GFP-LC3 puncta and their colocalization with mitochondria are increased in stable PINK1-deficient SH-SY5Y cells.22 To investigate whether loss of PINK1 promotes effective autophagy, we transfected PINK1+/+ and PINK1−/− mouse embryonic fibroblasts (MEFs) with tandem-tagged mCherry-GFP-LC3B, which displays green-red fluorescence in APs whereas the GFP fluorescence is quenched by the acidic pH of the autolysosomes (ALs) (Fig. 1A).23 In addition, cells were co-transfected with pmTurquoise2-mito, which labels mitochondria, to determine whether mitophagy occurred. PINK1−/− MEFs presented a higher number of exclusive mCherry-LC3B–positive puncta (78.39 ± 14.00% mCherry-LC3B (+) puncta/cell, compared to 15.88 ± 12.88% in PINK1+/+ MEFs; Fig. 1B and D), as well as enhanced localization of mCherry-LC3B in mitochondria (Mander's colocalization coefficient 0.111 ± 0.006 in PINK1−/− MEFs and 0.002 ± 0.002 in PINK1+/+ MEFs; Fig. 1C and E). These findings were confirmed by Western-blotting, which showed an increase in LC3-II levels in PINK1-deficient cells (Fig. S1A and D). Next, we examined whether the autophagy observed in PINK1−/− MEFs is regulated by a mTOR-dependent pathway. In this regard, there were no significant differences in the phosphorylation of mTOR at residue Ser2448 between the two cell lines (Fig. S1A and C).
Figure 1.
Autophagy and mitophagy induction in PINK1−/− MEFs. (A) Double mCherry-GFP tags were fused to LC3B or p62. The mCherry tag is acid-insensitive, whereas GFP is acid-sensitive. Consequently, phagophores and autophagosomes are visualized as yellow (green + red), and autolysosomes are labeled red. (B) Higher magnification images of the immunofluorescence of mCherry-GFP-LC3B (cells represented in the insets) in MEFs transiently cotransfected for 24 h and treated with EBSS, Baf. A1, or Baf. A1 + EBSS for 4 h. Arrows indicate the pattern of each condition. Scale bars: 10 μm. (C) Representative confocal microphotographs showing the localization of mCherry-LC3B (red) on mitochondria (pmTurquoise2-mito; blue). Arrows indicate LC3B puncta, highlighting the pattern of each condition. Scale bars: 10 μm. (D) Percentage of mCherry-LC3B (+) puncta per cell. (E) Confocal microscopy analysis of the mitochondrial localization of mCherry-LC3B using Mander's colocalization coefficient.
To confirm that autophagy is exacerbated in PINK1−/− MEFs, we examined selective p62 turnover by expression of mCherry-GFP-p62 in our model system. We observed that loss of PINK1 induced higher rates of mCherry-p62–positive puncta (69.09 ± 10.96% mCherry-p62–positive puncta/cell, compared to 15.87 ± 9.16% in PINK1+/+ MEFs; Fig. 2A and C). Similarly, PINK1−/− MEFs exhibited an increased number of GFP-LC3 puncta (110.10 ± 23.31 in PINK1−/− MEFs versus 8.10 ± 4.72 in PINK1+/+ MEFs; Fig. 2B and D) and Lysotracker Red (LTR)-stained puncta (160.10 ± 10.59 in PINK1−/− MEFs versus 24.00 ± 6.65 in PINK1+/+ MEFs; Fig. 2B and E). However, when we analyzed CatD levels, we did not observe differences in the precursor isoforms (data not shown) or the fully active mature isoform (mat-CatD) between these cell lines (Fig. S1B and E), indicating that loss of PINK1 does not affect expression levels of this lysosomal enzyme. As expected, the red fluorescence of LTR disappeared (Fig. 2B and E) and mat-CatD levels were reduced (Fig. S1B and E) after treatment with bafilomycin A1 (Baf. A1) as a result of lysosomal pH alkalinization. Baf. A1 is a specific inhibitor of vacuolar H+-ATPase that blocks the fusion of APs and lysosomes.
Figure 2.
Increased p62 turnover and GFP-LC3 and LTR puncta in PINK1−/− MEFs. (A) Higher magnification images showing immunofluorescence of mCherry-GFP-p62 (cells represented in the insets) on MEFs transiently transfected for 24 h and treated as described in Figure 1A. Arrows indicate the pattern of each condition. Scale bars: 10 μm. (B) Higher magnification images of immunofluorescence of GFP-LC3 and LTR (cells represented in the insets) on MEFs transiently transfected for 24 h, stained with LTR and treated as described in Figure 1A. Arrows indicate the pattern of each condition. Scale bars: 10 μm. (C) Percentage of mCherry-p62 (+) puncta per cell. (D) Number of GFP-LC3 puncta per cell. (E) Number of LTR puncta per cell.
To further reinforce these findings, all of the previous experiments were also performed in cells treated with Baf. A1 and/or under nutrient-deprivation conditions using Earle's Balanced Salt Solution (EBSS) medium. First, we incubated both cell lines in EBSS medium for different time points (2, 4, and 6 h) with no differences in autophagic response (data not shown), therefore we established a EBSS treatment time of 4 h for further experiments. The results obtained suggest that autophagy and mitophagy are enhanced in PINK1+/+ MEFs under conditions of starvation (as evidenced by increased levels of mCherry-LC3B (+), mCherry-p62 (+), GFP-LC3, and LTR puncta, and higher rates of mitochondrial mCherry-LC3B; Figs. 1, 2, and S1), although these processes are not further increased in PINK1−/− MEFs. Moreover, exposure to Baf. A1 blocked the different autophagic hallmarks analyzed in this study (low levels of mCherry-LC3B and mCherry-p62 (+) puncta and mitochondrial mCherry-LC3B localization, and LC3 accumulation) as observed by immunofluorescence (IF) and Western-blotting (Figs. 1, 2, and S1).
Several reports have shown that different models of PINK1 deficiency share certain characteristics, such as impaired mitochondrial function and increased sensitivity to multiple stressors.24-29 To study mitochondrial shape and the presence of AVs (phagosomes, APs, and ALs), we analyzed cells by transmission electron microscopy. We found that PINK1−/− MEFs exhibit fragmented mitochondria, dramatically reducing the mitochondrial area (13.59 ± 7.30 μm2 in PINK1−/− MEFs versus 26.81 ± 10.60 μm2 in PINK1+/+ MEFs; Fig. 3B), in addition to other morphologic parameters such as reduced perimeter (15.54 ± 4.81 μm in PINK1−/− MEFs versus 20.51 ± 3.86 μm in PINK1+/+ MEFs; Fig. 3C), Feret's diameter (5.90 ± 1.98 μm in PINK1−/− MEFs versus 7.34 ± 1.5 μm in PINK1+/+ MEFs; Fig. 3D), and circularity (0.69 ± 0.14 in PINK1−/− MEFs versus 0.77 ± 0.10 in PINK1+/+ MEFs; Fig. 3E). A remarkably lower mitochondrial mass was also observed in PINK1−/− MEFs (Fig. 3F and G). Furthermore, loss of PINK1 slightly increased the number of AVs (6.33 ± 4.03 AVs/cell, compared to 3.16 ± 3.00 AVs/cell in PINK1+/+ MEFs). In line with the results shown in previous figures (Figs. 1, 2 and S1), treatment with Baf. A1 and EBSS provides a clue to how this process is exacerbated in PINK1−/− MEFs; cells treated with Baf. A1 showed significant accumulation of AVs (19.88 ± 11.52 AVs/cell), whereas EBSS treatment did not dramatically enhance AV levels (EBSS-treated [9.86 ± 3.29 AVs/cell] versus non-treated cells [6.33 ± 4.03 AVs/cell] and Baf. A1 plus EBSS-treated [17.83 ± 10.68 AVs/cell] versus Baf. A1-treated cells [19.88 ± 11.52 AVs/cell]; Fig. 3A and H).
Figure 3.
Ultrastructural mitophagic/autophagic hallmarks are upregulated in PINK1−/− MEFs. (A) Higher magnifications views of the boxed regions are shown in insets. Scale bar: 10 μm. Mitochondria (M) and autophagic vacuoles (AVs) are shown in the enlarged view. Scale bar insets: 2 μm. (B–E) Analysis of mitochondrial parameters (number of mitochondria, 197 for PINK1+/+ MEFs and 130 for PINK1−/− MEFs) including area (B), perimeter (C), Feret's diameter (D) and circularity (E). (F) Representative western blot analysis of COX IV. Molecular mass is indicated in kilodaltons (kDa) next to the blots. (G) Graph showing relative amount of COX IV, in arbitrary units of intensity, determined by optical densitometry. Data were normalized to expression of α-tubulin. (H) Number of AVs per cross-sectioned cell. n = 25 (PINK1+/+ MEFs); 8 (PINK1+/+ MEFs + Baf. A1 + EBSS); 9 (PINK1−/− MEFs); 7 (PINK1−/− MEFs + EBSS); 8 (PINK1−/− MEFs + Baf. A1); 6 (PINK1−/− MEFs + Baf. A1 + EBSS). (I) Electron micrograph detail, showing different vesicles considered in this study as AVs.
PINK1 WT and kinase mutants restore autophagic response to basal levels in PINK1−/− MEFs
To elucidate the role of PINK1 in autophagy, we cotransfected PINK1+/+ and PINK1-/- MEFs with the mCherry-GFP-LC3B plasmid and the following PINK1 constructs: WT, K219M (artificial kinase dead mutant), E240K (PD-related mutant), L489P (PD-related mutant), and E240K/L489P (E/L, heterozygous mutant) (Fig. S2).30 We observed that the number of mCherry-LC3B–positive puncta in PINK1+/+ MEFs expressing the different constructs was similar to that in cells transfected with empty vector (pcDNA): pcDNA, 11.74 ± 1.83% mCherry-LC3B–positive puncta/cell; PINK1 WT, 7.58 ± 12.08%; K219M, 10.22 ± 9.08%; E240K 9.14 ± 6.19%; L489P, 7.08 ± 5.61%; E/L, 9.98 ± 8.07%. Interestingly, PINK1-deficient MEFs transfected with the PINK1 constructs showed fewer mCherry-LC3B–positive puncta (PINK1 WT, 8.86 ± 10.04%; K219M, 13.46 ± 8.75%, E240K, 11.61 ± 4.64%; L489P, 10.00 ± 11.55%; E/L, 11.36 ± 10.92%) than those transfected with pcDNA (54.38 ± 10.66%), which was significantly higher than the value for PINK1+/+ MEFs transfected with pcDNA and concordant with the results shown in Figure 1 (Fig. 4A and B). We analyzed mCherry-GFP-LC3B puncta in the empty vector (pcDNA) condition to show that differences in autophagic flux are due only to overexpression of PINK1 constructs. Moreover, we confirmed that transfection vehicle (Attractene Transfection Reagent) and pcDNA did not affect cell viability (data not shown).
Figure 4.

Transfection of PINK1−/− MEFs with PINK1 WT or kinase mutants reduces autophagy response to basal levels. (A) Representative immunofluorescence images of mCherry-GFP-LC3B and V5-tagged PINK1 (represented in the insets) in MEFs after transient cotransfection for 24 h. Arrows indicate the pattern of each condition. Scale bars: 10 μm. (B) Percentage of mCherry-LC3B (+) puncta per cell.
Discussion
Based on epidemiological studies, one of the greatest risk factors for the development of PD is aging.31 Mitochondrial integrity may be the underlying link in this close relationship. Abundant evidence implicates oxidative stress in PD pathogenesis, and mitochondrial dysfunction is one of the primary sources of such stress.32,33 Mitochondrial alterations accumulate progressively and include mutations in mitochondrial DNA and decreased mitochondrial respiration.34,35 Several PD-related proteins have been associated with mitochondria, with PINK1 and Parkin being the best studied because mutations in these genes are the most common causes of recessive PD.
Autophagy is implicated in the clearance of damaged organelles and misfolded proteins. A growing body of evidence shows that monitoring LC3 and p62 fluxes in the autophagy pathway is a powerful tool for analyzing its activity, as recently demonstrated in vivo in the nervous system of mice.36 Previous studies have suggested analysis of mitophagy induction by immunofluorescence, but this was necessarily accompanied by ultrastructural analysis to demonstrate mitophagic flux.37 Recently, other studies have developed tandem mCherry–GFP tags to monitor mitophagy.38 Here, we have developed a cutting-edge method to characterize autophagic and mitophagic efficiencies by immunofluorescence.
Increased GFP-LC3 puncta and LC3-II protein levels have been described in PINK1-deficient cells and animals, suggesting that autophagy is upregulated.22,39,40 Here, we provide the first lines of evidence for autophagy and mitophagy fluxes in a model of loss of PINK1 and for autophagy flux in PINK1 kinase mutants by immunofluorescence. Our data suggest that autophagy in PINK1−/− MEFs occurs by an mTOR-independent pathway. Remarkably, this process is so exacerbated in these cells that EBSS treatment does not increase its induction. Interestingly, in PINK1-deficient cells basal and starvation-induced autophagic levels are quite similar, meaning that the autophagy pathway might be initially saturated.
The PINK1/Parkin-dependent mitophagy pathway is well known.15-18 Furthermore, it has been shown that PINK1 kinase mutants and variants lacking the MTS compromise the recruitment of Parkin to mitochondria and, therefore, the removal of mitochondria by mitophagy.15,41 However, in Drosophila melanogaster, Parkin exhibits residual activity in the absence of PINK1, which could be sufficient to maintain basal mitophagy. In this regard, a recent study has revealed that the Beclin-1/Parkin interaction facilitates Parkin translocation to mitochondria, and also occurs in PINK1-deficient MEFs.42 Moreover, cytosolic cleaved PINK1 interacts with Parkin, repressing its mitochondrial translocation and subsequent mitophagy.43 These discoveries could reinforce our proposed hypothesis that the absence of PINK1 (and thus also cytosolic cleaved PINK1) could not prevent the translocation of Parkin, thus allowing the elimination of damaged mitochondria.
It is also possible that the selective removal of mitochondria by mitophagy might be mediated by other proteins that target this organelle for its degradation. Consistent with this possibility, a recent study has suggested that a compensatory mechanism of protein turnover masks mitophagy defects in PINK1 mutants.44 Furthermore, it has been shown that mitophagy might also occur by a PINK1/Parkin-independent pathway.38
A previous report showed that WT PINK1 reduces neuronal apoptosis, whereas the same PINK1 mutants that we used in our study (K219M, E240K, L489P, and E/L) abrogated this cytoprotective effect.45 However, the present data indicate that the presence of PINK1 (either WT or kinase mutants) was sufficient to abolish the upregulation of the autophagic machinery and restore basal levels in PINK1−/− MEFs (Fig. 5), suggesting that this degradative mechanism is controlled by PINK1 in a kinase-independent manner. Future studies are required to understand whether mitophagy is modulated by different kinase mutants in our cell model, as well as whether mitochondrial PINK1 localization is essential for autophagy induction.
Figure 5.

Proposed model of autophagy in PINK1−/− MEFs. PINK1 loss-of-function results in exacerbated autophagy, characterized by mCherry-LC3B and mCherry-p62 accumulation, an increase in GFP-LC3 and LTR puncta, and increased levels of LC3-II protein and AVs. Moreover, PINK1 deficiency results ineffective mitophagy, as determined by increased colocalization of mCherry-LC3B with mitochondria and fragmented mitochondria. Overexpression of PINK1 (WT or kinase mutants) in PINK1−/− MEFs restores autophagy to basal levels.
Based on our data, we hypothesize that PINK1 is a key molecular sensor, and that its activity might be critical for mitochondrial quality control and therefore cellular homeostasis, both of which are highly vulnerable during aging. Autophagy upregulation as a strategy to treat neurodegenerative diseases has been tested in several cellular and animal models with satisfactory results, such as reduced protein aggregation and cell death.46-48 Currently, the autophagy inducers tamoxifen and rilmenidine are in Phase II/I and Phase I clinical trials for ALS and HD, respectively.49 Thus, PINK1 deficiency contributes to compromised cellular homeostasis, and autophagy may be increased as an adaptive response for cell survival. Further investigations are needed for consideration of autophagy upregulation as an effective therapeutic target for the treatment of neurodegenerative disorders, including PD. Moreover, autophagy promotes oncosuppressive functions, preventing malignant transformation in healthy cells. Conversely, once malignant transformation occurs, there is substantial evidence that autophagy supports tumor progression and resistance to therapy. In this regard, there are several ongoing Phase II/I clinical trials based on a combination of the autophagy inhibitor hydroxychloroquine and anticancer therapy (radiation therapy or chemotherapy).14 It is important to mention that cancer cells are highly dependent on functional mitophagy to maintain optimal energetic requirements. In this scenario, modulation of PINK1 activity could be considered as a potential target to interfere with tumor progression.
In summary, PINK1 is involved in several signaling networks that are essential for cellular homeostasis, thus increasing the risk of developing separate disorders.50 Because of its dual role, PINK1 is a promising therapeutic strategy for PD and cancer, but further information about its function is required in order to better understand both diseases and ultimately decrease their progression.
Materials and Methods
Cell culture and treatments
PINK1+/+ and PINK1−/− MEFs were cultured in Dulbecco's Modified Eagle Medium (DMEM)-High Glucose (Sigma-Aldrich, D6546) supplemented with 10% fetal bovine serum (Sigma-Aldrich, F7524), 1% L-glutamine (Sigma-Aldrich, G7513), and penicillin–streptomycin (Hyclone, Thermo Fisher Scientific, SV30010). Cells were seeded at a density of 5 × 105 in 175-cm2 tissue culture flasks (Sigma-Aldrich, CLS431080) and incubated at 37°C under saturating humidity in 5% CO2/95% air. Cells were plated at 2.5 × 105 cells/mL 24 h before starting the different treatments. For starvation experiments, MEFs were washed and incubated with EBSS medium (Sigma-Aldrich, E2888) for 4 h. To block autophagy flux, cells were pretreated with 100 nM Baf. A1 (LC Laboratories, B-1080) for 1 h.
Plasmid transfection
MEFs were transiently transfected using Attractene Transfection Reagent (Qiagen, 301005) according to the manufacturer's protocol. The constructs used were as follows: mCherry-GFP-LC3B and mCherry-GFP-p62 (Dr. Terje Johansen, gift), GFP-LC3 (Dr. Tamotsu Yoshimori, gift), pmTurquoise2-mito (Addgene plasmid 36208), and V5-tagged PINK1 (WT, K219M, E240K, L489P, and E240K/L489P).23,45,51,52 Transfection efficiency was 75–80% in both types of MEFs.
Western-blotting
Cells were washed with 1×PBS and lysed in buffer containing 100 mM Tris-HCl (pH 7.4), 300 mM NaCl, and 0.5% NP-40 (Roche, 11332473001), supplemented with protease inhibitors cocktail (cOmplete, Mini, EDTA-free, Roche, 11836170001) and phosphatase inhibitors cocktail (PhosSTOP, Roche, 04906837001). Proteins were resolved by 12% SDS-gel electrophoresis as previously described.19 Blots were probed with antibodies against p-mTOR (Ser2448) (Cell Signaling Technology, #2971), mTOR (Cell Signaling Technology, #2972), LC3B (Cell Signaling Technology, #2775), catD (Santa Cruz Biotechnology, clone C-20), subunit IV of cytochrome c oxidase (COX IV, Abcam, ab14744), and α-tubulin (Santa Cruz Biotechnology, clone TU-02).
Immunofluorescent staining
For lysosome staining, cells were incubated with LTR (1 μM, Molecular Probes®, L-7528) for 15 minutes (min) at 37°C. Slides were viewed using an inverted fluorescence microscope (IX51, Olympus) or a confocal scanning laser (Nikon A1) coupled to an inverted microscope (Eclipse Ti, Nikon). For the detection of V5-tagged PINK1, cells were incubated with monoclonal anti-V5 antibody (Novex®, Life Technologies, R960–25) and then stained with Alexa Fluor® 633 anti-mouse secondary antibody (Molecular Probes®, Life Technologies, A-21052). The autophagic flux was measured using tandem-tagged mCherry-GFP constructs (LC3B and p62) by determining the percentage of mCherry-positive puncta (ALs) among the total number of mCherry puncta (mCherry-GFP [APs] and mCherry [ALs]). Quantitative analyses of mCherry-GFP-LC3B puncta, mCherry-GFP-p62 puncta, GFP-LC3 puncta, and LTR puncta were performed using Ifdotmeter® software. At least 150 cells were counted per condition.
Transmission electron microscopy
Cells were fixed with freshly prepared 2% glutaraldehyde in 0.4 M HEPES pH 7.2 for 2 h. After post-fixation in 1% OsO4 for 1 h at 4°C, the cells were dehydrated in a graded acetone series, embedded in EPON 812 resin, and polymerized for 48 h at 60°C. Ultrathin sections (60–70 nm) were cut using an ultramicrotome (EM UC6, Leica) and deposited on formvar-carbon coated grids. Sections were contrasted with 2% uranyl acetate for 40 min at 4°C and examined using a scanning electron microscope (S-3600N, Hitachi). Mitochondrial parameters (area, perimeter, Feret's diameter, and circularity) were determined using ImageJ software.
Statistical analyses
Each experiment was repeated at least 3 times, with a satisfactory correlation between the results of individual experiments. The data shown are those of a representative experiment. Data were evaluated with 2-tailed unpaired Student's t-test and ANOVA test, and all comparisons with a p value less than 0.05 (p < 0.05) were considered statistically significant. p < 0.001, p < 0.01, and p < 0.05 are indicated with triple, double, or single asterisks, respectively. Non-significant results are not indicated in the figures. The data are expressed as the mean ± the standard error of the mean (SEM).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank R. Ronco, P. Delgado, and G. Martínez for invaluable technical assistance and C. Patiño (Servicio de Microscopía Electrónica, Centro Nacional de Biotecnología, Madrid, Spain) for the preparation of electron microscopy samples. The authors also thank FUNDESALUD. We thank George Auburger (Experimental Neurology, Goethe University Medical School, Frankfurt am Main, Germany) for PINK1+/+ and PINK1−/− cells. Moreover, we thank Dr. Terje Johansen (Molecular Cancer Research group, Institute of Medical Biology, University of Tromsø, Norway) and Tamotsu Yoshimori (Department of Genetics, Osaka University Graduate School of Medicine, Japan) for kindly providing the corresponding constructs.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by Instituto de Salud Carlos III [PI11/00040, PI12/02280, PI14/00170 (co-financed by European Union FEDER funds), CB06/05/0041], Gobierno de Extremadura [GR10054]. R.G-S. was supported by an “Acción III” postdoctoral contract (Universidad de Extremadura, Spain), E.P-E. was supported by a predoctoral fellowship (CIBERNED, Instituto de Salud Carlos III, Spain), M.R-A. was supported by a FPU predoctoral fellowship (Ministerio de Educación, Cultura y Deporte, Spain), F.E.M-C. was supported by a RETICEF [RD12–0043–0016], BFU2011–24365 and GR 10009 JEX, A.T. was supported by operating grants from the Parkinson Society of Canada and Canadian Institutes of Health Research, and R.A.G-P. was supported by a talent research contract (Gobierno de Extremadura, Spain).
References
- 1.Wood-Kaczmar A, Gandhi S, Yao Z, Abramov AY, Miljan EA, Keen G, Stanyer L, Hargreaves I, Klupsch K, Deas E, et al.. PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS One 2008; 3:e2455; PMID:18560593; http://dx.doi.org/ 10.1371/journal.pone.0002455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moisoi N, Fedele V, Edwards J, Martins LM. Loss of PINK1 enhances neurodegeneration in a mouse model of Parkinson disease triggered by mitochondrial stress. Neuropharmacology 2014; 77:350-7; PMID:24161480; http://dx.doi.org/ 10.1016/j.neuropharm.2013.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang HL, Chou AH, Wu AS, Chen SY, Weng YH, Kao YC, Yeh TH, Chu PJ, Lu CS. PARK6 PINK1 mutants are defective in maintaining mitochondrial membrane potential and inhibiting ROS formation of substantia nigra dopaminergic neurons. Biochim Biophys Acta 2011; 1812:674-84; PMID:21421046; http://dx.doi.org/ 10.1016/j.bbadis.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 4.Amo T, Sato S, Saiki S, Wolf AM, Toyomizu M, Gautier CA, Shen J, Ohta S, Hattori N. Mitochondrial membrane potential decrease caused by loss of PINK1 is not due to proton leak, but to respiratory chain defects. Neurobiol Dis 2011; 41:111-8; PMID:20817094; http://dx.doi.org/ 10.1016/j.nbd.2010.08.027 [DOI] [PubMed] [Google Scholar]
- 5.Exner N, Treske B, Paquet D, Holmstrom K, Schiesling C, Gispert S, Carballo-Carbajal I, Berg D, Hoepken HH, Gasser T, et al.. Loss-of-function of human PINK1 results in mitochondrial pathology and can be rescued by parkin. J Neurosci 2007; 27:12413-8; PMID:17989306; http://dx.doi.org/ 10.1523/JNEUROSCI.0719-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abramov AY, Gegg M, Grunewald A, Wood NW, Klein C, Schapira AH. Bioenergetic consequences of PINK1 mutations in Parkinson disease. PLoS One 2011; 6:e25622; PMID:22043288; http://dx.doi.org/ 10.1371/journal.pone.0025622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ong EL, Goldacre R, Goldacre M. Differential risks of cancer types in people with Parkinson's disease: a national record-linkage study. Eur J Cancer 2014; 50:2456-62; PMID:25065294; http://dx.doi.org/ 10.1016/j.ejca.2014.06.018 [DOI] [PubMed] [Google Scholar]
- 8.Unoki M, Nakamura Y. Growth-suppressive effects of BPOZ and EGR2, two genes involved in the PTEN signaling pathway. Oncogene 2001; 20:4457-65; PMID:11494141; http://dx.doi.org/ 10.1038/sj.onc.1204608 [DOI] [PubMed] [Google Scholar]
- 9.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol 2005; 7:591-600; PMID:15864305; http://dx.doi.org/ 10.1038/ncb1258 [DOI] [PubMed] [Google Scholar]
- 10.Martin SA, Hewish M, Sims D, Lord CJ, Ashworth A. Parallel high-throughput RNA interference screens identify PINK1 as a potential therapeutic target for the treatment of DNA mismatch repair-deficient cancers. Cancer Res 2011; 71:1836-48; PMID:21242281; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-2836 [DOI] [PubMed] [Google Scholar]
- 11.O'Flanagan CH, Morais VA, Wurst W, De Strooper B, O'Neill C. The Parkinson gene PINK1 regulates cell cycle progression and promotes cancer-associated phenotypes. Oncogene 2015; 34:1363-74; PMID:24681957; http://dx.doi.org/ 10.1038/onc.2014.81 [DOI] [PubMed] [Google Scholar]
- 12.He LQ, Lu JH, Yue ZY. Autophagy in ageing and ageing-associated diseases. Acta Pharmacol Sin 2013; 34:605-11; PMID:23416930; http://dx.doi.org/ 10.1038/aps.2012.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anglade P, Vyas S, Javoy-Agid F, Herrero MT, Michel PP, Marquez J, Mouatt-Prigent A, Ruberg M, Hirsch EC, Agid Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson disease. Histol Histopathol 1997; 12:25-31; PMID:9046040 [PubMed] [Google Scholar]
- 14.Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V, et al.. Autophagy in malignant transformation and cancer progression. EMBO J 2015; 34:856-80; PMID:25712477; http://dx.doi.org/ 10.15252/embj.201490784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 2010; 12:119-31; PMID:20098416; http://dx.doi.org/ 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- 16.Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 2010; 8:e1000298; PMID:20126261; http://dx.doi.org/ 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, et al.. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 2010; 189:211-21; PMID:20404107; http://dx.doi.org/ 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al.. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A 2010; 107:378-83; PMID:19966284; http://dx.doi.org/ 10.1073/pnas.0911187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez-Sanchez R, Gegg ME, Bravo-San Pedro JM, Niso-Santano M, Alvarez-Erviti L, Pizarro-Estrella E, Gutierrez-Martin Y, Alvarez-Barrientos A, Fuentes JM, Gonzalez-Polo RA, et al.. Mitochondrial impairment increases FL-PINK1 levels by calcium-dependent gene expression. Neurobiol Dis 2014; 62:426-40; PMID:24184327; http://dx.doi.org/ 10.1016/j.nbd.2013.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet 2010; 19:4861-70; PMID:20871098; http://dx.doi.org/ 10.1093/hmg/ddq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iguchi M, Kujuro Y, Okatsu K, Koyano F, Kosako H, Kimura M, Suzuki N, Uchiyama S, Tanaka K, Matsuda N. Parkin-catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation. J Biol Chem 2013; 288:22019-32; PMID:23754282; http://dx.doi.org/ 10.1074/jbc.M113.467530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dagda RK, Cherra SJ 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem 2009; 284:13843-55; PMID:19279012; http://dx.doi.org/ 10.1074/jbc.M808515200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131-45; PMID:17580304; http://dx.doi.org/ 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- 24.Samann J, Hegermann J, von Gromoff E, Eimer S, Baumeister R, Schmidt E. Caenorhabditits elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. J Biol Chem 2009; 284:16482-91; PMID:19251702; http://dx.doi.org/ 10.1074/jbc.M808255200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 2006; 441:1162-6; PMID:16672981; http://dx.doi.org/ 10.1038/nature04779 [DOI] [PubMed] [Google Scholar]
- 26.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, et al.. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 2006; 441:1157-61; PMID:16672980; http://dx.doi.org/ 10.1038/nature04788 [DOI] [PubMed] [Google Scholar]
- 27.Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A 2008; 105:11364-9; PMID:18687901; http://dx.doi.org/ 10.1073/pnas.0802076105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haque ME, Mount MP, Safarpour F, Abdel-Messih E, Callaghan S, Mazerolle C, Kitada T, Slack RS, Wallace V, Shen J, et al.. Inactivation of Pink1 gene in vivo sensitizes dopamine-producing neurons to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and can be rescued by autosomal recessive Parkinson disease genes, Parkin or DJ-1. J Biol Chem 2012; 287:23162-70; PMID:22511790; http://dx.doi.org/ 10.1074/jbc.M112.346437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sallinen V, Kolehmainen J, Priyadarshini M, Toleikyte G, Chen YC, Panula P. Dopaminergic cell damage and vulnerability to MPTP in Pink1 knockdown zebrafish. Neurobiol Dis 2010; 40:93-101; PMID:20600915; http://dx.doi.org/ 10.1016/j.nbd.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 30.Rogaeva E, Johnson J, Lang AE, Gulick C, Gwinn-Hardy K, Kawarai T, Sato C, Morgan A, Werner J, Nussbaum R, et al.. Analysis of the PINK1 gene in a large cohort of cases with Parkinson disease. Arch Neurol 2004; 61:1898-904; PMID:15596610 [DOI] [PubMed] [Google Scholar]
- 31.Tanner CM, Goldman SM. Epidemiology of Parkinson's disease. Neurol Clin 1996; 14:317-35; PMID:8827174; http://dx.doi.org/ 10.1016/S0733-8619(05)70259-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenner P. Oxidative stress in Parkinson's disease. Ann Neurol 2003; 53 Suppl 3:S26-36; discussion S-8; PMID:12666096; http://dx.doi.org/ 10.1002/ana.10483 [DOI] [PubMed] [Google Scholar]
- 33.Brunk UT, Terman A. The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur J Biochem 2002; 269:1996-2002; PMID:11985575; http://dx.doi.org/ 10.1046/j.1432-1033.2002.02869.x [DOI] [PubMed] [Google Scholar]
- 34.Richter C. Oxidative damage to mitochondrial DNA and its relationship to ageing. Int J Biochem Cell Biol 1995; 27:647-53; PMID:7648420; http://dx.doi.org/ 10.1016/1357-2725(95)00025-K [DOI] [PubMed] [Google Scholar]
- 35.Hansford RG, Castro F. Age-linked changes in the activity of enzymes of the tricarboxylate cycle and lipid oxidation, and of carnitine content, in muscles of the rat. Mech Ageing Dev 1982; 19:191-200; PMID:6287124; http://dx.doi.org/ 10.1016/0047-6374(82)90010-0 [DOI] [PubMed] [Google Scholar]
- 36.Castillo K, Valenzuela V, Matus S, Nassif M, Onate M, Fuentealba Y, Encina G, Irrazabal T, Parsons G, Court FA, et al.. Measurement of autophagy flux in the nervous system in vivo. Cell Death Dis 2013; 4:e917; PMID:24232093; http://dx.doi.org/ 10.1038/cddis.2013.421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu J, Dagda RK, Chu CT. Monitoring mitophagy in neuronal cell cultures. Methods Mol Biol 2011; 793:325-39; PMID:21913110; http://dx.doi.org/ 10.1007/978-1-61779-328-8_21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen GF, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep 2013; 14:1127-35; PMID:24176932; http://dx.doi.org/ 10.1038/embor.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi Z, Yang W, Liu Y, Cui T, Gao H, Duan C, Lu L, Zhao C, Zhao H, Yang H. Loss of PINK1 function decreases PP2A activity and promotes autophagy in dopaminergic cells and a murine model. Neurochem Int 2011; 59:572-81; PMID:21672589; http://dx.doi.org/ 10.1016/j.neuint.2011.03.023 [DOI] [PubMed] [Google Scholar]
- 40.Liu S, Lu B. Reduction of protein translation and activation of autophagy protect against PINK1 pathogenesis in Drosophila melanogaster. PLoS Genet 2010; 6:e1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geisler S, Holmstrom KM, Treis A, Skujat D, Weber SS, Fiesel FC, Kahle PJ, Springer W. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy 2010; 6:871-8; PMID:20798600; http://dx.doi.org/ 10.4161/auto.6.7.13286 [DOI] [PubMed] [Google Scholar]
- 42.Choubey V, Cagalinec M, Liiv J, Safiulina D, Hickey MA, Kuum M, Liiv M, Anwar T, Eskelinen EL, Kaasik A. BECN1 is involved in the initiation of mitophagy: it facilitates PARK2 translocation to mitochondria. Autophagy 2014; 10:1105-19; PMID:24879156; http://dx.doi.org/ 10.4161/auto.28615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fedorowicz MA, de Vries-Schneider RL, Rub C, Becker D, Huang Y, Zhou C, Alessi Wolken DM, Voos W, Liu Y, Przedborski S. Cytosolic cleaved PINK1 represses Parkin translocation to mitochondria and mitophagy. EMBO Rep 2014; 15:86-93; PMID:24357652; http://dx.doi.org/ 10.1002/embr.201337294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, Pallanck LJ. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci U S A 2013; 110:6400-5; PMID:23509287; http://dx.doi.org/ 10.1073/pnas.1221132110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petit A, Kawarai T, Paitel E, Sanjo N, Maj M, Scheid M, Chen F, Gu Y, Hasegawa H, Salehi-Rad S, et al.. Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J Biol Chem 2005; 280:34025-32; PMID:16079129; http://dx.doi.org/ 10.1074/jbc.M505143200 [DOI] [PubMed] [Google Scholar]
- 46.Pan T, Rawal P, Wu Y, Xie W, Jankovic J, Le W. Rapamycin protects against rotenone-induced apoptosis through autophagy induction. Neuroscience 2009; 164:541-51; PMID:19682553; http://dx.doi.org/ 10.1016/j.neuroscience.2009.08.014 [DOI] [PubMed] [Google Scholar]
- 47.Xiong N, Jia M, Chen C, Xiong J, Zhang Z, Huang J, Hou L, Yang H, Cao X, Liang Z, et al.. Potential autophagy enhancers attenuate rotenone-induced toxicity in SH-SY5Y. Neuroscience 2011; 199:292-302; PMID:22056603; http://dx.doi.org/ 10.1016/j.neuroscience.2011.10.031 [DOI] [PubMed] [Google Scholar]
- 48.Malagelada C, Jin ZH, Jackson-Lewis V, Przedborski S, Greene LA. Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson's disease. J Neurosci 2010; 30:1166-75; PMID:20089925; http://dx.doi.org/ 10.1523/JNEUROSCI.3944-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kroemer G. Autophagy: a druggable process that is deregulated in aging and human disease. J Clin Invest 2015; 125:1-4; PMID:25654544; http://dx.doi.org/ 10.1172/JCI78652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Triplett J, Zhang Z, Sultana R, Cai J, Klein JB, Bueler H, Butterfield DA. Quantitative Expression Proteomics and Phosphoproteomics Profile of Brain from PINK1 Knockout Mice: Insights into Mechanisms of Familial Parkinson Disease. J Neurochem 2015; 133(5):750-65 [DOI] [PubMed] [Google Scholar]
- 51.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720-8; PMID:11060023; http://dx.doi.org/ 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goedhart J, von Stetten D, Noirclerc-Savoye M, Lelimousin M, Joosen L, Hink MA, van Weeren L, Gadella TW Jr., Royant A. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93%. Nat Commun 2012; 3:751; PMID:22434194; http://dx.doi.org/ 10.1038/ncomms1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.