ABSTRACT
Adeno-associated virus type 2 (AAV2) is a defective DNA virus that was previously considered to be non-pathogenic. We identified somatic AAV2 integration in a subset of 11 hepatocellular carcinomas (HCC) that mainly developed in normal liver without known etiology through recurrent insertional mutagenesis in cancer driver genes such as telomerase reverse transcriptase (TERT), cyclin A2 (CCNA2), cyclin E1 (CCNE1), tumor necrosis factor (ligand) superfamily, member 10 (TNFSF10), and lysine (K)-specific methyltransferase 2B (KMT2B).
KEYWORDS: Adeno-associated virus type 2, gene therapy, hepatocellular carcinoma, insertional mutagenesis, TERT
Abbreviations
- 3′ ITR
3′ inverse tandem repeat
- 3′ UTR
3′ untranslated region
- AAV2
adeno-associated virus type 2
- EBV
Epstein-Barr virus
- HBV
hepatitis B virus
- CCNA2
cyclin A2
- CCNE1
cyclin E1
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HHV8
human herpes virus 8
- HPV
human papilloma virus
- HTLV1
human T-cell leukemia/lymphoma virus type 1
- MLL4
myeloid/lymphoid or mixed-lineage leukemia 4
- mTOR
mammalian target of rapamycin
- TERT
telomerase reverse transcriptase
- TNFSF10
tumor necrosis factor (ligand) superfamily, member 10
- TRAIL
tumor-necrosis-factor related apoptosis inducing ligand.
Hepatocellular carcinoma (HCC) mainly develops in the context of cirrhosis, and is a cancer field standing at the crossroad of genetic predisposition and well-known viral and toxic exposures to hepatitis B virus (HBV), hepatitis C virus (HCV), alcohol intake, metabolic syndrome, and hemochromatosis. However, a small subset of HCC occurs in normal liver without any of the classic known risk factors of cirrhosis. HCC is the result of accumulation of several somatic genetic alterations in tumor hepatocytes, with an average of 40 functional genetic defects in each tumor.1 Next-generation sequencing has identified the crucial signaling pathways that are altered in HCC, namely telomere maintenance, WNT/β-catenin pathway, cell cycle genes, epigenetic modifier genes, mitogen-activated kinases, mammalian target of rapamycin (mTOR) pathway, and oxidative stress pathway.2,3 Moreover, viral infection with HBV and HCV in HCC has been associated with direct or indirect mechanisms of carcinogenesis.1,4
Our knowledge of virus-induced oncogenesis dates back more than 100 years to the identification of the Rous sarcoma retrovirus that causes sarcoma in chickens.5 For a long time there was no evidence of a direct involvement of viruses in human carcinogenesis, until the identification of a link between Epstein-Barr virus (EBV) and Burkitt's lymphoma in 1964. At the present time 6 viruses have been associated with tumor development in humans: HBV with HCC; HCV with HCC and lymphoma; EBV with Burkitt lymphoma, nasopharyngeal carcinoma, and lymphoproliferative disease; human papilloma virus (HPV) with cervical and head and neck cancer; Merckel cell polyomavirus with Merckel cell carcinoma; human herpes virus 8 (HHV8) with Kaposi sarcoma; and human T-cell leukemia/lymphoma virus type 1 (HTLV1) with T-cell leukemia.5 HBV promotes malignant transformation not only through a direct oncogenic effect of viral proteins like Hbx, but also through a mechanism of insertional mutagenesis.4 HBV was shown to integrate into human DNA and sometimes, if this integration occurred near cancer genes, promote cell survival and proliferation and lead to tumor development.4 Recurrent HBV clonal integrations in HCC have been described in telomerase reverse transcriptase (TERT), cyclin E1 (CCNE1), or lysine (K)-specific methyltransferase 2B (KMT2B, also known as myeloid/lymphoid or mixed-lineage leukemia 4 [MLL4]), leading to overexpression of these targeted genes in most cases.6
Recently, we identified somatic recurrent clonal insertion of adeno-associated virus type 2 (AAV2) in human HCC.7 AAV2 is a defective DNA virus that requires co-infection by a helper virus such as adenovirus to replicate and infect other cells. AAV2 is transmitted through the air and is generally considered non-pathogenic. AAV2 persists in an episomal form in the cell but also integrates into human DNA and stays quiescent until new infection with a helper virus occurs.8 The AAV2 genome is composed of 4,759 base pairs and encodes non-structural rep proteins that are involved in replication and genome packaging and structural cap proteins that compose the viral capsid.8 The 5′ and 3′ extremities of the genome contain inverse tandem repeats (ITRs) that are organized in a T-shaped hairpin structure and are involved in AAV2 replication and integration in human DNA.8
We identified clonal integration of AAV2 in 11 tumors among 193 HCCs, an overall frequency of 5%.7 Clonal integrations were located in the cyclin A2 (CCNA2) gene in 4 cases, CCNE1 in 3 cases, TERT in 1 case, KMT2B/MLL4 in 1 case, and tumor necrosis factor (ligand) superfamily, member 10 (TNFSF10) in 2 cases. CCNA2 and CCNE1 respectively code for cyclins A and E, 2 key proteins involved in cell cycle progression; KMT2B codes for MLL4, an histone methyltransferase; TERT codes for the telomerase reverse transcriptase that is overexpressed in 90% of HCCs and functions to allow telomere synthesis and avoid cell senescence; and TNFSF10 codes for tumor necrosis factor-related apoptosis inducing ligand (TRAIL), which is known to activate the apoptotic pathway but also activates proliferative pathways under specific circumstances.
All the tumors with AAV2 insertion harbored overexpression of the inserted genes compared to the normal liver and corresponding non-tumor liver tissues. Moreover, we modeled AAV2 insertions in vitro to investigate the effect of AAV2 insertion in targeted genes. To this aim, we introduced the exact AAV2 insertions observed in tumors into the TERT promoter and the 3′ untranslated region (3′ UTR) of TNFSF10 in a luciferase expression vector. In all cases, AAV2 insertion increased luciferase activity whereas scramble inserted sequences had no effect. Interestingly, RNA sequencing analysis of HCC that harbored CCNA2 and TNFSF10 insertions showed overexpression of the mature transcript and premature ending at the viral polyA instead of the human polyA leading to immature transcripts in some cases of AAV2 insertion in CCNA2. Analysis of the AAV2 inserted sequences revealed a minimal common inserted region composed of the 3′ inverse tandem repeat region of the virus in 10 of the 11 insertions.7 These results highlight that AAV2 sequences located at the 3′ ITR act as a direct activator of the expression of genes targeted by the viral insertion (Fig. 1).
Figure 1.
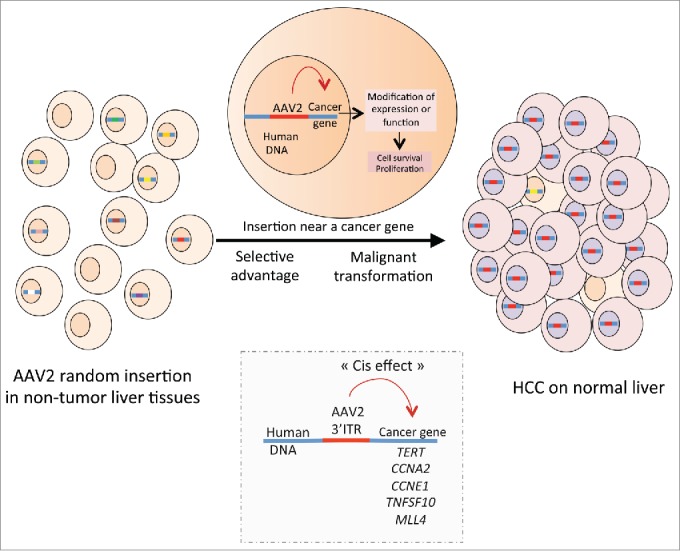
Insertional mutagenesis induced by AAV2 promotes the development of hepatocellular carcinoma. A small part of AAV2 including the 3′ inverse tandem repeat region integrates near cancer genes such as telomerase reverse transcriptase (TERT), cyclin A2 (CCNA2), cyclin E1 (CCNE1), tumor necrosis factor (ligand) superfamily, member 10 (TNFSF10), and lysine (K)-specific methyltransferase 2B (KMT2B), inducing gene expression and promoting cell proliferation and malignant transformation.
Interestingly, HCCs with AAV2 insertion were mostly identified in patients with normal liver without known etiology or chronic liver disease, suggesting a strong role of AAV2 infection in HCC development in these cases. AAV family members have been used for several years as vectors for gene therapy in mice and humans because of their non-pathogenic feature and their efficacy to transduce cells, particularly hepatocytes.9 However, 2 different mouse models infected with AAV2 vectors used for gene therapy developed liver tumors with AAV integration in the genome as the mechanism of HCC induction.10 These findings raised new questions about the pathogenicity of AAV2 infection. Despite the high prevalence of AAV2 in the general population (40 to 60% positivity for AAV2 antibody), HCC development with AAV2 integration is a very rare event. This situation is similar to the infrequent induction of tumors by the common EBV infection.5,8 It remains to be understood whether genetic or environmental factors might synergize with viral infection to promote tumor development. Moreover, the consequence for AAV-mediated gene therapy and future clinical trials remains a subject of debate. At the very least, patients treated by AAV-related gene therapy should be closely followed up to assess the potential risk of tumor development.
In conclusion, the seminal findings of HCC occurrence in mice treated by AAV-mediated gene therapy together with the current identification of AAV2-related insertional mutagenesis in humans underline the role of AAV2 in liver carcinogenesis in normal liver. Moreover, after human papilloma virus, Merkel cell polyomavirus, and hepatitis B virus, AAV2 is the fourth virus to show evidence of insertional mutagenesis in human tumors.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Acknowledgments
This work was supported by INCa with the ICGC and the PAIR-CHC project NoFLIC (also funded by ARC). The group is supported by the Ligue Nationale contre le Cancer. J.-C.N. and A.F. were supported by a fellowship from INCa and Ligue Nationale contre le Cancer respectively.
References
- 1.Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. The genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015; 149(5):1226–1239; PMID:26099527; http://dx.doi.org/25822088 10.1053/j.gastro.2015.05.061 [DOI] [PubMed] [Google Scholar]
- 2.Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, et al.. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015; 47:505–11; PMID:25822088; http://dx.doi.org/ 10.1038/ng.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nault JC, Mallet M, Pilati C, Calderaro J, Bioulac-Sage P, Laurent C, Laurent A, Cherqui D, Balabaud C, Zucman-Rossi J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun 2013; 4:2218; PMID:23887712; http://dx.doi.org/ 10.1038/ncomms3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neuveut C, Wei Y, Buendia MA. Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol 2010; 52:594–604; PMID:20185200; http://dx.doi.org/ 10.1016/j.jhep.2009.10.033 [DOI] [PubMed] [Google Scholar]
- 5.Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer 2010; 10:878–89; PMID:21102637; http://dx.doi.org/ 10.1038/nrc2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C, et al.. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 2012; 44:765–9; PMID:22634754; http://dx.doi.org/ 10.1038/ng.2295 [DOI] [PubMed] [Google Scholar]
- 7.Nault JC, Datta S, Imbeaud S, Franconi A, Mallet M, Couchy G, Letouze E, Pilati C, Verret B, Blanc JF, et al.. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet 2015; 47(10):1187–93; PMID:26301494; http://dx.doi.org/ 10.1038/ng.3389 [DOI] [PubMed] [Google Scholar]
- 8.Goncalves MA. Adeno-associated virus: from defective virus to effective vector. Virol J 2005; 2:43; PMID:15877812; http://dx.doi.org/ 10.1186/1743-422X-2-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, et al.. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med 2011; 365:2357–65; PMID:22149959; http://dx.doi.org/ 10.1056/NEJMoa1108046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donsante A, Miller DG, Li Y, Vogler C, Brunt EM, Russell DW, Sands MS. AAV vector integration sites in mouse hepatocellular carcinoma. Science 2007; 317:477; PMID:17656716; http://dx.doi.org/ 10.1126/science.1142658 [DOI] [PubMed] [Google Scholar]


