ABSTRACT
Histone variants are attracting attention in the field of cancer epigenetics. Our study has established a novel role for the uncharacterized histone variant H2A.Z.2 as a driver of malignant melanoma. H2A.Z.2 promotes cellular proliferation by recruiting BRD2 and E2F1 to E2F target genes and facilitating their transcription. High H2A.Z.2 expression correlates with poor survival in patients, and its depletion sensitizes cells to chemotherapy and targeted therapies.
KEYWORDS: BET, BRD2, chromatin, E2F, H2A.Z, histone variants, melanoma
Malignant melanoma is the most lethal form of skin cancer and shows an increasing incidence. Despite the emergence of effective therapies, metastatic melanoma remains a largely incurable disease. Although recent advances in targeted or immune therapies have raised hope for a “melanoma cure”, they are only effective in distinct subsets of patients, cause significant toxicity, and/or are overcome by the rapid emergence of drug resistance.1,2 Thus, investigation of alternative approaches is essential. Although much effort has been expended in characterizing and targeting the genetic alterations in melanoma,3 studies have only just begun to shed light on the importance of epigenetic regulation in melanoma pathogenesis.
Histone variants, which are sequence and structural variants of canonical histones, replace their conventional counterparts within the nucleosome through the action of dedicated factors known as histone chaperones.4 We have previously identified the histone variant macroH2A as a tumor suppressor in melanoma that directly regulates expression of the cyclin-dependent kinase CDK8.5 In addition, the chromatin remodeler ATRX, a negative regulator of macroH2A, can be used as a prognostic marker of melanoma.6 Recently, we have reported a role for another variant of the H2A family, namely H2A.Z.2, in promoting proliferation of melanoma cells.7
H2A.Z is a highly conserved H2A variant with a well-established role in transcriptional regulation. Two distinct H2A.Z isoforms, H2A.Z.1 and H2A.Z.2, have been identified in the vertebrate genome as products of 2 non-allelic genes that differ by only 3 amino acids. Isoform-specific functions remain unclear but H2A.Z.1 mouse knockout studies suggest that the 2 genes are non-redundant. Previous studies have shown H2A.Z to be overexpressed and play a role in other tumor types; however, these studies either focused solely on H2A.Z.1 or did not clearly distinguish between the isoforms.5
We have demonstrated that both H2A.Z isoforms are highly expressed in melanoma and that high expression levels correlate with shorter survival of patients. Although we do not exclude a role for H2A.Z.1 in melanoma pathogenesis, we identified a distinct role for H2A.Z.2 as a mediator of melanoma cell proliferation. To our knowledge, this is the first report to define a biologic function for H2A.Z.2 in any tumor type. Through loss-of-function approaches we showed that H2A.Z.2 exerts a key role in promoting cell cycle progression by controlling the transcriptional output of E2F target genes. Our integrated genomic and transcriptomic analyses further revealed that these genes are highly expressed and display a unique signature of H2A.Z occupancy—they are highly enriched for H2A.Z at the promoter and depleted across the gene body. Notably, this specific pattern of H2A.Z distribution has been previously observed in plants, yeast, and worms, where it also correlates with high gene expression levels.
To further dissect H2A.Z function in melanoma, we performed quantitative mass spectrometry and identified the bromodomain and extraterminal domain (BET) protein BRD2 as an H2A.Z interacting protein. BET proteins (BRD2, BRD3, BRD4, and BRDT) bind to acetylated lysine residues in histones and function as scaffolds to recruit chromatin modifying enzymes and transcription factors, thereby coupling histone acetylation to transcription.8 BRD2 is also overexpressed in melanoma, and its depletion causes a G1/S arrest, leading us to hypothesize that H2A.Z.2 and BRD2 work together to promote expression of E2F target genes. Indeed, chromatin immunoprecipitation experiments revealed that H2A.Z, BRD2, and E2F1 co-occupy the promoters of H2A.Z.2-regulated cell cycle genes and, furthermore, that BRD2 and E2F1 are recruited in an H2A.Z.2-dependent manner (Fig. 1). Importantly, we found evidence of hyperactivation of the H2A.Z.2-BRD2-E2F1 axis in a subset of melanoma patients, but not in benign nevi. Based on our observation that H2A.Z.2 loss has a dramatic effect on histone acetylation and BRD2 cellular levels, we queried whether its deficiency could cooperate with BET inhibition.9 Intriguingly, whereas in control cells the BET inhibitor JQ110 exerts a cytostatic effect, cells depleted of H2A.Z.2 that are treated with the same doses of JQ1 die by apoptosis. In addition, H2A.Z.2 deficiency sensitizes melanoma cells to chemotherapy and to MEK inhibitors.
Figure 1.
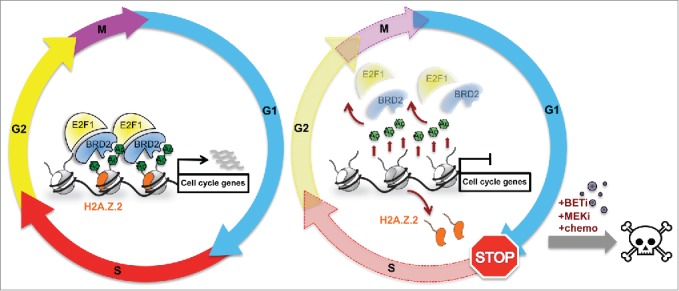
A model for the function of the H2A.Z.2-BRD2-E2F1 axis in melanoma cells. H2A.Z.2 stabilizes acetylation of histones in chromatin, thus facilitating recruitment of BRD2 and its associated E2F1 to E2F target genes. This in turn promotes expression of cell cycle regulatory genes and progression through the cell cycle. Loss of H2A.Z.2 results in destabilization of acetylated histones and consequently BRD2 and E2F1 are no longer bound to chromatin. E2F target genes are downregulated and cell cycle progression is impaired. Combination of H2A.Z.2 depletion and targeted therapy (BETi, MEKi) or chemotherapy results in cell death by apoptosis.
Collectively, our findings implicate H2A.Z.2 as a mediator of cell proliferation and drug sensitivity in malignant melanoma. Because histone modification and deposition are reversible processes, our study holds invaluable therapeutic potential for this highly intractable cutaneous neoplasm.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Kaufman HL, Kirkwood JM, Hodi FS, Agarwala S, Amatruda T, Bines SD, Clark JI, Curti B, Ernstoff MS, Gajewski T, et al.. The Society for Immunotherapy of Cancer consensus statement on tumour immunotherapy for the treatment of cutaneous melanoma. Nat Rev Clin Oncol 2013; 10, 588-98; PMID:23982524; http://dx.doi.org/ 10.1038/nrclinonc.2013.153 [DOI] [PubMed] [Google Scholar]
- 2.Lito P, Rosen N, Solit DB. Tumor adaptation and resistance to RAF inhibitors. Nat Med 2013; 19:1401-9; PMID:24202393; http://dx.doi.org/ 10.1038/nm.3392 [DOI] [PubMed] [Google Scholar]
- 3.The Cancer Genome Atlas Network . Genomic classification of cutaneous melanoma. Cell 2015; 161(7):1681-96; PMID:26091043; http://dx.doi.org/ 10.1016/j.cell.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vardabasso C, Hasson D, Ratnakumar K, Chung CY, Duarte LF, Bernstein E. Histone variants: emerging players in cancer biology. Cell Mol Life Sci 2013; 71(3):379-404; PMID:23652611; http://dx.doi.org/ 10.1007/s00018-013-1343-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, Menendez S, Vardabasso C, Leroy G, Vidal CI, et al.. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nat 2010; 68(7327)pp. 1105-9; PMID:21179167; http://dx.doi.org/ 10.1038/nature09590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qadeer ZA, Harcharik S, Valle-Garcia D, Chen C, Birge MB, Vardabasso C, Duarte LF, Bernstein E. Decreased expression of the chromatin remodeler ATRX associates with melanoma progression. J Invest Dermatol 2014; 134(6):1768-72; PMID:24468746; http://dx.doi.org/ 10.1038/jid.2014.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vardabasso C, Gaspar-Maia A, Hasson D, Pünzeler S, Valle-Garcia D, Straub T, Keilhauer EC, Strub T, Panda T, Dong J, et al.. Histone variant H2A.Z.2 mediates proliferation and drug sensitivity of malignant melanoma. Mol Cell 2015; 59(1):75-88; PMID:26051178; http://dx.doi.org/ 10.1016/j.molcel.2015.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer 2012; 12, 465-77; PMID:22722403; http://dx.doi.org/ 10.1038/nrc3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segura MF, Fontanals-Cirera B, Gaziel-Sovran A, Guijarro MV, Hanniford D, Zhang G, Gonzalez-Gomez P, Morante M, Jubierre L, Zhang W, et al.. BRD4 Sustains Melanoma Proliferation and Represents a New Target for Epigenetic Therapy. Cancer Res 2013; 73:6264-76; PMID:23950209; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-0122-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al.. Selective inhibition of BET bromodomains. Nature 2010; 468:1067-73; PMID:20871596; http://dx.doi.org/ 10.1038/nature09504 [DOI] [PMC free article] [PubMed] [Google Scholar]


