Abstract
We employed next generation RNA sequencing analysis to reveal dysregulated long non-coding RNAs (lncRNAs) in lung cancer utilizing 461 lung adenocarcinomas (LUAD) and 156 normal lung tissues from 3 separate institutions. We identified 281 lncRNAs with significant differential-expression between LUAD and normal lung tissue. LINC00857, a top deregulated lncRNAs, was overexpressed in tumors and significantly associated with poor survival in LUAD. knockdown of LINC00857 with siRNAs decreased tumor cell proliferation, colony formation, migration and invasion in vitro, as well as tumor growth in vivo. Overexpression of LINC00857 increased cancer cell proliferation, colony formation and invasion. Mechanistic analyses indicated that LINC00857 mediates tumor progression via cell cycle regulation. Our study highlights the diagnostic/prognostic potential of LINC00857 in LUAD besides delineating the functional and mechanistic aspects of its aberrant disease specific expression and potentially using as a new therapeutic target.
Keywords: non-coding RNA, LINC00857, lung adenocarcinoma, prognosis, diagnosis
INTRODUCTION
Lung cancer is a molecularly-heterogeneous disease and the leading cause of cancer mortality [1]. The molecular basis for this clinical heterogeneity remains incompletely understood. Over the past decade, research has primarily focused on the deregulation of protein-coding genes to identify oncogenes and tumor suppressors that could serve as diagnostic and therapeutic targets for lung cancer such as EGFR mutation and EML4-ALK gene fusion [2]. Identification of oncogenic driver mutations have helped improve the outcomes in specific subtypes of patients with non-small cell lung cancer (NSCLC), however the majority of the patients with lung cancer do not have an actionable molecular aberration [3, 4]. Hence, there is an urgent need for reliable biomarkers and identification of alternative treatment options. Increasing appreciation of the role of long non-coding RNAs (lncRNAs) in cancer progression has fostered efforts to characterize their role in disease biology and to evaluate them as novel biomarkers, as well as potential therapeutic targets [5–9].
LncRNAs are RNA transcripts that lack an open reading frame encoding a protein. LncRNAs are generally polyadenylated, greater than 200 bp in length and distinct from small RNAs and microRNAs [10–12]. In the past few years, lncRNAs have emerged as novel mechanisms in mediating cancer biology [13–18]. LncRNAs could act as an oncogene or tumor suppressor in tumor progression by affecting cell proliferation [19], differentiation [20], migration [15], immune response [13], and apoptosis [21]. A variety of mechanisms are involved in these tumor biological process such as remodeling of chromatin (HOTAIR, XIST, ANRIL) [22–24], transcriptional co-activation or co-repression (H19, LincRNA-p21) [21, 25], protein inhibition (TERRA) [26] post-transcriptional modifiers (MALT1) [27] or decoy elements (PTENP1) [28].
Genome-wide analyses indicate that IncRNAs frequently demonstrate restricted tissue-specific and cancer-specific expression patterns, distinguishing them from most miRNAs and protein-coding mRNAs [11, 29]. Moreover, lncRNA expression may confer clinical information about patient outcomes and have utility in diagnostic tests [22, 30, 31]. For example, prostate cancer gene 3 (PCA3) has been used as a biomarker in clinical practice for predicting prostate cancer volume [32]. HULC is reportedly detected in peripheral blood cells of HCC patients [33]. HOTAIR might be a biomarker for lymph node metastasis in HCC [34]. lncRNAs may also represent good candidates as therapeutic targets [35, 36]. HOTAIR can reduce the sensitivity of lung adenocarcinoma cells to chemotherapeutic drugs such as cisplatin [36]. Down regulation of MALAT1 expression reduced tumor growth in vivo [37]. The characterization of the RNA species, their function, and their clinical applicability has therefore become an area of biological and clinical importance in cancer research.
High-throughput RNA sequencing (RNA-Seq) in human cancer shows remarkable potential to identify both novel markers of disease and uncharacterized aspects of tumor biology, particularly lncRNA species [12, 29]. We analyzed the RNA-Seq data on a large cohort of lung cancer tissues and cells lines to discover lncRNAs with diagnostic or prognostic use in lung cancer. We identified 281 differently expressed lncRNAs in LUAD and present our results from an in depth characterization of our top candidate LINC00857. We validated the differential expression in multiple independent datasets and established the prognostic importance of LINC00857 expression. We next generated both cell line and xenograft models representing knockdown and overexpression of LINC00857 to delineate its functions.
RESULTS
Differentially expressed lung lncRNAs discovery and cross-validation
We recently performed RNA-Seq on a large cohort of lung cancer samples [4] (UM cohort) including 113 lung cancer tissues (67 LUADs, 36 SCCs and 10 large cell lung cancers), 6 matched normal lung tissues, and 26 lung cancer cell lines (Supplementary Figure S1A, Supplementary Table S1). In that study we also compiled two large RNA-Seq datasets then available, to perform a comprehensive gene fusion analysis. In the current study we perform a comprehensive analysis on the gene expression data-matrix from these three cohorts to discover differentially expressed lncRNAs in LUAD. The three cohorts are the UM cohort described above and two large publically available RNA-Seq data namely the Korean cohort (Seo) [38] including 85 LUADs and 77 normal, and finally The Cancer Genome Atlas (TCGA) LUAD data [39] including 309 LUADs and 73 normal lung samples (Supplementary Figure S2). Mate-pair reads were aligned using TopHat against the Ensembl GRCh37 human genome and initial transcripts elucidated with Cufflinks. Expression levels of transcripts were represented as Fragments Per Kilobase, Per Million mapped reads (FPKM). A total of 55,400 transcripts were mapped and classified as protein-coding genes, pseudogenes, lncRNAs, etc. according to their overlap with known transcripts in the Ensembl 66 database.
In order to find transcripts having higher expression value in lung tissues, we filtered the dataset using the following criteria; transcript FPKM value > 0 in at least 4 samples and a minimum of one sample with value > 4 among the 119 UM lung tissue samples (113 cancers and 6 normal lung tissues). Filtering excluded 33,480 genes from further analysis and indicated that a significant portion of the transcriptome has either very low to no expression in lung tissues. The remaining 21, 560 Ensembl genes belonged to various classes that include 16,017 protein-coding genes (74%), 1,726 pseudogenes (8%), and 3,136 lncRNAs (15%) (Figure 1A, left). The 3,136 lncRNAs include: 1,145 anti-sense, 951 lncRNAs, and 659 processed transcripts (Figure 1A, right). We found that protein coding genes had a higher expression level than non-coding RNAs (Supplementary Figure S1B).
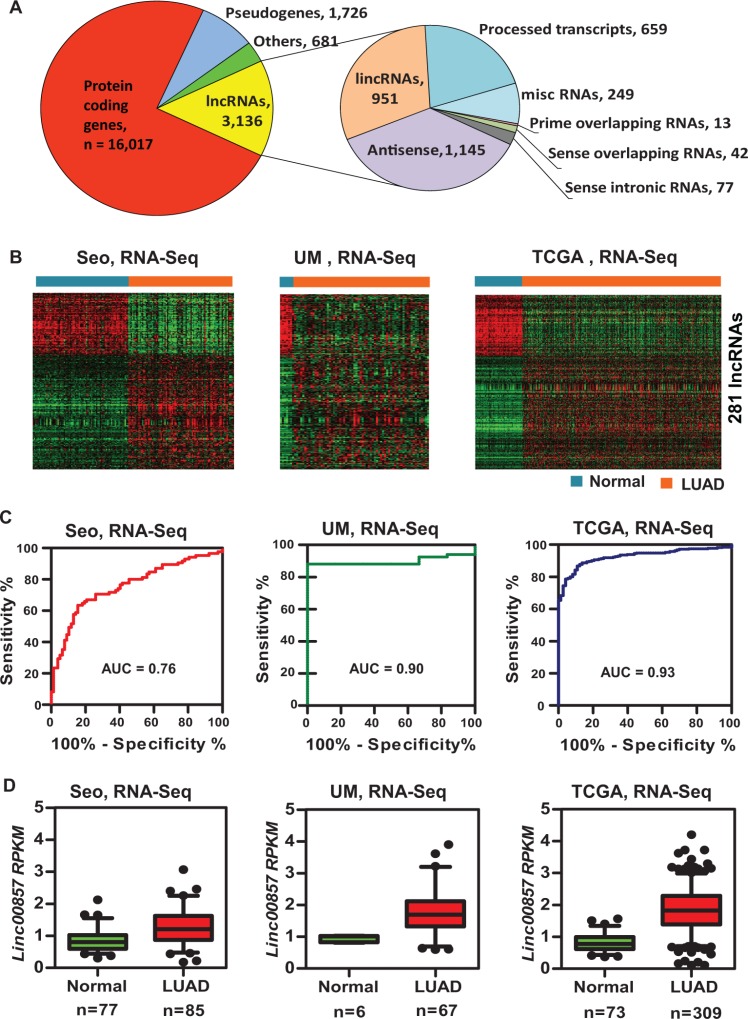
Figure 1. Differentially-expressed lncRNAs including LINC00857 in lung AD and normal lung tissues.
(A) Classification of all transcripts observed in this study from RNA-Seq data on 119 lung samples. The left pie chart shows transcript distribution in lung cancer. LncRNA accounts for 3,136 of all 21,560 expressed transcripts. The right pie chart shows further subclassification of 3,136 lncRNAs. Antisense, lncRNAs and processed transcripts are the major type of lncRNAs expressed in lung cancer. (B) Heat maps showing 281 different expressed lncRNAs from UM (67 Ad vs 6 N), Seo (85 AD vs 77 N) and TCGA (309 AD vs 73 N) RNA-Seq data sets. Columns represent samples, and rows lncRNAs, red is high expression and green low expression. (C) ROC curves of LINC00857 in Seo (85 AD vs 77 N), UM (67 AD vs 6 N) and TCGA (309 AD vs 73 N) RNA-Seq data sets. (D) Boxplots of LINC00857 expression levels in AD and normal tissue samples in Seo, UM and TCGA RNA-Seq datasets (AD vs. N, p < 0.001 in all 3 data sets).
Our analysis primarily aimed at identifying lncRNAs whose expression pattern had significant clinical utility. Towards this we developed a methodology to analyze existing high throughput RNA sequencing data, accounting for robust differential expression in cancer samples, stringent cross validation across multiple independent datasets/platforms and finally significant association with prognostic index (Supplementary Figure S2). Accordingly we performed Receiver Operating Characteristic (ROC) curve analysis, and the area under the curve (AUC) values was used to select the top list of differently expressed lncRNAs in LUAD. There were a total of 281 lncRNAs which had an AUC value larger than 0.7 (182 lncRNAs) or less than 0.25 (99 lncRNAs) in all 3 data sets (Figure 1B, Supplementary Figure S3A and S3B and Supplementary Table S2). Several known lung cancer-associated lncRNAs, such as HOTAIR, GAS5, PVT1, and UCA1 [40] were found in this top list (Table S2, Figure S3C) which further supported the power of this analysis. Importantly 34 of these lncRNAs correlated to patient survival (Cox model) in lung cancer (Supplementary Table S3). We also observed that most of these lncRNAs were deregulated in large cell or squamous cell lung cancer (Supplementary Figure S4). These lncRNAs demonstrated unique expression patterns and suggested potential novel diagnostic or prognostic markers in lung cancer.
LINC00857 is overexpressed in lung AD and predicts poor patient survival
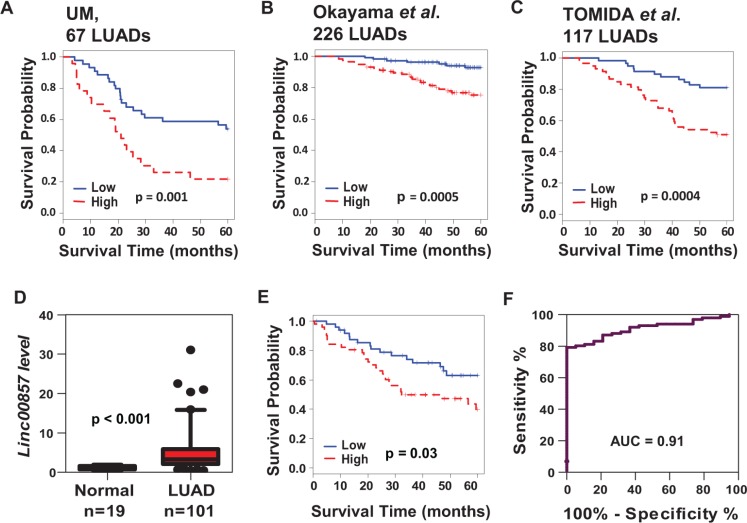
Among the most overexpresssed lncRNAs, LINC00857 was found to be significantly increased in LUADs (Figure 1C, 1D and Supplementary Figure S5) and was associated with worse survival (p < 0.001) in our patient cohort (UM 67 LUADs) (Figure 2A). We next validated this association in two independent published LUAD microarray data sets where survival information was available namely, Okayama et al., p < 0.0005 (226 LUADs, stage 1 and 2) [41] and Tomida et al. p < 0.0004 (117 LUADs, stage 1 to 3) [42] (Figure 2B, 2C). To further confirm the diagnostic and prognostic significance of LINC00857 in LUAD, we examined it's expression in an expanded UM cohort (101 lung ADs and 19 normal lung tissues) by an independent qRT-PCR assay. We found that LINC00857 expression levels were significantly higher in LUAD as compared to normal lung tissues (Figure 2D) with an excellent performance (AUC = 0.91) for classifying the tumors from normal lung based on LINC00857 expression (Figure 2F), and higher expression of LINC00857 was significantly related to worse patient survival (Figure 2E). LINC00857 expression levels and association with other clinical variables from this validation set are shown in Supplementary Table S3. In addition, we did not find any evidence for association between LINC00857 expression and KRAS or EGFR mutation status in our analysis (data not shown).
Figure 2. LINC00857 was increased and significantly associated with poor patient survival in lung AD.
(A–C) Kaplan-Meier curves and log-rank test of LINC00857 in UM (67 ADs), Okayama (226 ADs) and TOMIDA (117ADs) data sets indicating higher LINC00857 expression was associated to poorer patient survival. (D–F) qRT-PCR validation of LINC00857 expression in an independent data set including 19 normal and 101AD tissue samples. Boxplot (D) indicated LINC00857 expression was increased in tumor (vs. normal). Kaplan-Meier curve (E) indicated higher LINC00857 expression was unfavorable for patient survival, and ROC curve (F) indicated an excellent (AUC = 0.91) of classifying the 101 tumors from 19 normal based on LINC00857 expression.
To explore if LINC00857 is expressed in other types of cancer, we downloaded 6,220 cancer RNA-Seq expression data from MiTranscriptome [29]. We found that higher expression (FRKM log2 value > 0.5) of LINC00857 was presented in bladder, gastric, lung AD, cervix and pancreas cancers, and lower in prostate, thyroid, brain, blood and skin cancers (FRKM log2 value < 0.5) (Supplementary Figure S6A). When we compared to tumor vs. normal in 10 types of cancer which have normal tissues, we found that bladder, gastric, head & neck, liver, thyroid and lung cancers were significantly increased (vs. normal), whereas breast, kidney and prostate cancer were decreased (vs. normal) (Supplementary Figure S6B top panel). Surprisingly, we found that lung AD was the most significant one in term of p value by tumor vs. normal among these 10 types of cancer (Supplementary Figure S6B bottom panel). These indicated that LINC00857 was highly expressed in some types of cancer especially in lung AD (vs. normal) and could be potential useful as lung AD specific marker.
In order to confirm the FPKM value of LINC00857 detected by RNA-Seq and to select tumor cells for functional analyses, we measured LINC00857 expression in 33 lung cancer cell lines using qRT-PCR. We found a strong correlation of LINC00857 expression measured by RNA-Seq and qRT-PCR (Pearson r = 0.83, p < 0.001, Supplementary Figure S7). While, high LINC00857 expression was observed in 29 of the 33 cell lines tested, it was not expressed in 3 non-adherent lung cancer cell lines including 2 small cell lung cancers (H82 and H526) and one NSCLC derived from a metastatic lymph node (H1155) showed no expression.
The bi-exonic LINC00857 gene is located in the forward strand on chromosome 10q22.3 and is 2,171 bases long (Supplementary Figure S8A). To determine if LINC00857 overexpression arises from genomic amplification, we examined potential DNA copy number alterations in 90 lung ADs by Affymetrix SNP6.0 array (unpublished data). Except for one tumor with genomic gain we did not find any aberrations in this region in any other sample (Supplementary Figure S8B). We also observed that knockdown of LINC00857 using LINC00857-specific siRNA did not affect the expression of neighboring genes (Supplementary Figure S8C) indicating LINC00857 expression is not involved in cis-regulation of its neighbors. Taken together, LINC00857 is significantly overexpressed in LUAD across multiple studies and is a predictor of poor patient survival. Our data supports further examination of LINC00857's clinical utility in larger multi-institutional cohorts with outcome data to examine whether this gene has diagnostic and/or prognostic importance for lung cancer.
LINC00857 promotes cancer cell proliferation and invasion
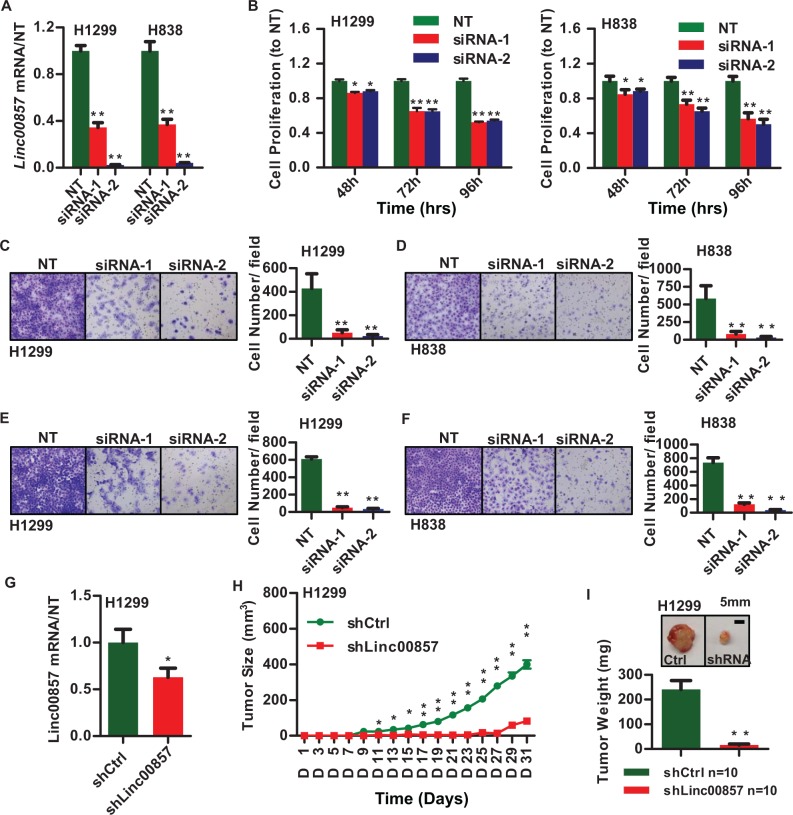
To examine the functional role of LINC00857, we performed small interfering RNA (siRNA)-mediated knockdown in H1299 and H838, two lung LUAD cancer cells, which contains high levels of the endogenous LINC00857 transcript (Supplementary Figure S7A and S9A). These two cell lines also exhibit high invasive potential among a panel of lung cell lines when assessed using the Boyden chamber assay (data not shown). To minimize the possibility of off-target effects, we employed two independent gene-specific siRNAs whose knockdown efficiency were 60 and 90% respectively, as estimated by qRT-PCR (Figure 3A). A significant decrease in cell proliferation in both H1299 and H838 cell lines (Figure 3B) and reduced cancer cell invasion (as measured by Boyden chamber matrigel invasion assays) was noted when LINC00857 was knocked down (Figure 3C and 3D). Similarly LINC00857 abrogation resulted in a significant loss in cell migration (Figure 3E and 3F) and colony formation (Supplementary Figure S9B).
Figure 3. LINC00857 is involved in tumor progression.
(A) LINC00857 siRNA knockdown efficiency (48 h) in H1299 and H838 measured by qRT-PCR. (B) Cell proliferation decreased after LINC00857 siRNAs treatment in H1299 and H838 cell lines. (C–D) LINC00857 siRNA knockdown impaired cellular invasion ability in H1299 and H838 cell lines (10X). (E–F) LINC00857 siRNA knockdown impaired cellular migration ability in H1299 and H838 cell lines (10 ×). (G) LINC00857 shRNA knockdown efficiency in H1299 measured by qRT-PCR. (H) Mouse in vivo xenograft tumor growth curves of H1299 cells expressing control and LINC00857 shRNA. (I) Tumor weight of H1299 cells expressing control and LINC00857 shRNA. **p < 0.01, n = 10 mice/group.
Having demonstrated a role for LINC00857 in cell growth and migration using the knockdown model, we next focused on probing the overexpression phenotype. Towards this end, we generated a full length LINC00857 expression construct in a pcDNA-DEST53 vector and transfected the lung cancer cell line SK-LU-1, which possesses low endogenous levels of LINC00857 as compared to H1299 and H838 (Supplementary Figure S9A). Contrary to knockdown experiments, overexpression of LINC00857 promoted cancer cell colony formation (Supplementary Figure S9C and S9D), cell proliferation and invasion in SK-LU-1 cells (Figure S9E and S9F). Furthermore subcutaneous injections in nude mice (n = 10 mice/group) with LINC00857 stable knock down H1299 cells showed impaired tumor growth (Figure 3G–3I). These results reaffirm our findings that LINC00857 plays a critical role in lung cancer cell proliferation, migration, invasion and colony formation. Thus LINC00857 may not only be a potential biomarker for diagnosis and prognosis of LUAD, but also play an important role in lung cancer progression.
We next looked at cellular localization of this lncRNA in H1299 and H838 cells. Quantitative RT-PCR of total RNA isolated from total, nuclear and cytoplasmic fraction of these two cell lines revealed enrichment in the cytoplasmic fraction (Supplementary Figure S10A). Interestingly, the A549 RNA-Seq data from the Encode database also shows cytoplasmic enrichment of LINC00857 providing further support (Supplementary Figure S10B). Further identification of the LINC00857 direct binding proteins using RNA pull-down assay or identifying the functional domains of LINC00857 using domain-specific chromatin isolation by RNA purification (dChIRP) [43] are currently being investigated.
LINC00857 knockdown causes cell cycle arrest at G1/S via CCNE1 regulation
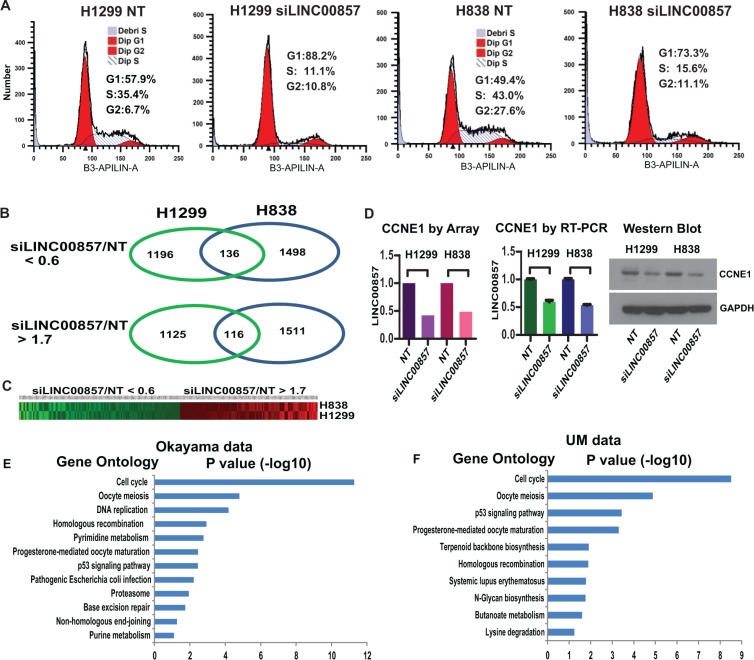
To help determine how LINC00857 affects tumor growth, we utilized flow cytometry analysis to examine the effect of LINC00857 knockdown on the cell cycle. As shown in Figure 4A, knockdown of LINC00857 using siRNA, induced cell cycle arrest at the G1/S phase indicating that G1 arrest might be one mechanism for how LINC00857 knockdown decreased lung tumor cell growth.
Figure 4. LINC00857 regulates cell cycle at the G1/S phase and potentially through CCNE1 and CDK2 regulation.
(A) Knockdown using LINC00857 siRNA caused G1 arrest in both H1299 and H838 lung cells. (B–C) LINC00857 siRNA knockdown induced 136 down and 116 up-regulated genes in both H1299 and H838 cells. (D) CCNE1 was decreased after LINC00857 siRNA treatment measured by both microarray and qRT-PCR for mRNA, and protein by Western blot. (E–F) The LINC00857 positively correlated genes in primary tumors were significantly involved in cell cycle regulation analyzed by gene ontology from DAVID.
Next, in order to investigate potential genes specifically regulated by LINC00857, we performed whole transcriptome analysis using Affymetrix ST2.1 exon arrays after LINC00857 siRNA knockdown in H1299 and H838 cell lines. As shown in Figure 4B and 4C, 1196 and 1498 genes were down-regulated at least 40% (relative to non-target siRNA control) in H1299 and H838 cells, respectively, and 136 genes were down-regulated in both cell lines (Supplementary Table S4) and we independently validated several dysregulated genes using qRT-PCR (Supplementary Figure S11A and S11B). The expression of cyclin E1 (CCNE1) (Figure 4D) and cyclin-dependent kinase 2 (CDK2) (Supplementary Figure S11C) were decreased after LINC00857 knockdown supporting the premise that LINC00857 may affect the cell cycle at G1/S through regulation of CCNE1 and CDK2 genes (Supplementary Figure S11D). Other cell cycle related genes regulated by LINC00857 knockdown are listed in Supplementary Table S5.
Next, we performed DAVID Gene Ontology analysis on genes positively correlated (Pearson correlation) with LINC00857 based on primary tumor data using Affymetrix microarrays [41] and UM RNA-Seq data [4]. Surprisingly, we found that LINC00857 positively correlated genes including CCNE1 and CDK2 (Supplementary Table S6) in primary tumors were significantly involved in cell cycle regulation (Figure 4E and 4F, and Supplementary Figure S12) which further supports LINC00857 involvement in cell cycle regulation.
There were 1125 and 1511 genes up-regulated at least 1.7-fold in H1299 and H838 cells, respectively, with 116 genes up-regulated in both cells (Figure 4B and 4C).
DISCUSSION
To date, only a few lncRNAs (including MALAT1, HOTAIR, H19, MEG3, GAS5, ANRIL, and SOX2OT etc.) have been reported to be dysregulated and functionally characterized in lung cancer and most of the studies were based on microarray expression data [40, 44, 45]. In comparison whole transcriptome sequencing provides a more comprehensive quantitative assessment of expressed transcripts without any probe limitations (associated with microarray analysis) and allows identification of novel and important lncRNAs which may have diagnostic, prognostic and functional potential in lung cancer [12, 29, 46].
In this study, there were 56,369 transcripts that could be matched to the Ensembl database, but only 3,136 lncRNAs were expressed in lung tissues indicating the tissue specific expression pattern of lncRNAs (Figure 1A). Based on three large RNA-Seq data sets containing a total of 461 LUADs and 156 normal lung tissues, we have identified 281 lncRNAs which were differently expressed in LUADs vs normal with an AUC large than 0.7 (higher in tumor) or less than 0.25 (lower in tumor) in all three data sets (Supplementary Figures S2, S3, Figure 1B). Some of the lncRNAs were also related to patient survival in lung cancer although some survival related lncRNAs may have been missed due to the selection criterion using AUC (tumor vs normal) rather than survival analysis by the Cox model. These lncRNAs could potentially serve as diagnostic or prognostic biomarkers in lung cancer. One of the top dysregulated lncRNAs LINC00857 was further validated for its diagnostic or prognostic potential using 3 independent data sets with different platforms including microarray and qRT-PCR (Figure 2).
Functional characterization indicated that LINC00857 plays an important role in tumor proliferation, migration and invasion. We also found that LINC00857 regulated the cell cycle by causing G1/S arrest through regulating CCNE1 and CDK2 expression. The LINC00857 genomic locus was not amplified (Supplementary Figure S8B), thus we hypothesize that the cancer specific LINC00857 expression may be mediated by specific transcription factors coupled with changes in histone and DNA modification (H3K27 acetylation) in its gene promoter. Preliminary in silico analysis of a 1.2 kb LINC00857 promoter region (−1000 to +200 of transcription start site) using MatInspector tool, reveals the presence of DNA binding elements for transcription factors such as CREB, CTCF, NFKB and STAT (Supplementary Figure S13A and S13C). The existing ENCODE data on DNA methylation and transcription factor binding/histone marks from Chip-Seq includes the lung cancer cell line A549 (which has high levels of LINC00857 expression). The ENCODE Infinium 450K methylation data reveals that the LINC00857 promoter is unmethylated in A549 (compared to prostate cell line LNCaP where it is fully methylated). While the Chip-Seq data suggests binding of CTCF and CREB (these binding elements were noted in our in silico analysis of LINC00857 promoter) along with increased histone acetylation marks in A549 cells (Supplementary Figure S13B). The insights gained from these observations will help inform future detailed LINC000857 promoter characterization by generating promoter-reporter constructs (with luciferase as the reporter gene) of various lengths and studying their activity by transfecting them into lung cancer and normal cell lines. This will help us narrow down the critical regions in the promoter and potential transcription factors that regulate LINC000857 expression. In addition we will also look at both DNA and histone modifications of this promoter region in cell lines that exhibit varying levels of LINC00857 expression.
Taken together, our study provides a comprehensive analysis of newly characterized LINC00857 in lung cancer. It establishes the role of LINC00857 as potential driver of lung cancer pathogenesis and a diagnostic or prognostic biomarker of disease.
MATERIALS AND METHODS
Cell lines
H1299, H838, SK-LU-1 cells were maintained in RPMI 1640 or EMEM supplemented with 10% FBS and 1% antibiotic-antimycotic. All cell lines were cultured at 37°C in a 5% CO2 cell culture incubator. All cell lines were obtained from the American Type Culture Collection and maintained using standard media and conditions. All cell lines were genotyped for identity at the University of Michigan Sequencing Core and were tested routinely for Mycoplasma contamination.
Tissue samples
The lung cancer and paired non-tumor lung tissues were obtained from patients undergoing curative cancer surgery during the period from 1991 to 2012 at the University of Michigan Health System. None of the patients included in this study received any preoperative radiation or chemotherapy. All the patients provided informed consent. This project received approval from the University of Michigan Institutional Review Board and Ethics Committee. Resected specimens were frozen in liquid nitrogen and then stored at −80°C until use. Another portion of the tissues were fixed in 10% formalin and embedded in paraffin for histopathological analysis. Frozen tissues for regions containing a minimum of 70% tumor cellularity defined by cryostat sectioning, were utilized for RNA isolation. The median follow-up time was 8.12 years among the patients that remained alive. The clinical information of LUAD samples used in RNA-Seq (67 LUADs) and qRT-PCR validation (101 LUADs) was in Supplementary Tables S1 and S3.
RNA isolation, RNA sequencing and quantitative RT-PCR
Total RNA was isolated from tissue samples and cell lines followed by column purification using miRNeasy Mini-kit (Qiagen) according to the manufacturers' instructions. RNA quality was analyzed by 2100 Bioanalyzer. Samples with RNA integrity number (RIN) > 8.0 were subjected to RNA-Seq. Polyadenylated mRNAs were used to generate strand specific, bar-coded, paired-end Illumina sequencing libraries and sequenced on an Illumina Hi-Seq 2000. cDNA was prepared from RNA samples using High Capacity cDNA Reverse Transcription kit (Applied Biosytems) according to manufacturer's instructions. Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) was prepared using Power SYBR Green master Mix (Life Technology Inc.) and was performed with an ABI StepOne Real-Time PCR System (Applied Biosystems). Each sample had a final volume of 15 μL containing approximately 20 ng of cDNA. The oligonucleotide primers for LINC00857 were in Supplementary Table S7. The housekeeping genes GAPDH and ACTB were used as loading controls. Fold-changes were calculated relative to housekeeping genes and were normalized to the median value in normal samples.
RNA sequencing data analysis
Mate-pair reads were aligned using TopHat [47] against the Ensembl GRCh37 human genome and initial transcripts elucidated with Cufflinks [47, 48]. Expression levels of transcripts were represented as Fragments Per Kilobase Per Million mapped reads (FPKM) [11, 47]. Transcripts were then classified as protein-coding genes, pseudogenes, lncRNAs, etc. according to their overlap with transcripts in known Ensembl database. There were 56,369 transcripts which could be matched to the Ensembl database with many having very low or no expression value. Because a 250–300bp size selection was employed in the RNA-Seq library generation protocol, transcripts that are less than 250bp in length (such as miRNA, snoRNA or snRNA), with background noise or low-level expressed transcripts were removed from further analysis [11].
Published microarray and RNA sequencing data collections
Two published microarray data sets representing 343 primary lung AD tissues were utilized. These included Okayama et al., 226 LUADs with stage 1 and 2 [41], and Tomida et al., with 117 stage 1 to 3 LUADs [42]. The CEL files of microarray data were normalized using Robust Multi-array Average (RMA) method [49]. We also obtained Seo and TCGA RNA-Seq data sets [38, 39] consisting of a total of 394 ADs, 212 SCCs and 150 normal lung tissues. Expression levels of transcripts were represented as FPKM [47]. Our primary outcome was overall survival, censored at five years. The information concerning adjuvant chemotherapy or radiation therapy was provided in the original papers.
Immunoblot analysis
For immunoblot analysis (Western blot analysis), lung cancer cell lysates were boiled in sample buffer, separated by polyacrylamide gel electrophoresis, and transferred onto polyvinylidene difluoride membranes. After blocking for 1 h with 5% non-fat milk, the membranes were incubated with primary monoclonal antibodies against human CCNE1 (1:1000 dilution), GAPDH (1:10,000 dilution) overnight at 4°C. After incubation with HRP-conjugated secondary antibody (at a 1:2000 dilution) for 1 h at room temperature, the membranes were developed using ECL and exposed to X-ray film.
siRNA-mediated knockdown
Cells were plated in 60 mm plates at a desired concentration and transfected with 10 nM experimental siRNA oligonucleotides or non-targeting controls 24 h after plating. Knockdown was performed with Lipofectamine® RNAiMax Reagent (Invitrogen, USA) in OptiMEM medium according to the manufacturer's instructions. Knockdown efficiency was determined by qPCR. LINC00857 siRNA sequences for knockdown experiments are provided in Supplementary Table S8.
Overexpression
Full-length LINC00857 transcript was bought from Thermo and cloned into the Gateway pcDNA-DEST53 vector (Invitrogen) along with control sequence according to the manufacturer's instructions. Insert sequences were confirmed by Sanger sequencing at the University of Michigan Sequencing Core. The lung cancer cell lines SK-LU-1 with low expression of LINC00857 was used and isolated cells generated by selection with Geneticin (Gibco).
Cell proliferation assay
Cells were plated in 96 well plates at a desired concentration and transfected with 10 nM experimental siRNA oligonucleotides or non-targeting controls at 24 h after plating. Knockdown was performed with Lipofectamine® RNAiMax Reagent in OptiMEM medium. At 48 h, 72 h, 96 h after transfection with siRNA, the proliferation rates were measured by Cell Proliferation Reagent (WST-1) (Roche) according to manufacturer's instructions. The cell viability percentages were calculated by normalizing to the survival fraction of the non-target siRNA group.
Basement membrane matrix invasion assays
For invasion assays, cells were treated with the indicated siRNAs. After 48 h transfection, cells were trypsinized, counted with a Coulter counter and diluted to a desired concentration (H1299: 0.5 × 105; H838: 1 × 105. 0.5 ml cell suspension per well). Cells were seeded onto basement membrane matrix Boyden chambers (8-mm pore size, BD) present in the insert of a 24-well culture plate (Matrigel was purchased from BD Company). 20% FBS was added to the lower chamber as a chemoattractant. After 48 h, the non-invading cells and EC matrix were gently removed with a cotton swab. Invasive cells located on the lower side of the chamber were stained with Diff-QuikTM Stain Set (SIEMENS), air dried and photographed.
mRNA expression detection using gene expression array
After siRNA treatment for 48 h on H1299 and H838 cells, the RNAs were collected for gene expression array. Non-target siRNA was used as the control group. Affymetrix Human Gene ST2.1 exon arrays were utilized to detect the RNA expression processed in the University of Michigan Genomics Core.
Cell cycle analysis by flow cytometry
Forty-eight hours after treatment with LINC00857 siRNA and/or control siRNA, H1299 and H838 cells were collected and then fixed with 70% ice-cold ethanol, washed with PBS, re-suspended in 1 ml of propidium iodide (PI) staining solution (0.1% (v/v) Triton X-100, 10 μg/mL PI, and 100 μg/mL DNase-free RNase A in PBS), and then incubated for 30 min at room temperature in the dark. Samples were transfered to the flow cytometer and used to measure the cell cycle in University of Michigan flow cytometry core.
shRNA-mediated stable knockdown
The lung cancer cell line H1299 was seeded at 50–60% confluency and allowed to attach overnight. Cells were transfected with pTRIPZ LINC00857 shRNA or with non-targeting shRNA using Thermo Scientific™ TurboFect™ in vitro Transfection Reagent for 48 h. RFP-positive cells were selected with 1.5 μg/ml puromycin for 1 week. At 48 h after the start of selection, cells were collected for both protein and RNA using RIPA buffer or TRIzol, respectively, as described above.
Mouse xenograft model
All experimental procedures were approved by the University Committee for the Use and Care of Animals (UCUCA). To evaluate the role of LINC00857 in tumor formation and growth, we propagated stable LINC00857 knockdown H1299 pools using LINC00857 shRNAs and non-targeting shRNA control cells and inoculated 1 × 106 cells into the dorsal flank of 5–7 week-old nude mice (CB-17 SCID), using a Matrigel scaffold (BD Matrigel Matrix, BD Biosciences) (n = 10 mice/group). Tumor size was measured every 2 days, and tumor volumes were calculated using the formula (π/6) (L × W2), where L = length of tumor and W = width and represented as mm3/tumor. Post-monitoring, tumors were dissected from the animals, photographed and weighed. Tumor weight is represented by mg/tumor. Drinking water containing 2 mg/ml Doxycycline [50] was changed every two days.
Statistical analyses for experimental studies
Data were analyzed using GraphPad Prism 6 (GraphPad software) and R software. To select the top list of differently expressed lncRNAs in LUAD (vs. normal), we performed Receiver Operating Characteristic (ROC) curve analysis. It showed the tradeoff between sensitivity and specificity (any increase in sensitivity will be accompanied by a decrease in specificity) for the different possible cut-points of a diagnostic test. The diagnostic accuracy was measured by the area under the curve (AUC). An AUC of 1 represented a perfect test; an AUC of 0.5 represented an imprecise test. Survival curves were plotted using the Kaplan-Meier method and survival differences were assessed by the log-rank test using the median of LINC00857 as cutoff value. Univariate or multivariate (adjusted by age, gender, stage and differentiation) Cox proportional hazards models were calculated considering lncRNAs as a continuous variable. To identify pathway/gene expression patterns, an unsupervised hierarchical cluster analysis using un-centered average linkage was performed using Cluster v3.0 after mean-centering genes and arrays and heat maps were visualized using Tree View software [51]. The other data such as proliferation were evaluated by unpaired Student's t-test. A two-tailed p value < 0.05 was considered significant. To determine potential underlying biological processes associated with LINC00857 correlated or regulated genes, Gene Ontology enrichment analysis was performed based on significantly correlated genes using DAVID bioinformatics website [52].
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Acknowledgments
The authors thank Drs. Jiejun Fu and Chunlin Zou for helpful discussions and Ms. Xiaolin Zhou and Xin Huang for editing.
Footnotes
GRANT SUPPORT
This work was supported, in whole or in part, by the National Institutes of Health (grant R01CA154365 to D.G.B and A.M.C.), the University of Michigan's Cancer Center Support Grant (P30 CA46592), University of Michigan's Cancer Center Thoracic Oncology Program Research Grant (G.C.) and a University of Michigan's Department of Surgery RAC grant (G.C.), the National Natural Science Foundation of China (NSFC) Key Project (81130046 to J.Z., NSFC81171993 and NSFC81272415 to Y.L.), Guangxi Key Projects 2013GXNSFEA053004 (J.Z.) and 2012GXNSFCB053004 (Y.L.).
CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nat Med. 2012;18:349–351. doi: 10.1038/nm.2697. [DOI] [PubMed] [Google Scholar]
- 3.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhanasekaran SM, Balbin OA, Chen G, Nadal E, Kalyana-Sundaram S, Pan J, Veeneman B, Cao X, Malik R, Vats P, Wang R, Huang S, Zhong J, et al. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes. Nature communications. 2014;5:5893. doi: 10.1038/ncomms6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang LP, Jean S, Li C, Huang Q, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S, Ostensson M, Akyurek LM, Abrahamsson J, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Chang HY. Physiological roles of long noncoding RNAs: insight from knockout mice. Trends Cell Biol. 2014;24:594–602. doi: 10.1016/j.tcb.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balbin OA, Malik R, Dhanasekaran SM, Prensner JR, Cao X, Wu YM, Robinson D, Wang R, Chen G, Beer DG, Nesvizhskii AI, Chinnaiyan AM. The landscape of antisense gene expression in human cancers. Genome Res. 2015;25:1068–1079. doi: 10.1101/gr.180596.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O'Neill LA, Moore MJ, Caffrey DR, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y, Liu N, Wang JP, Wang YQ, Yu XL, Wang ZB, Cheng XC, Zou Q. Regulatory long non-coding RNA and its functions. J Physiol Biochem. 2012;68:611–618. doi: 10.1007/s13105-012-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Molecular cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, Evans CP, Rosenfeld MG. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 18.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS genetics. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, et al. large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15 (INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. The FEBS journal. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 26.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic acids research. 2010;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Molecular cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Kok JB, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, Swinkels DW, Schalken JA. DD3 (PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695–2698. [PubMed] [Google Scholar]
- 31.Meseure D, Drak Alsibai K, Nicolas A, Bieche I, Morillon A. Long Noncoding RNAs as New Architects in Cancer Epigenetics, Prognostic Biomarkers, and Potential Therapeutic Targets. BioMed research international. 2015;2015:320214. doi: 10.1155/2015/320214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roobol MJ, Schroder FH, van Leeuwen P, Wolters T, van den Bergh RC, van Leenders GJ, Hessels D. Performance of the prostate cancer antigen 3 (PCA3) gene and prostate-specific antigen in prescreened men: exploring the value of PCA3 for a first-line diagnostic test. European urology. 2010;58:475–481. doi: 10.1016/j.eururo.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 33.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 34.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. The Journal of international medical research. 2011;39:2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 35.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, Zornig M, MacLeod AR, Spector DL, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W, De W, Wang Z, Wang R. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PloS one. 2013;8:e77293. doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A, Hillejan L, Wiebe K, Berdel WE, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. Journal of thoracic oncology. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 38.Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, Lee J, Jung YJ, Kim JO, Shin JY, Yu SB, Kim J, Lee ER, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–2119. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.TCGA Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth A, Diederichs S. Long Noncoding RNAs in Lung Cancer. Curr Top Microbiol Immunol. 2015 Jun 3; doi: 10.1007/82_2015_444. [DOI] [PubMed] [Google Scholar]
- 41.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S, Sakamoto H, Kumamoto K, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 42.Tomida S, Takeuchi T, Shimada Y, Arima C, Matsuo K, Mitsudomi T, Yatabe Y, Takahashi T. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol. 2009;27:2793–2799. doi: 10.1200/JCO.2008.19.7053. [DOI] [PubMed] [Google Scholar]
- 43.Quinn JJ, Ilik IA, Qu K, Georgiev P, Chu C, Akhtar A, Chang HY. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol. 2014;32:933–940. doi: 10.1038/nbt.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sang H, Liu H, Xiong P, Zhu M. Long non-coding RNA functions in lung cancer. Tumour Biol. 2015;36:4027–37. doi: 10.1007/s13277-015-3449-4. [DOI] [PubMed] [Google Scholar]
- 45.Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15:429. doi: 10.1186/s13059-014-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 50.Centlivre M, Zhou X, Pouw SM, Weijer K, Kleibeuker W, Das AT, Blom B, Seppen J, Berkhout B, Legrand N. Autoregulatory lentiviral vectors allow multiple cycles of doxycycline-inducible gene expression in human hematopoietic cells in vivo. Gene Ther. 2010;17:14–25. doi: 10.1038/gt.2009.109. [DOI] [PubMed] [Google Scholar]
- 51.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.