ABSTRACT
Epigenetic machinery have become a major focus for new targeted cancer therapies. Our previous report described the discovery and biological activity of a potent, selective, orally bioavailable, irreversible inhibitor of Lysine Demethylase 1 (LSD1), GSK2879552. A proliferation screen of cell lines representing a number of tumor types indicated that small cell lung carcinoma (SCLC) was sensitive to LSD1 inhibition. The SCLC lines that undergo growth inhibition in response to GSK2879552 exhibit DNA hypomethylation of a signature set of probes suggesting this may be used as a predictive biomarker of activity. This targeted mechanism coupled with a novel predictive biomarker make LSD1 inhibition an exciting potential therapy for SCLC.
KEYWORDS: Demethylase, DNA methylation, epigenetic, LSD1, SCLC
Lung cancer is one of the most prevalent cancers and has extremely poor prognosis; it is the leading cause of cancer-related death in the US with a 5-year survival rate of less than 20%. Small cell lung cancer (SCLC), accounting for approximately 15% of all lung cancers, is one of the most aggressive subtypes and has a 5-year survival rate of less than 10%.1 SCLC can be distinguished by its neuroendocrine features and is morphologically distinct from non-small cell lung cancer (NSCLC). SCLC has also been described as poorly differentiated with rapid growth, and the majority of patients present at diagnosis with extensive stage disease characterized by distant metastases.2 Although response to chemotherapy and radiotherapy is observed in the majority of patients, most relapse within 6–12 months and are then resistant to further therapy.2 Despite numerous clinical trials over the past several decades no major advances have been made to alter the current therapeutic landscape for this disease.2
Epigenetic dysregulation has emerged as an important mechanism in many cancer types, including SCLC.3 The machinery that controls DNA and histone modifications has become a major focus for targeted therapies.4 Our recent report describes the discovery and biological activity of a cyclopropylamine-containing inhibitor of the histone modifying enzyme, lysine demethylase 1 (KDM1A, best known as LSD1).5 GSK2879552 is a potent, selective, orally bioavailable, mechanism-based irreversible, small-molecule inactivator of LSD1. The monoamine oxidase (MAO) family of enzymes that are mechanistically related to LSD1 have been successfully inhibited in the clinic by cyclopropylamine-containing small molecules. This mechanism of enzyme inactivation was exploited in the development of both GSK2879552 and an LSD1 inhibitor developed by Oryzon Genomics (ORY-1001). Both GSK and Oryzon's compounds have undergone extensive preclinical validation and are currently in Phase I clinical trials. Other LSD1 inhibitors, including additional cyclopropylamines as well as compounds with distinct modes of action such as polyamines and other reversible inhibitors, have been described; however, most have not yet progressed to clinical development.6
LSD1 has emerged as an interesting cancer therapeutic target for several reasons. LSD1 is overexpressed in many human cancers including lung, breast, prostate, and blood cancers.7 Loss of LSD1 expression decreases the growth of cancer cells and LSD1 is required for normal differentiation in embryonic as well as adult cells.7 The activity of LSD1 is essential for the maintenance of pluripotency in embryonic stem cells.8 In the adult setting, LSD1 is necessary for normal hematopoiesis, and loss of LSD1 results in impaired maturation of hematopoietic progenitors through a block in differentiation.9 The importance of LSD1 in normal differentiation suggests that aberrant gene expression resulting from dysregulation of LSD1 may result in alterations in pathways associated with a stem-cell like phenotype. In acute myeloid leukemia (AML), LSD1 is most highly expressed in less differentiated subtypes and knockdown or inhibition of LSD1 has a pro-differentiation effect.7,10 Cells without active LSD1 demonstrate increases in gene expression signatures associated with differentiation, alterations in myeloid cell surface markers, and morphologic changes.10 Together, studies in developmental systems and cancer cells indicate a critical role for LSD1 in stem cell biology and highlight a potential tumor-promoting effect in cancer.
GSK2879552 was used to screen a panel of more than 150 cancer cell lines for effects on proliferation. These studies indicated that AML and SCLC were uniquely sensitive to LSD1 inhibition. This growth inhibition was predominantly cytostatic and not all SCLC cell lines tested were sensitive to the antiproliferative effects of GSK2879552. Genomic analyses of SCLC cell lines revealed a differential pattern of DNA methylation in sensitive versus resistant lines. This potential biomarker was also found in primary SCLC samples and its utility in predicting sensitivity to LSD1 inhibition was tested in patient-derived xenograft (PDX) models. Only PDX models with the sensitivity-associated DNA methylation signature responded to LSD1 inhibition in vivo. Although a mechanistic link between the methylation state of the signature probe set and drug sensitivity will require additional study, the hypomethylation status of this signature set of probes may allow the stratification of patients that might respond to the inhibition of LSD1 by GSK2879552 and might provide a predictive biomarker for use in SCLC.
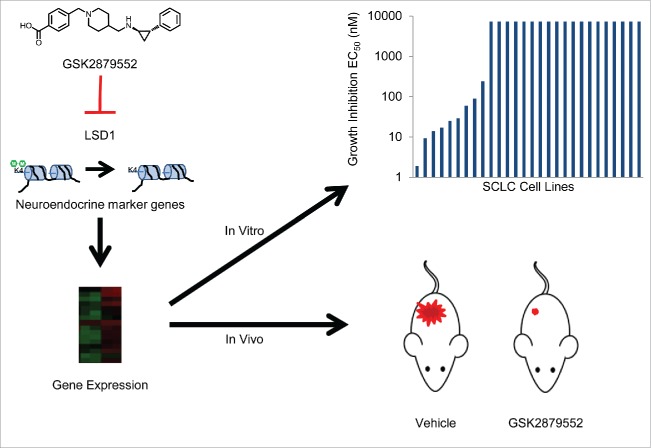
Unlike NSCLC, which has a number of known genetic alterations that have been characterized as oncogenic drivers, the molecular mechanisms that are critical for SCLC have remained elusive. Despite many recent studies to characterize the genetic and epigenetic alterations in SCLC, the identification and efficacy of targeted therapies for this disease have continued to pose a significant challenge in addressing this unmet medical need.1,2 SCLC is a neuroendocrine tumor and expresses molecular features associated with this cell type. A change in neuroendocrine marker expression in SCLC cells may therefore indicate an altered cell state. A survey of neuroendocrine marker gene expression in a panel of SCLC lines revealed that expression of many genes was altered in response to LSD1 inhibition. Chromatin immunoprecipitation (ChIP)-sequencing studies examining localization of the LSD1 protein revealed that genes important for neuron differentiation and cell development were among the most strongly bound by LSD1, further underscoring a potential role for LSD1 in differentiation of SCLC (Fig. 1). Together, the cytostatic nature of growth inhibition by GSK2879552, the altered expression of neuroendocrine markers in response to inhibitor treatment, and genomic localization of LSD1 to differentiation-associated genes indicate that GSK2879552 may affect the neuroendocrine cell state of SCLC (Fig. 1). Overall, the mode of action of LSD1 inactivation in SCLC appears to reflect a mechanism similar to the observed pro-differentiation effect in leukemia upon loss or inhibition of LSD1.10 Pro-differentiation agents have not been tested to date in SCLC, therefore GSK2879552 provides an interesting mechanism of action in a tumor type in which targeted therapies have so far largely failed.1,2
Figure 1.
Putative mechanism of action of LSD1 inhibitors. GSK2879552 inhibits the demethylation of histones by LSD1, leading to changes in the expression of neuroendocrine marker genes. Alterations in gene expression may give rise to in vitro and in vivo growth inhibition of small cell lung cancer cells.5
GSK2879552 is a small molecule inhibitor of LSD1 with excellent physicochemical properties that demonstrates efficacy in preclinical models of SCLC. The irreversible mechanism of GSK2879552 may provide a significant advantage given that maximal gene expression responses and efficacy require durable inhibition.5 Inactivation of LSD1 provides a targeted mechanism of antitumor activity in SCLC, potentially through promotion of differentiation of this tumor type in a manner similar to a reported mechanism in leukemia cells.10 A DNA hypomethylation signature may allow stratification of patients to reveal those that might respond, therefore GSK2879552 may represent a targeted therapy with a predictive biomarker strategy for SCLC patients.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev 2015 Jul; 29(14):1447-62; PMID:26220992; http://dx.doi.org/ 10.1101/gad.263145.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sgambato A, Casaluce F, Maione P, Rossi A, Sacco PC, Panzone F, Ciardiello F, Gridelli C. Medical treatment of small cell lung cancer: state of the art and new development. Expert Opin Pharmacother 2013 Oct; 14(15):2019-31; PMID:23901936; http://dx.doi.org/ 10.1517/14656566.2013.823401 [DOI] [PubMed] [Google Scholar]
- 3.Poirier JT, Gardner EE, Connis N, Moreira AL, de SE, Hann CL, Rudin CM. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene (2015) 34; 5869-5878; PMID:25746006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helin K, Dhanak D. Chromatin proteins and modifications as drug targets. Nature 2013 Oct; 502(7472):480-8; PMID:24153301; http://dx.doi.org/ 10.1038/nature12751 [DOI] [PubMed] [Google Scholar]
- 5.Mohammad HP, Smitheman KN, Kamat CD, Soong D, Federowicz KE, Van Aller GS, Schneck JL, Carson JD, Liu Y, Butticello M, et al.. A DNA Hypomethylation Signature Predicts Antitumor Activity of LSD1 Inhibitors in SCLC. Cancer Cell 2015 Jul; 28(1):57-69; PMID:26175415; http://dx.doi.org/ 10.1016/j.ccell.2015.06.002 [DOI] [PubMed] [Google Scholar]
- 6.Zheng YC, Ma J, Wang Z, Li J, Jiang B, Zhou W, Shi X, Wang X, Zhao W, Liu HM. A Systematic Review of Histone Lysine-Specific Demethylase 1 and Its Inhibitors. Med Res Rev 2015 Sep; 35(5):1032-71; PMID:25990136; http://dx.doi.org/ 10.1002/med.21350 [DOI] [PubMed] [Google Scholar]
- 7.Lynch JT, Harris WJ, Somervaille TC. LSD1 inhibition: a therapeutic strategy in cancer? Expert Opin Ther Targets 2012 Dec; 16(12):1239-49; PMID:22957941; http://dx.doi.org/ 10.1517/14728222.2012.722206 [DOI] [PubMed] [Google Scholar]
- 8.Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol 2011 Jun; 13(6):652-9; PMID:21602794; http://dx.doi.org/ 10.1038/ncb2246 [DOI] [PubMed] [Google Scholar]
- 9.Sprussel A, Schulte JH, Weber S, Necke M, Handschke K, Thor T, Pajtler KW, Schramm A, Konig K, Diehl L, et al.. Lysine-specific demethylase 1 restricts hematopoietic progenitor proliferation and is essential for terminal differentiation. Leukemia 2012 Sep; 26(9):2039-51; PMID:22699452; http://dx.doi.org/ 10.1038/leu.2012.157 [DOI] [PubMed] [Google Scholar]
- 10.Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Ciceri F, Blaser JG, Greystoke BF, Jordan AM, et al.. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 2012 Apr; 21(4):473-87; PMID:22464800; http://dx.doi.org/ 10.1016/j.ccr.2012.03.014 [DOI] [PubMed] [Google Scholar]