ABSTRACT
Recently, we suggested the microRNA (miR) landscape defining metastasis. The first miR-driven network orchestrating invasion, intravasation, and metastasis was confirmed independently across several malignancies, suggesting a rather general principle for metastasis regulation. We hope that our data will stimulate the field in terms of further hypothesis generation, metastasis prediction, and metastasis prevention.
KEYWORDS: Cancer, EMT, FOXN3, microRNA, metastasis, stem cell
Recently, we published our results on a metastasis-specific microRNA (miR) signature derived from systematic hypothesis generation by microRNA (miR) and mRNA profiling of resected colorectal cancer metastasis compared to matched primary tumor and corresponding normal tissues and subsequent extensive validation.1 We identified an exclusive miR signature that is differentially expressed in metastases. This signature includes both established miRs (e.g., the miR-34, let-7, and miR-200 families) and novel candidate miRs (such as miR-552, -218, -135, -210 and -654). Resulting mechanistic studies suggested the first miR-orchestrated network in which 3 of these miRs—miR-218 (acting, at least in part, synergistically with miR-200), miR-135b, and miR-210—act via 5 novel targets—zinc finger E-box binding homeobox 2 (ZEB2), N-cadherin (neuronal cadherin, also known as CDH2), siah E3 ubiquitin protein ligase 1 (SIAH1), SET domain containing 2 (SETD2), and forkhead box N3 (FOXN3) transcription factor—to significantly orchestrate epithelial-mesenchymal transition (EMT), migration, invasion, intravasation, and metastasis (Fig. 1). This novel network could be validated independently in our own patient series and across several databases of up to 15 different tumor entities in up to 6,000 patients (www.oncomine.org),2 suggesting the relevance of our network for metastasis of several cancer entities.
Figure 1.
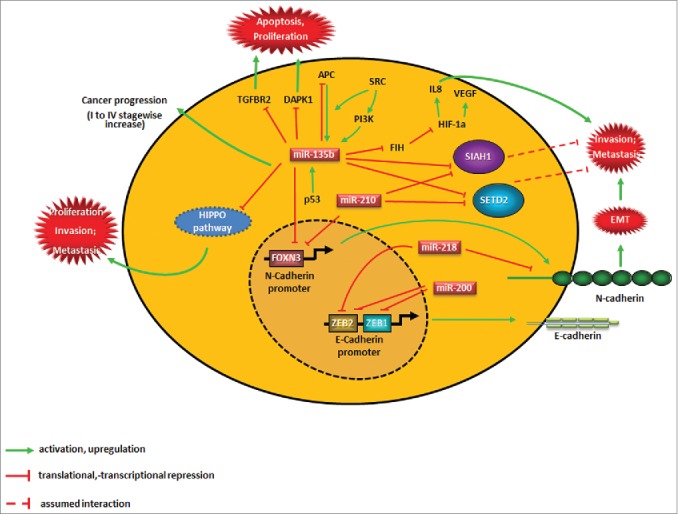
Novel network implicating microRNAs (miRs) in epithelial-mesenchymal transition (EMT) and metastasis. This schematic summary shows the major players leading to the development of colorectal cancer metastasis as described in our network and incorporating other key molecules published by other groups. The details of these interactions are described in part in this manuscript and completely in the original publications.1,3 APC, adenomatous polyposis coli; CASR, calcium-sensing receptor (also known as FIH); CXCL8, chemokine (C-X-C motif) ligand 8 (commonly known as IL 8); DAPK1, death associated protein kinase 1; E-cadherin, epithelial cadherin, also known as CDH1); FOXN, forkhead box N3; HIF1A, hypoxia inducible factor 1, α subunit; N-cadherin, neuronal cadherin (also known as CDH2); PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; SETD2, SET domain containing 2; (SIAH1) siah E3 ubiquitin protein ligase 1; TGFBR2, transforming growth factor, β receptor II; TP53, tumor protein p53 (best known as p53); SRC, SRC proto-oncogene, non-receptor tyrosine kinase; VEGF, vascular endothelial growth factor; ZEB, zinc finger E-box binding homeobox.
The validity of our translational hypothesis generation process and the resulting data is supported by a number of studies.1 Strikingly, while our paper was under consideration for publication an exciting paper from Croce's lab was published, showing that miR-135b expression in colon cancer was induced upon loss of adenomatous polyposis coli (APC) and phosphatase and tensin homolog (PTEN) or mutation and/or activation of phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K, also known as PIK3CA) and SRC proto-oncogene, non-receptor tyrosine kinase (SRC).3 Specifically, PI3K mutations resulted in the phosphorylation of forkhead box O1 (FOXO1) and forkhead box O3 (FOXO3A), followed by an increase in miR-135b expression. Although the exact molecular mechanisms of this interaction warrant further investigation, this work, together with our results identifying miR-135b as a member of a prometastatic network inducing FOXN3, which is a novel negative regulator of N-cadherin, (Fig. 1) underlines the important function of this miR in colon carcinogenesis and progression. It furthermore suggests that the functional interplay between particular miRs and FOX transcription factors definitely deserves further attention when studying metastasis.
Interestingly, miR-135b also regulates members of the Hippo pathway that have been shown to determine stemness-like properties,4 providing a potential link between the cancer stem cell hypothesis and our current view on the metastatic cascade. This hypothesis is supported by other findings in our paper, in which we noted that members of our metastasis-regulating miR network were also deregulated in primary tumors of patients with advanced tumor stages, suggesting the presence of a metastatically relevant subclone with a critical cell number that was detectable in the molecular profiles. Interestingly, when we analyzed miR profiles from early-stage tumor samples in the GEO database (GSE35982 and GSE10259) our signature was less visible, signifying that the metastatically capable clone becomes evident only after a certain advanced tumor stage has been attained. A comparable finding had previously been reported by the Croce group on miR-135b in stage I-IV colorectal cancer patients.3
We observed some significantly deregulated miRs in the primary tumor that were not visible in the matched metastasis tissue, signifying that their important cancer-promoting functions were limited to early disease stages. This also implies that these miRs might regulate important factors capable of priming the pre-metastatic niche, since it is increasingly evident that the primary tumor has an amazing function in initiating this process.5
A breast cancer-specific miR landscape that correlates with clinical classification and follow-up predictions has recently been defined in a large series of resected primary breast cancers.6 Although this study did not include analysis of the corresponding metastases, a number of miRs in the breast cancer signature are common to those in our colorectal cancer metastasis-associated signature and were also predicted to drive similar pathways to those implicated in our study, such as Wnt and hepatocyte growth factor (HGF) signaling. As the breast study was conducted using a different solid tumor entity and with a completely different intention (defining the breast cancer miR landscape with attention to particular subtypes and clinical outcome), these observations again support the notion that analysis of resected patient tissues can give highly relevant information about molecular networks that are functionally important for cancer progression and metastasis across different tumor types.
With very recent progress regarding the potential therapeutic benefits of miRs, we believe that these molecules are currently receiving a second peak of attention in the scientific and clinical community. Supporting this claim are the eagerly awaited results of the Phase 1 study for miR-34 (MRX34), due to be completed at the end of the year (https://clinicaltrials.gov/ct2/show/NCT01829971?term=MRX34&rank=1). 7 As stated above, the miR-34 family was among the top candidates in our list of significantly deregulated miRs in metastasis,1 which makes us confident that our data might stimulate many other colleagues into hypothesis generation not only from a mechanistic viewpoint, but also on the miRs that have the highest likelihood to become successful therapeutics in the future, either alone if they suppress metastasis or as their (chemically modified) anti-miRs in the case of prometastatic miRs. Thus, our data might not only to lead to interesting and easy-to-measure biomarkers that indicate a certain likelihood of a patient developing future metastasis, but might also assist in the development of therapeutic tools able to prevent metastasis.
Taken together, we hope that with our systematic approach, we can stimulate basic, translational, and clinical research in terms of further hypothesis generation and investigations on important miR-driven networks as key regulators of metastasis and markers for metastasis prediction, and the advancement of tools for metastasis prevention.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
HA is supported by Deutsche Krebshilfe/Dr. Mildred-Scheel-Stiftung (together with MA), the Alfried Krupp von Bohlen und Halbach Foundation (Award for Young Full Professors) (Essen, Germany), Hella Bühler Foundation (Heidelberg, Germany), Dr. Ingrid zu Solms Foundation, (Frankfurt/Main, Germany), the Walter Schulz Foundation (Munich, Germany), the German-Israeli Project Cooperation DKFZ-MOST (Ministry of Science and Technology), the Wilhelm-Sander Foundation, Munich, Germany (together with JHL), and 2 grants of the HIPO/POP-Initiative, DKFZ Heidelberg.
References
- 1.Mudduluru G, Abba M, Batliner J, Patil N, Scharp M, Lunavat TR, Leupold JH, Oleksiuk O, Juraeva D, Thiele W, Rothley M, Benner A, Ben-Neriah Y, Sleeman J, Allgayer H.. A systematic approach to defining the microRNA landscape in metastasis. Cancer Res 2015. Aug 1;75(15):3010-9. doi: 10.1158/0008-5472.CAN-15-0997. Epub 2015 Jun 11. PubMed PMID: 26069251.17356713 [DOI] [PubMed] [Google Scholar]
- 2.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al.. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007; 9:166-80; PMID:17356713; http://dx.doi.org/ 10.1593/neo.07112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat F, et al.. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell 2014; 25:469-83; PMID:24735923; http://dx.doi.org/ 10.1016/j.ccr.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin CW, Chang YL, Chang YC, Lin JC, Chen CC, Pan SH, Wu CT, Chen HY, Yang SC, Hong TM, et al.. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat Commun 2013; 4:1877; PMID:23695671; http://dx.doi.org/ 10.1038/ncomms2876 [DOI] [PubMed] [Google Scholar]
- 5.Sceneay J, Smyth MJ, Moller A. The pre-metastatic niche: finding common ground. Cancer Metast Rev 2013; 32:449-64; PMID:23636348; http://dx.doi.org/ 10.1007/s10555-013-9420-1 [DOI] [PubMed] [Google Scholar]
- 6.Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, et al.. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 2013; 497:378-82; PMID:23644459; http://dx.doi.org/ 10.1038/nature12108 [DOI] [PubMed] [Google Scholar]
- 7.Agostini M, Knight RA. miR-34: from bench to bedside. Oncotarget 2014; 5:872-81; PMID:24657911 [DOI] [PMC free article] [PubMed] [Google Scholar]


