Crystal structures of the C-terminal domains of the Ebolavirus nucleoproteins (NPCt) from the Bundibugyo and Taï Forest species (BDBV and TAFV, respectively) have been determined. The structures show high similarity to that reported for the Zaire Ebolavirus NPCt. However, NMR data revealed that the corresponding domain from the NP of the related MARV species of Marburgvirus is distinctly different.
Keywords: viral proteins, Ebolavirus, Marburgvirus, nucleoprotein, Filoviridae
Abstract
The Filoviridae family of negative-sense, single-stranded RNA (ssRNA) viruses is comprised of two species of Marburgvirus (MARV and RAVV) and five species of Ebolavirus, i.e. Zaire (EBOV), Reston (RESTV), Sudan (SUDV), Taï Forest (TAFV) and Bundibugyo (BDBV). In each of these viruses the ssRNA encodes seven distinct proteins. One of them, the nucleoprotein (NP), is the most abundant viral protein in the infected cell and within the viral nucleocapsid. It is tightly associated with the viral RNA in the nucleocapsid, and during the lifecycle of the virus is essential for transcription, RNA replication, genome packaging and nucleocapsid assembly prior to membrane encapsulation. The structure of the unique C-terminal globular domain of the NP from EBOV has recently been determined and shown to be structurally unrelated to any other known protein [Dziubańska et al. (2014 ▸), Acta Cryst. D70, 2420–2429]. In this paper, a study of the C-terminal domains from the NP from the remaining four species of Ebolavirus, as well as from the MARV strain of Marburgvirus, is reported. As expected, the crystal structures of the BDBV and TAFV proteins show high structural similarity to that from EBOV, while the MARV protein behaves like a molten globule with a core residual structure that is significantly different from that of the EBOV protein.
1. Introduction
Most viruses of the Ebolavirus and Marburgvirus genera, which comprise the Filoviridae family within the order Mononegavirales, cause severe hemorrhagic fever in humans. The ongoing outbreak of Ebola virus disease (EVD) in West Africa demonstrates the grave threat that filoviruses pose globally to human health. While the EVD outbreak is slowly losing momentum, it is still unprecedented, resulting in over 23 000 cases and more than 9000 deaths when this manuscript was in preparation (according to Center for Disease Control data). The epidemic spread more extensively and rapidly than in past outbreaks, beginning in Guinea in December 2013 and then spreading throughout Guinea, Liberia and Sierra Leone (Baize et al., 2014 ▸). The current epidemic is caused by a variant of the Zaire strain (EBOV), one of five currently known Ebolavirus species. The other four are Taï Forest (TAFV), Bundibugyo (BDBV), Reston (RESTV) and Sudan (SUDV) (Bukreyev et al., 2014 ▸). EBOV, SUDV and BDBV have all caused large outbreaks of EVD in the past (Roddy et al., 2012 ▸; Feldmann & Geisbert, 2011 ▸), while TAFV has so far been reported in only one patient (Formenty et al., 1999 ▸). The five Ebolavirus species vary with respect to their corresponding fatality rates: RESTV is nonpathogenic to humans and SUDV and BDBV have fatality rates of about 40%, while EBOV has had a fatality rate of up to 90% in the past, although in the current outbreak it has been significantly lower. The fatality rate in the only two significant outbreaks of Marburg fever, caused by MARV infection, was also ∼90% (Brauburger et al., 2012 ▸). The molecular basis underlying this variation in fatality rate is not known.
Like other viruses in the Mononegavirales order, Ebolavirus and the closely related Marburgvirus are membrane-enveloped viruses which contain negative-sense, single-stranded RNA (ssRNA) encoding seven genes that owing to co-transcriptional and post-translational processes generate more than seven proteins. Two of them are associated with the membrane: the glycoprotein (GP), which is a transmembrane protein, and VP40, which is associated with the inner surface of the membrane. The remaining five principal proteins [i.e. the nucleoprotein (NP), VP35, VP30, VP24 and RNA polymerase (L)] all interact with ssRNA to form the nucleocapsid (Beniac et al., 2012 ▸; Bharat et al., 2012 ▸; Mühlberger et al., 1999 ▸). The critical protein responsible for the assembly of the nucleocapsid is the NP. It is a 739-residue (EBOV) single polypeptide chain, with an N-terminal region (approximately 1–400) that is engaged in packaging the ssRNA (Watanabe et al., 2006 ▸) and a C-terminal region that is largely unstructured and which is implicated in several protein–protein interactions (Noda et al., 2007 ▸; Shi et al., 2008 ▸; Licata et al., 2004 ▸). We have recently shown that this region contains a unique globular domain (NPCt; residues 641–739) and we have determined its crystal structure (Dziubańska et al., 2014 ▸). Crystallographic studies have also been reported for the N-terminal globular domain with and without a peptide derived from VP35 (PDB entries 4ypi, 4z9p and 4ztg; Leung et al., 2015 ▸; Dong et al., 2015 ▸; Kirchdoerfer et al., 2015 ▸).
Interestingly, the NPCt contains one of the most divergent amino-acid sequences among the proteins encoded by the Filoviridae. Pairwise sequence comparisons between the five Ebolavirus species show amino-acid identity ranging from 60 to 85%, while the NPCt from MARV contains only 12 residues found in the Ebolavirus consensus sequence. In this context, it is interesting to note that the nucleocapsids of Ebolavirus and Marburgvirus exhibit differences in their structures, suggesting functional variation among the respective nucleoproteins (Watanabe et al., 2006 ▸; Kolesnikova et al., 2000 ▸; Mavrakis et al., 2002 ▸; Huang et al., 2002 ▸; Noda et al., 2006 ▸). In an effort to gain more understanding of the sequence–function relationships in the nucleoproteins from Filoviridae, we have undertaken a study of the NPCt domains from other species of Ebolavirus and from MARV. In this paper, we report the crystal structures of the NPCt from BDBV and TAFV, and the surprising results of NMR studies of the MARV NPCt, attesting to critical differences between the Ebolavirus and Marburgvirus nucleoproteins.
2. Materials and methods
2.1. Preparation of recombinant proteins
2.1.1. Cloning, expression and purification
cDNA constructs coding for the 641–739 fragments of TAFV, BDBV and RESTV NP, as well as the 641–738 fragment of SUDV NP and the 600–695 fragment of MARV NP, were synthesized commercially (GENEWIZ) using optimized codon frequencies for Escherichia coli. The constructs were cloned into the MBP-His6-Parallel1 vector with an rTEV cleavage site downstream of His6 (Sheffield et al., 1999 ▸).
Briefly, BL21-CodonPlus (DE3)-RIPL E. coli cells (Stratagene) were grown in Terrific Broth at 37°C. Induction was carried out at an OD600 of ∼2.5 with the addition of 0.5 mM IPTG after the temperature of the culture was decreased to 16°C, and growth continued for 18 h. The cells were harvested by centrifugation and the pellet was frozen at −20°C. The pellet was resuspended in 5 ml lysis buffer (50 mM Tris–HCl, 300 mM NaCl, 5 mM β-mercaptoethanol pH 7.5) per gram of pellet. Cells were disrupted with a Dounce homogenizer and sonication (Branson Sonifier 450) and were centrifuged at 35 000 rev min−1 (45 Ti rotor) for 45 min. The supernatant was applied onto an Ni–NTA gravitational column (5 ml resin; Qiagen). The column was washed with 400 ml lysis buffer and the recombinant protein was eluted with 50 mM Tris–HCl, 300 mM NaCl, 5 mM β-mercaptoethanol, 250 mM imidazole pH 7.5. The affinity tags were removed by incubating the recombinant protein with rTEV protease with concomitant dialysis against 4 l dialysis buffer (50 mM Tris–HCl, 300 mM NaCl, 5 mM β-mercaptoethanol pH 7.5) overnight. The protein solution was then applied onto an Ni–NTA column and the flowthrough containing NPCt was collected, followed by a 30 ml wash with the same buffer. Samples concentrated using Amicon microconcentrators with a 3000 Da cutoff were subjected to size-exclusion chromatography on a Superdex 75 column connected to a GE Healthcare ÄKTA system and equilibrated with 50 mM Tris–HCl, 150 mM NaCl, 5 mM β-mercaptoethanol pH 7.5 at 4°C. Fractions containing NPCt were pooled. The BDBV NPCt fusion protein (MBP-His6-BDBV) was also purified using an amylose resin column (Qiagen) according to the manufacturer’s instructions to achieve higher purity prior to cleavage.
2.1.2. Preparation of 15N-labeled and 13C,15N-labeled MARV NPCt
15N-Labeled and 13C,15N-labeled MARV NPCt samples were expressed as described previously for the EBOV NPCt (Dziubańska et al., 2014 ▸). Labeled proteins were purified in exactly the same manner as unlabeled MARV NPCt, except that the buffer for size-exclusion chromatography consisted of 40 mM HEPES, 150 mM NaCl, 5 mM β-mercaptoethanol pH 7.5. For assignment experiments, a sample of 500 µM 15N-NPCt and 950 µM 13C,15N-NPCt in 40 mM HEPES, 150 mM NaCl, 5 mM β-mercaptoethanol pH 7.5 buffer supplemented with 5% D2O was prepared.
2.2. Thermal stability assays
The midpoint melting temperature (T m) of protein samples was determined by monitoring the fluorescence of SYPRO Orange dye (Life Technologies) in the presence of the protein as a function of temperature (Ericsson et al., 2006 ▸). Assays were performed in 20 µl containing 20 µg protein, with the concentration of the dye as recommended by the manufacturer (i.e. 1000× dilution of the DMSO stock, followed by 5× dilution in the final sample) and buffer (50 mM Tris–HCl, 150 mM NaCl, 5 mM β-mercaptoethanol pH 7.5). Fluorescence was recorded as a function of temperature from 20 to 90°C using an Applied Biosystems StepOnePlus Real-Time PCR System (Life Technologies). This instrument uses wavelengths of 488 nm for excitation and 586 nm for emission. The StepOne software (v.2.1) was used for data processing and figure preparation.
2.3. Crystallization of NPCt
All crystallization experiments were carried out using the sitting-drop vapor-diffusion method in crystallization screens set up with a Mosquito robot (TTP Labtech). Protein samples were concentrated to ∼10–20 mg ml−1. Three commercial screens were used, JCSG+, PEG/Ion and SaltRX, in a canonical setting or using the alternative reservoir approach (Newman, 2005 ▸). For each crystallization condition, 1:1 and 2:1 ratios of precipitant to protein solution were used, with a drop volume of 200–250 nl.
2.4. Data collection and structure determination
Crystals were cryoprotected under a range of conditions and then screened for diffraction quality. The crystals of TAFV NPCt used for final data collection were flash-cooled directly from the crystallization drop. Those of BDBV NPCt that subsequently gave the best diffraction were soaked in 1.5 M LiSO4, 0.075 M Tris pH 8.5, 20% glycerol. All crystals were flash-cooled by immersion into liquid nitrogen. X-ray data were collected at ∼100 K on the SER-CAT beamlines (Southeast Regional Collaborative Access Team) at the Advanced Photon Source, Argonne National Laboratory. Data were indexed, integrated and scaled with HKL-3000 (Minor et al., 2006 ▸). Table 1 ▸ gives details of the data-processing statistics.
Table 1. Crystallization data and refinement statistics for the NPCt domain from TAFV and BDBV.
Values in parentheses are for the highest resolution shell.
| TAFV NPCt | BDBV NPCt | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.9792 | 0.9900 |
| Space group | P1 | P6422 |
| Z | 8 | 12 |
| Unit-cell parameters (Å, °) | a = 57.85, b = 60.08, c = 73.55, α = 69.15, β = 68.94, γ = 89.9 | a = b = 60.08, c = 82.74 |
| Resolution (Å) | 50.00–2.10 (2.14–2.10) | 80.00–2.30 (2.38–2.30) |
| Completeness (%) | 96.1 (84.6) | 94.3 (64.7) |
| Total observations | 103077 | 184210 |
| Mean I/σ(I) | 12.4 (1.9) | 31.3 (1.9) |
| CC1/2 | (0.789) | (0.981) |
| Multiplicity | 2.1 (2.0) | 14.8 (7.1) |
| R merge † (%) | 0.070 (0.316) | 0.084 (0.432) |
| Structure refinement | ||
| Unique reflections | 48012 | 12396 |
| Reflections in R free set | 2235 | 604 |
| R ‡ (%) | 19.1 | 16.1 |
| R free ‡ (%) | 26.0 | 20.2 |
| R.m.s.d., bond lengths (Å) | 0.007 | 0.008 |
| R.m.s.d., bond angles (°) | 1.1 | 1.3 |
| No. of atoms | ||
| Protein atoms | 6738 | 870 |
| O atoms from waters | 664 | 113 |
| Ligand/ion atoms | 73 | 13 |
| Mean B value (Å2) | ||
| Overall | 42 | 82 |
| Protein atoms (Å2) | 42 | 82 |
| O atoms from waters (Å2) | 45 | 79 |
| Ligand/ion atoms (Å2) | 47 | 116 |
| Clashscore | 0.08 | 0.00 |
| Clashscore percentile (%) | 100 | 100 |
| Rotamer outliers (%) | 1.76 | 1.09 |
| Ramachandran outliers (%) | 0.00 | 0.00 |
| Ramachandran favored (%) | 99.61 | 99.01 |
R
merge = 
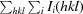 , where 〈I(hkl)〉 is the mean of i observations I
i(hkl) of reflection hkl.
, where 〈I(hkl)〉 is the mean of i observations I
i(hkl) of reflection hkl.
R factor and R
free = 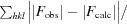
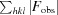 , where F
obs and F
calc are the observed and calculated structure factors, respectively, calculated for recorded data (R factor) and for 5% of the data which were omitted in refinement (R
free).
, where F
obs and F
calc are the observed and calculated structure factors, respectively, calculated for recorded data (R factor) and for 5% of the data which were omitted in refinement (R
free).
The structure of TAFV NPCt was solved at 2.1 Å resolution by molecular replacement (MR) using MOLREP (Vagin & Teplyakov, 2010 ▸) operated within the HKL-3000 interface (Minor et al., 2006 ▸). The crystal structure of the NPCt from EBOV (PDB entry 4qb0; Dziubańska et al., 2014 ▸) was used as a search model to obtain the initial phases. The structure of the BDBV NPCt was determined at 2.3 Å resolution using the same search model and Phaser (Bunkóczi et al., 2013 ▸) as implemented in PHENIX (Adams et al., 2010 ▸). Refinement of both structures was performed with REFMAC5 (Murshudov et al., 2011 ▸) within HKL-3000, with Coot (Emsley et al., 2010 ▸), allowing manual intervention, and with the CCP4 suite (Winn et al., 2011 ▸). TLS (translation/libration/screw) refinement was used in the next stage of refinement, and TLS groups were determined using the TLSMD server (Painter & Merritt, 2006 ▸). Individual isotropic displacement parameters were used from this point onwards. Water molecules were added in several stages during the refinement automatically and were reviewed visually. The standalone version of the MolProbity server (Chen et al., 2010 ▸) and the PDB Validation Server were both used for structure validation (Berman et al., 2000 ▸). Crystal contacts were analyzed using the PISA server (Krissinel & Henrick, 2007 ▸). The final electron-density maps are of high quality and are easily interpretable (Fig. 1 ▸). Refinement statistics are summarized in Table 1 ▸.
Figure 1.
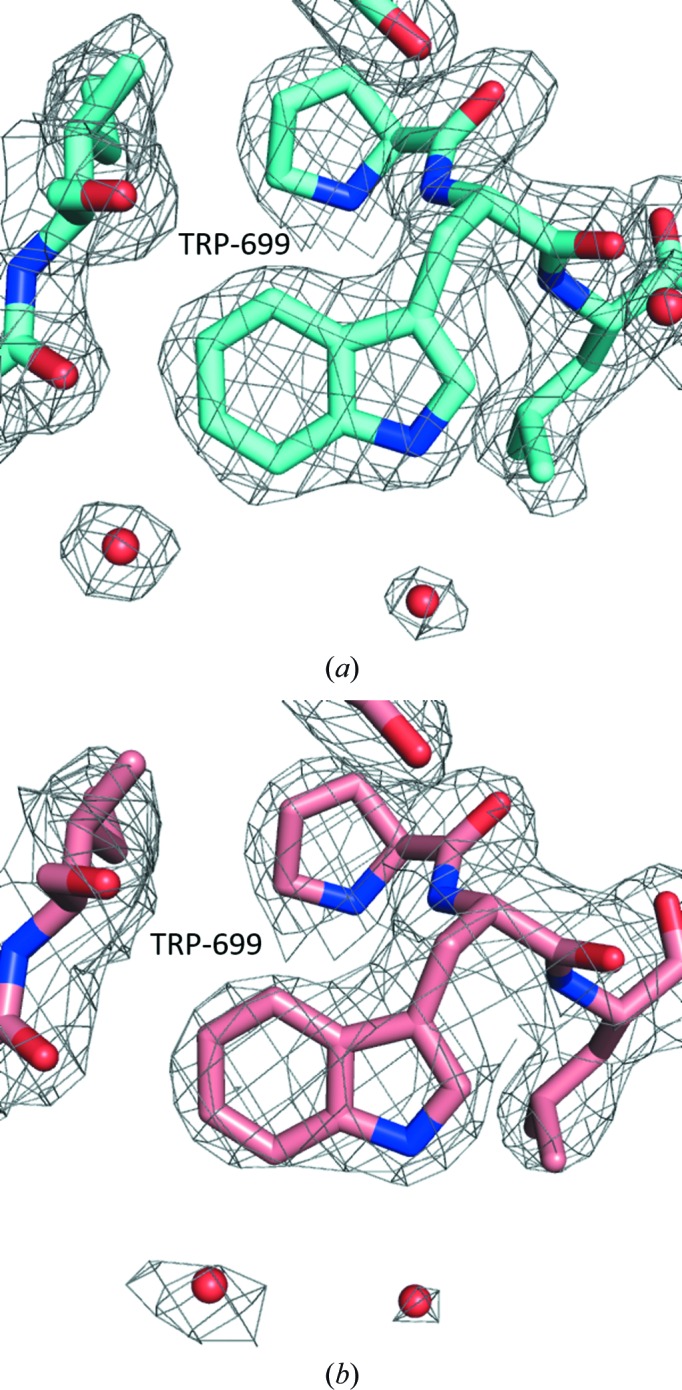
Representative electron-density map (2F o − F c) centered on Trp669 for TAFV NPCt. (a) Analogous electron density for the same structural element in BDBV NPCt. (b) The electron-density map is contoured at the 1σ level; O atoms are shown in red, N atoms in blue and C atoms in magenta and pink (for TAFV and BDBV, respectively).
2.5. Heteronuclear NMR
A Bruker Avance III 600 MHz spectrometer equipped with a cryoprobe was used to obtain NMR spectra at 25°C. NMRPipe (Delaglio et al., 1995 ▸) was used to process the spectral data. CcpNmr Analysis v.2 (Vranken et al., 2005 ▸) and Sparky 3 (T. D. Goddard & D. G. Kneller, University of California, San Francisco, USA) were used for spectrum visualization and sequential assignment of backbone and Cβ, 1H (except Hα), 13C and 15N resonances. TALOS-N was used for used for estimation of secondary structure (Shen & Bax, 2013 ▸).
2.6. Sequence and structure analysis
PyMOL (Schrödinger) was used to align structures and to calculate r.m.s.d. values between superposed structures with cycles and transform parameters set to zero. PyMOL was also used to generate figures. The STRIDE server (Heinig & Frishman, 2004 ▸) was used for assignment of two-dimensional structure elements from atomic resolution structures.
3. Results and discussion
3.1. Expression and purification
All five C-terminal domains of NP under study, i.e. from TAFV, BDBV, SUDV, RESTV and MARV, were overexpressed in E. coli as fusion proteins with an N-terminal MBP-His6 double tag, cleaved from the tag with rTEV and purified to homogeneity as described above (Supplementary Fig. S1). The final yields were approximately 10–15 mg pure protein per litre of culture.
3.2. Thermal stability
To ascertain that the expressed proteins are folded into stable modules in solution, we tested all five using a thermal shift assay which utilizes the dye SYPRO Orange. Table 2 ▸ lists the values for the temperature midpoints of cooperative unfolding determined for the five strains of EBOV. All fall within a relatively narrow range between 48.3 and 52.8°C, consistent with previous results obtained for the EBOV protein (Dziubańska et al., 2014 ▸). Unexpectedly, no thermal unfolding was observed for the MARV NPCt (Supplementary Fig. S2).
Table 2. Thermal stability of NPCt domains from various Ebola strains.
| Species of Ebolavirus | Midpoint of unfolding (°C) |
|---|---|
| ZEBOV | 56.9 |
| TAFV | 52.8 |
| BDBV | 50.8 |
| SUDV | 48.5 |
| RESTV | 48.3 |
3.3. Crystallization
Crystallization screens were set up as described above. Of the five proteins screened, the NPCt domains from TAFV and BDBV both yielded X-ray-quality single crystals, while those from RESTV, SUDV and MARV did not.
For the TAFV NPCt, initial crystals appeared in the JCSG+ screen in a range of solution including those consisting of (i) 20% PEG 8000, 0.1 M CHES pH 9.5; (ii) 10% PEG 6000, 0.1 M HEPES pH 7; and (iii) 20% PEG 8000, 0.2 M MgCl2, 0.1 M HEPES pH 7. Based on these observations, crystallization was optimized using a 1:1 ratio of precipitant (10% PEG 3350, 0.027 M HEPES/0.073 M Tris pH 8.6) to protein solution, with a protein concentration of ∼5.0 mg ml−1 and a 1.5 M NaCl reservoir. The crystals belonged to space group P1 and diffracted to 2.1 Å.
In the case of the BDBV NPCt, initial crystals appeared in a 1:1 mixture with 1.5 M lithium sulfate monohydrate, 0.1 M Tris pH 8.5 (SaltRx) equilibrated against a reservoir containing the screen solution. These conditions were used as a starting point for optimization, which led to single crystals suitable for X-ray experiments, which were grown using a 1:1 ratio of reservoir (1.33 M LiSO4, 0.1 M Tris pH 8.5) to protein solution with a protein concentration of 13.9 mg ml−1. The crystals exhibited the symmetry of space group P6222 or P6422 and diffracted to 2.3 Å resolution.
The SUDV protein crystallized in a number of conditions containing PEG 3350, but the crystals did not diffract well in spite of extensive efforts to improve them. Neither the RESTV nor the MARV proteins formed crystals in any of the screens. Efforts to crystallize MARV were discontinued in favor of an heteronuclear NMR study, which is described below.
3.4. The crystal structures of the Taï Forest and Bundibugyo NPCt domains
3.4.1. Overview
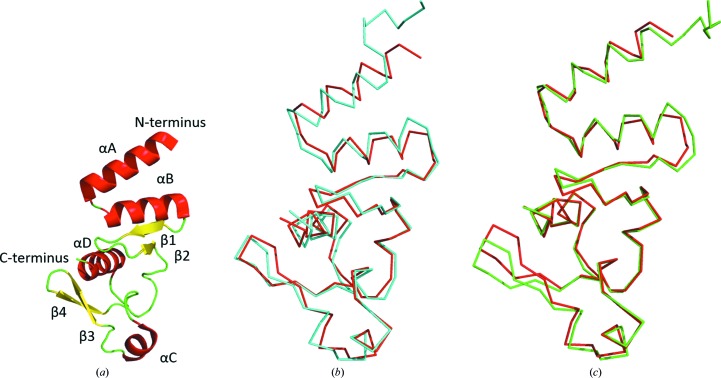
In general terms, the two crystal structures determined in our study show high similarity to the NPCt domain from EBOV, the structure of which has been reported by us previously (Dziubańska et al., 2014 ▸). This was of course expected given the 82% sequence identity between the BDBV and EBOV proteins, the 79% sequence identity between the TAFV and EBOV proteins, and the 86% sequence identity between the BDBV and TAFV proteins. The tertiary fold is the same: there are two α-helices at the N-terminus (αA and αB) folded into an antiparallel hairpin, followed by two antiparallel β-strands (β1 and β2), a coil fragment containing another short α-helix (αC) and another pair of antiparallel β-strands (β3 and β4); finally, the C-terminal α-helix (αD) which inserts itself between the two β-hairpins provides a number of hydrophobic interactions at the core (Fig. 2 ▸). The superposition of the Cα models presented here on EBOV NPCt (PDB entry 4qb0) shows that the core fragments, composed of αB, β1, β2 and αD, are virtually identical, as illustrated by low pairwise r.m.s.d. values of 0.3–0.9 Å (Cα only; see Fig. 2 ▸ and Table 3 ▸). Nevertheless, the domain shows some intrinsic flexibility, particularly with respect to the two C-terminal β-strands (β3 and β4) and the N-terminal α-helix. A comparison of crystal structures and molecular packing suggests that the slight conformational differences are more likely to be caused by packing forces and distortions owing to crystal contacts rather than genuine variation between strains. In each of the two crystal structures described here there are interesting aspects of molecular packing, and we describe those briefly below.
Figure 2.
(a) The EBOV NPCt structure in cartoon representation; α-helices are shown as red ribbons and β-sheets as yellow arrows. (b, c) Superposition of EBOV NPCt (red) on TAFV NPCt (cyan) and BDBV NPCt (green), respectively. Protein models are shown in Cα-trace representation.
Table 3. Comparison of structural changes in the characterized Ebolavirus NPCt structures expressed by Cα r.m.s.d. value.
Left, Cα r.m.s.d. of the NPCt structure (fragment 645–739); right, Cα r.m.s.d. of the core of the NPCt domain (αB, β1, β2 and αD).
3.4.2. The Bundibugyo NPCt structure
The molecular-replacement calculation identified P6422 as the correct space group. The atomic model of the BDBV NPCt includes the entire polypeptide comprising residues 641–739, as well as three additional residues remaining after removal of the MBP-His6 tag from the N-terminus (Ala-Met-Ala). There is only one molecule in the asymmetric unit, with an exceptionally high solvent content of 79%, which rationalizes the high mean isothermal displacement (B-factor) parameter for protein atoms of 72 Å2. In spite of this, the structure is relatively well resolved, with only six side chains (Lys684, Glu702, Lys703, Met706, Asp716 and Arg739) showing a significant degree of disorder.
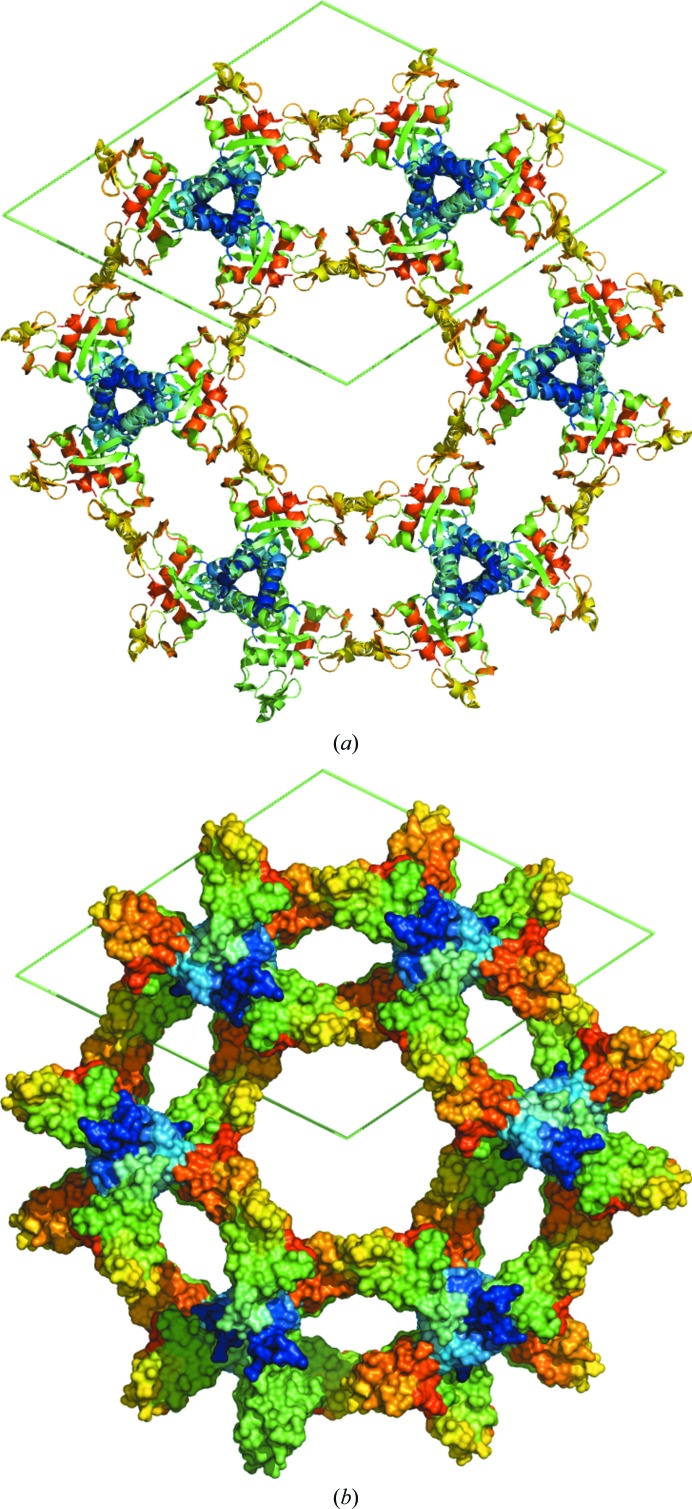
The BDBV NPCt structure is a good example of ‘minimal packing’, as it shows only three intermolecular contacts, which is the smallest number required for three-dimensional packing, except for the P212121 space group where only two are sufficient (Wukovitz & Yeates, 1995 ▸). The first contact involves the N-terminal fragment, which includes the tripeptide from the expression vector followed by the N-terminal five amino acids that in the Zaire isoform are unstructured in the crystal. This fragment folds into an extended conformation and aligns itself next to the β1 strand of the adjacent monomer in an antiparallel fashion. This homotypic interaction creates a crystallographic dimer with a total interface of over ∼1600 Å2. The surface buried by this contact contains significant large hydrophobic patches, and the critical residues are Met639, Ala640, Gln643, Tyr668, Glu674 and Ile677 (Supplementary Fig. S3). The propensity of unstructured N-terminal fragments to mediate crystal contacts has been noted by us before and runs contrary to the assumption that one should always remove even short tags to facilitate crystallization. The second contact is mediated by the N-terminal α-helical hairpin, and also creates a crystallographic dimer, burying over 1000 Å2 of surface in both molecules. Most of this surface is hydrophobic and is contributed by Met651, Gln659, Tyr667, Met670 and Met671. Together, these two primary contacts generate a linear ensemble of molecules lying along a threefold screw axis. The three-dimensional packing is made possible by the third contact, also homotypic, related by a twofold axis and burying over 800 Å2. It is formed by a back-to-back packing by the C-terminal β-hairpin, with the most surface contributed by Asp709, Phe712, Gln718, Gln719 and Tyr721. As a result, six strands of molecules related by contacts 1 and 2, all lying along the threefold screws, are related by the sixfold screw axis, along which runs an enormous solvent channel providing the 79% solvent content (Fig. 3 ▸).
Figure 3.
Molecular packing in the crystals of the BDBV NPCt domain looking down the sixfold screw axis. (a) Cartoon representation; coloring follows the RAINBOW option in PyMOL, starting with blue at the N-terminus. (b) Surface representation, visualizing the solvent channels; colored as in (a).
Given that the NPCt appears to be predominantly monomeric in solution, it is unlikely that the crystal packing has direct functional implications. However, the surfaces involved in crystal contacts, especially the distinctly hydrophobic surface of the N-terminal α-helical hairpin, may be involved in protein–protein interactions within the EBOV nucleocapsid, with the NPNt domain or perhaps between the NP and the VP40 matrix protein, the existence of which has been suggested by other studies (Licata et al., 2004 ▸; Noda et al., 2007 ▸; Liu et al., 2011 ▸; Bharat et al., 2012 ▸).
3.4.3. The Taï Forest NPCt structure
The TAFV NPCt crystal structure contains eight molecules in the asymmetric unit, although like other NPCt domains the protein is monomeric in solution as judged by gel filtration (data not shown). There is dynamic disorder that affects a number of surface residues (i.e. Lys641, His643–Ser647, Glu649, Glu650, Arg653, Glu657, Lys684, Glu695, Asp716, Asp717, Gln718, Gln736 and Lys739), so that there is at best a very low level of corresponding interpretable electron density for them either in one or more of the molecules. The mean isothermal displacement (B-factor) parameter for protein atoms in this structure is 42 Å2. It was not possible to build the N-terminal in chains E and F, where dynamic disorder makes it difficult to model a single conformation.
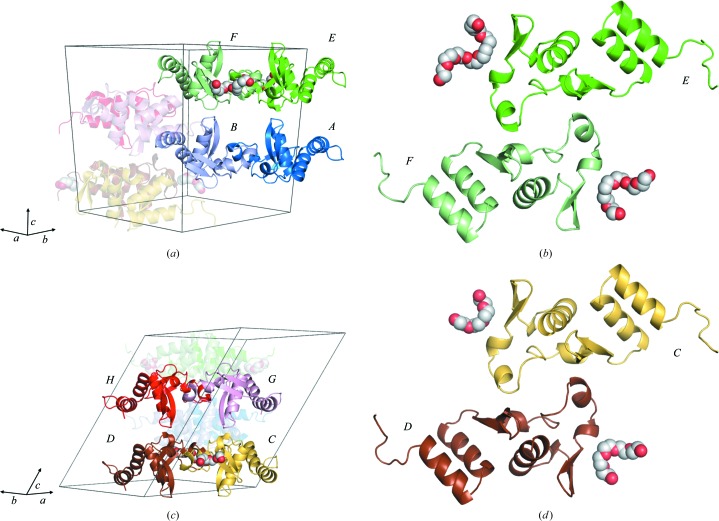
The asymmetric unit may be defined as two sets of four molecules, with each set affected by different packing forces: i.e. chains A, B, G and H and chains C, D, E and F (Fig. 4 ▸). Within each group, the molecules adopt nearly identical conformations. However, a comparison of the Cα coordinates between the two sets (e.g. between the A and E chains) shows an r.m.s.d. of 1.2 Å. This difference is primarily owing to the fact that the fragment including the β3–loop–β4 in one group of molecules rotates 15.9° when compared with the other group. When only the core structure is taken into account (Table 3 ▸) the r.m.s.d. between the two is 0.3 Å.
Figure 4.
Molecular packing and crystal contacts in the TAFV NPCt crystals. (a) A view visualizing the A/B and E/F crystallographic dimers related by a noncrystallographic translation vector. (b) A view of the A/B dimer together with bound fragments of PEG 3350 down the noncrystallographic twofold axis. (c) A view similar to that in (a) of the C/D and G/H dimers. (d) A view of the E/F dimer together with bound fragments of PEG 3350 down the noncrystallographic twofold axis
The eight molecules in the asymmetric unit are arranged into four nearly identical noncrystallographic dimers, i.e. A/B, C/D, E/F and G/H. The A/B and E/F pairs are related by a noncrystallographic translation along a vector perpendicular to the a*b plane; the same relationship occurs between the C/D and G/H pairs. Within each dimer, the monomers are related by a twofold axis parallel to the noncrystallographic translation vector (Fig. 4 ▸). The A/B/C/D and E/F/G/H sets of molecules, along with their symmetry-related partners, each form distinct layers within the crystal. The layers are stabilized by the antiparallel hydrogen-bonded interaction of the N-termini of the A and B molecules with the β2 strands of the symmetry-related B and A chains, respectively, and vice versa; a symmetric set of interactions occurs between the N-termini of the C and D molecules and the β2 strands of the symmetry-related D and C molecules. Analogous crystal contacts are observed in the layer formed by the E/F and G/H dimers. The only close crystal contact between the two layers of molecules, leading to three-dimensional ordering, is between the B and H molecules.
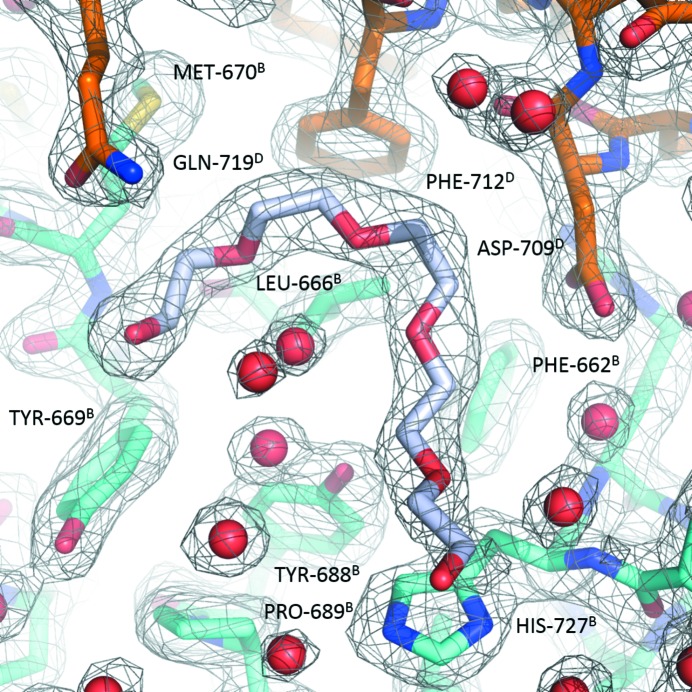
Such high-order NCS is intriguing, given no obvious reasons that differentiate the monomers. Somewhat unexpectedly, the answer appears to lie in part in the crystallization milieu, and specifically in the presence of PEG 3350, which is used as a precipitant. Fragments of four PEG molecules were identified in this crystal structure with interpretable electron density (Fig. 5 ▸). These four fragments associate with the polypeptide chains that form the A/B and G/H dimers. All four are located in the same position on the surface of the protein between helices αA and αB, and seem to cover a distinct hydrophobic patch created by the solvent-exposed side chains of Phe662, Leu666 and Tyr669. They are also wedged into crystal contacts: in the case of the A/B dimer the PEGs mediate crystal contacts with adjacent protein molecules C and D, respectively; for the G/H dimer the contacts involve the E and F molecules. The binding of the PEG molecules is also correlated with the enhanced order of the N-termini: the A/B and G/H dimers have ordered N-termini, while the C/D and E/F dimers have much weaker density.
Figure 5.
Representative 2F o − F c electron-density map contoured at the 1σ level for the PEG 3350 molecule; the PEG molecule is colored magenta and is associated with protein monomers B (cyan) and D (gold). All residues are labeled with both the number and the chain identifier. Waters are shown as red spheres.
3.5. The MARV NPCt
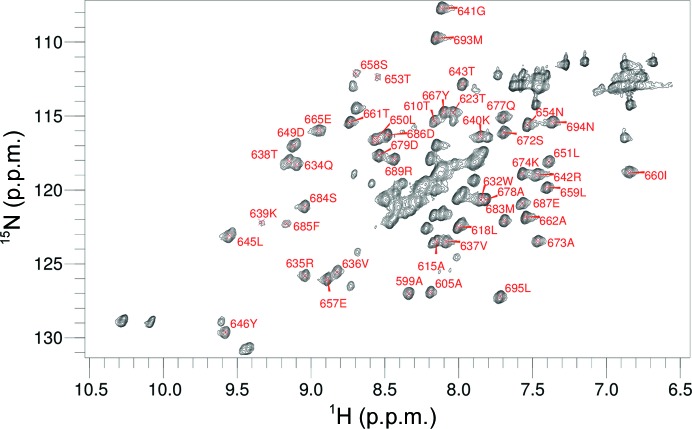
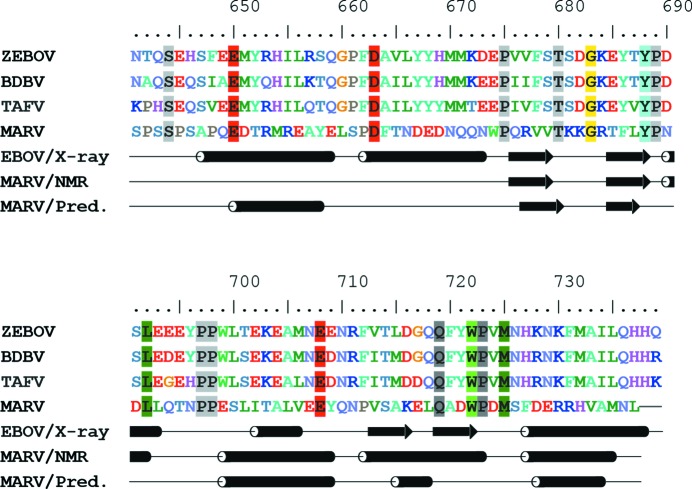
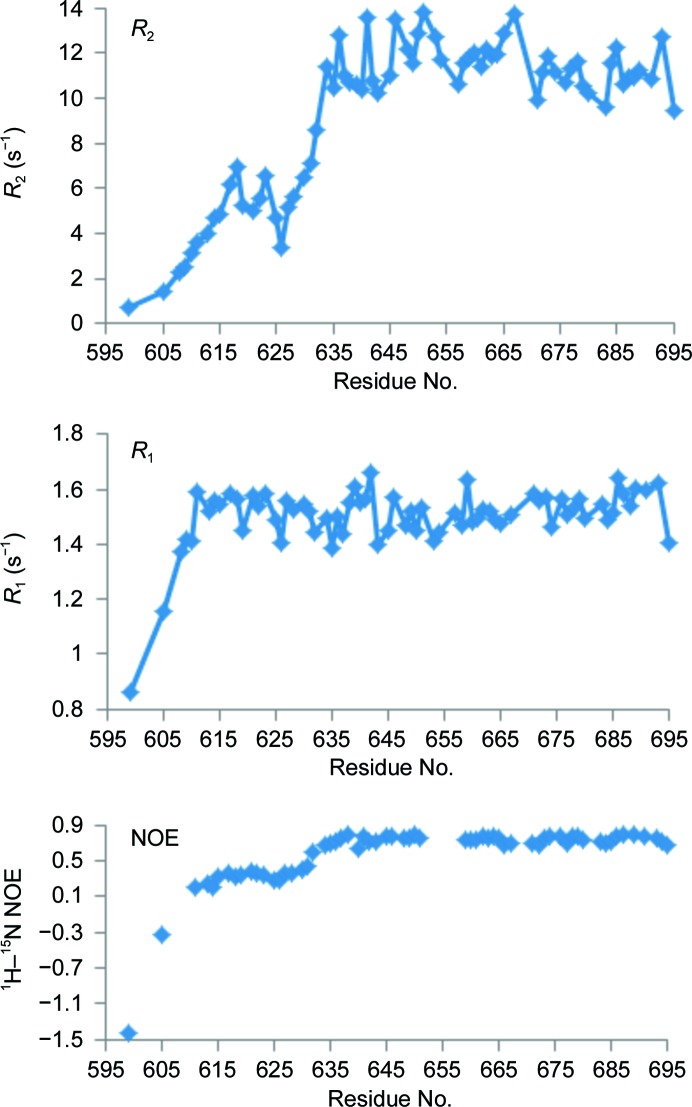
The lack of a detectable melting transition for this protein, along with the failure of all attempts to obtain crystals, prompted us to investigate the structure of MARV NPCt in solution using heteronuclear NMR. Although we expected the protein to be unfolded, the 15N–1H chemical shift correlation (HSQC) spectrum showed significant peak dispersion (Fig. 6 ▸). We were also able to obtain three-dimensional HNCO, HN(CA)CO, CBCA(CO)NH and HNCACB spectra and to determine the backbone (except Hα) and Cβ assignments for 82 out of 100 residues. Nine of the unassigned residues were within a 12-residue fragment at the N-terminus. Backbone N chemical shifts were not determined for prolines. The assigned chemical shifts were used in TALOS-N to define secondary-structure elements. Surprisingly, the results deviated significantly from the secondary structure established for the EBOV NPCt. The N-terminal stretch that corresponds to the α-helical hairpin in EBOV is clearly unstructured, and the second β-hairpin is replaced by a short α-helix, which evidently disrupts the tertiary structure (Fig. 7 ▸). Additional information on backbone order and dynamics was obtained by measuring N15 spin–lattice (R 1) and spin–spin (R 2) relaxation rates and H1–N15 nuclear Overhauser effects (NOE) (Fig. 8 ▸). The spin-relaxation and NOE results indicate that the C-terminal 62 residues are relatively well ordered and the N-terminal 38 residues have low order on the nanosecond to picosecond time scale. The average H1–N15 NOE for residues 634–693 was 0.75 ± 0.04. We used the average R 1 R 2 value for residues 634–693 (17.6 ± 1.7 s−2) to estimate an average backbone HN order parameter S 2 of 0.94 (Kneller et al., 2002 ▸). Nanosecond to picosecond order can also be estimated based on chemical shifts (Berjanskii & Wishart, 2005 ▸), and these estimates (Supplementary Fig. S4) are in semi-quantitative agreement with those based on the spin-relaxation and NOE data.
Figure 6.
1H–15N HSQC spectrum of NPCt with backbone amide assignments. The two peaks with the highest 15N shift are Trp indole NH. Unassigned peaks in the low-field 1H–15N region are owing to side chains.
Figure 7.
Sequence alignment of NPCt from EBOV, TAFV, BDBV and MARV. Conserved residues are highlighted. Secondary-structure elements are as determined by crystallographic analysis for EBOV (Dziubańska et al., 2014 ▸), TAFV and BDBV and by heteronuclear NMR for MARV (all from the present study). Helices are shown as tubes and β-strands as arrows. Also, an in silico prediction of two-dimensional elements for MARV as calculated by the consensus method using the GeneSilico Metaserver (Kurowski & Bujnicki, 2003 ▸) is shown at the bottom.
Figure 8.
Spin–spin (R 2; top) and spin–lattice (R 1; middle) 15N relaxation rates and 1H–15N nuclear Overhauser effects (NOE; bottom) versus residue number measured with a 600 MHz NMR spectrometer.
We conclude that the MARV NPCt domain is appreciably different from the same domain in other Ebolavirus species, and in isolation appears to exist as a molten globule. Such moieties are known to be relatively stable and yield reasonably dispersed NMR spectra (Redfield, 2004 ▸; Walczak et al., 2014 ▸). The possibility that the MARV NPCt fragment is a molten globule with significant helical content (e.g. a three-helix bundle) prompted us to investigate the thermal stability of the protein using circular dichroism and to monitor the ellipticity at 222 nm (characteristic of α-helices). We observed the expected unfolding of the helical structure at ∼50°C (not shown), clearly indicating the presence of α-helices below this temperature.
4. Conclusions
Until recently, of the seven proteins encoded by the Ebolavirus genome, two have not yet had their structures characterized: the nucleoprotein (NP) and the RNA polymerase (L). Both of these proteins are critical for the assembly and replication of the virus and consequently are recognized as suitable drug targets. Our previous work on the C-terminal domain of the EBOV nucleoprotein (Dziubańska et al., 2014 ▸), along with crystallographic analyses of the N-terminal, RNA-binding domain of the protein (Dong et al., 2015 ▸; Leung et al., 2015 ▸; Kirchdoerfer et al., 2015 ▸), provides a foundation for future studies of the NP.
In this paper, we show that the homologous C-terminal domains of NP from two related pathogenic species of Ebolavirus, i.e. Taï Forest and Bundibugyo, have structures that are highly similar to that of the Zaire variant, in spite of differences in the amino-acid sequence. Interestingly, the related NPCt domain from MARV has a structure that was significantly different from the Ebolavirus consensus structure. This was not completely surprising: our alignment of the amino-acid sequences revealed that of the seven amino acids that constitute the hydrophobic core in this domain in the Ebolavirus NPCt only one (Trp722) is conserved. All others are significantly different: Phe688 to Thr, Leu692 to Glu, Pro697 to Ser, Phe731 to Arg, Ala733 to Val and Ile734 to Ala. The loss of a number of these hydrophobic residues is most likely to be responsible for the lack of a stable globular core and rationalizes why the MARV NPCt domain appears to be in a molten globule state.
Importantly, the Ebolavirus NPCt has also been identified as a possible target for the development of species-specific diagnostic tests (Sherwood & Hayhurst, 2013 ▸; Changula et al., 2013 ▸; Niikura et al., 2003 ▸). Structural characterization of NPCt from the different Ebolavirus species is relevant to this potential application.
Supplementary Material
PDB reference: C-terminal domain of Ebola (Bundibugyo) nucleoprotein, 5dsd
PDB reference: C-terminal domain of Ebola (Taï Forest) nucleoprotein, 5e2x
Supporting Information.. DOI: 10.1107/S2059798315021439/gm5041sup1.pdf
Acknowledgments
The X-ray data-collection experiments were carried out at Argonne National Laboratory and specifically on beamlines operated by the Structural Biology Center (SCB) and South-Eastern Region Collaborative Access Team (SER-CAT) of the Advanced Photon Source. Argonne National Laboratory is operated by University of Chicago Argonne LLC for the US Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357. Supporting institutions of SER-CAT may be found at http://www.ser-cat.org/members.html. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy Office of Science by Argonne National Laboratory, was supported by US Department of Energy Contract DE-AC02-06CH11357. This research was supported by Research and Development and Ivy Foundation Biomedical Innovation grants from the University of Virginia (ZSD and DAE). We thank Ms Natalya Olekhnovich for technical assistance and Dr John H. Bushweller (UVA) for help with the NMR work.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Baize, S. et al. (2014). N. Engl. J. Med. 371, 1418–1425. [DOI] [PubMed]
- Beniac, D. R., Melito, P. L., deVarennes, S. L., Hiebert, S. L., Rabb, M. J., Lamboo, L. L., Jones, S. M. & Booth, T. F. (2012). PLoS One, 7, e29608. [DOI] [PMC free article] [PubMed]
- Berjanskii, M. V. & Wishart, D. S. (2005). J. Am. Chem. Soc. 127, 14970–14971. [DOI] [PubMed]
- Berman, H. M., Bhat, T. N., Bourne, P. E., Feng, Z., Gilliland, G., Weissig, H. & Westbrook, J. (2000). Nature Struct. Biol. 7, 957–959. [DOI] [PubMed]
- Bharat, T. A., Noda, T., Riches, J. D., Kraehling, V., Kolesnikova, L., Becker, S., Kawaoka, Y. & Briggs, J. A. (2012). Proc. Natl Acad. Sci. USA, 109, 4275–4280. [DOI] [PMC free article] [PubMed]
- Brauburger, K., Hume, A. J., Mühlberger, E. & Olejnik, J. (2012). Viruses, 4, 1878–1927. [DOI] [PMC free article] [PubMed]
- Bukreyev, A. A. et al. (2014). Arch. Virol. 159, 821–830. [DOI] [PMC free article] [PubMed]
- Bunkóczi, G., Echols, N., McCoy, A. J., Oeffner, R. D., Adams, P. D. & Read, R. J. (2013). Acta Cryst. D69, 2276–2286. [DOI] [PMC free article] [PubMed]
- Changula, K., Yoshida, R., Noyori, O., Marzi, A., Miyamoto, H., Ishijima, M., Yokoyama, A., Kajihara, M., Feldmann, H., Mweene, A. S. & Takada, A. (2013). Virus Res. 176, 83–90. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995). J. Biomol. NMR, 6, 277–293. [DOI] [PubMed]
- Dong, S., Yang, P., Li, G., Liu, B., Wang, W., Liu, X., Xia, B., Yang, C., Lou, Z., Guo, Y. & Rao, Z. (2015). Protein Cell, 6, 351–362. [DOI] [PMC free article] [PubMed]
- Dziubańska, P. J., Derewenda, U., Ellena, J. F., Engel, D. A. & Derewenda, Z. S. (2014). Acta Cryst. D70, 2420–2429. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Ericsson, U. B., Hallberg, B. M., DeTitta, G. T., Dekker, N. & Nordlund, P. (2006). Anal. Biochem. 357, 289–298. [DOI] [PubMed]
- Feldmann, H. & Geisbert, T. W. (2011). Lancet, 377, 849–862. [DOI] [PMC free article] [PubMed]
- Formenty, P., Boesch, C., Wyers, M., Steiner, C., Donati, F., Dind, F., Walker, F. & Le Guenno, B. (1999). J. Infect. Dis. 179, S120–S126. [DOI] [PubMed]
- Heinig, M. & Frishman, D. (2004). Nucleic Acids Res. 32, W500–W502. [DOI] [PMC free article] [PubMed]
- Huang, Y., Xu, L., Sun, Y. & Nabel, G. J. (2002). Mol. Cell, 10, 307–316. [DOI] [PubMed]
- Kirchdoerfer, R. N., Abelson, D. M., Li, S., Wood, M. R. & Saphire, E. O. (2015). Cell. Rep. 12, 140–149. [DOI] [PMC free article] [PubMed]
- Kneller, J. M., Lu, M. & Bracken, C. (2002). J. Am. Chem. Soc. 124, 1852–1853. [DOI] [PubMed]
- Kolesnikova, L., Mühlberger, E., Ryabchikova, E. & Becker, S. (2000). J. Virol. 74, 3899–3904. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Kurowski, M. A. & Bujnicki, J. M. (2003). Nucleic Acids Res. 31, 3305–3307. [DOI] [PMC free article] [PubMed]
- Leung, D. W. et al. (2015). Cell. Rep. 11, 376–389. [DOI] [PMC free article] [PubMed]
- Licata, J. M., Johnson, R. F., Han, Z. & Harty, R. N. (2004). J. Virol. 78, 7344–7351. [DOI] [PMC free article] [PubMed]
- Liu, Y., Stone, S. & Harty, R. N. (2011). J. Infect. Dis. 204, S817–S824. [DOI] [PMC free article] [PubMed]
- Mavrakis, M., Kolesnikova, L., Schoehn, G., Becker, S. & Ruigrok, R. W. (2002). Virology, 296, 300–307. [DOI] [PubMed]
- Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. (2006). Acta Cryst. D62, 859–866. [DOI] [PubMed]
- Mühlberger, E., Weik, M., Volchkov, V. E., Klenk, H. D. & Becker, S. (1999). J. Virol. 73, 2333–2342. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Newman, J. (2005). Acta Cryst. D61, 490–493. [DOI] [PubMed]
- Niikura, M., Ikegami, T., Saijo, M., Kurata, T., Kurane, I. & Morikawa, S. (2003). Clin. Diagn. Lab. Immunol. 10, 83–87. [DOI] [PMC free article] [PubMed]
- Noda, T., Ebihara, H., Muramoto, Y., Fujii, K., Takada, A., Sagara, H., Kim, J. H., Kida, H., Feldmann, H. & Kawaoka, Y. (2006). PLoS Pathog. 2, e99. [DOI] [PMC free article] [PubMed]
- Noda, T., Watanabe, S., Sagara, H. & Kawaoka, Y. (2007). J. Virol. 81, 3554–3562. [DOI] [PMC free article] [PubMed]
- Painter, J. & Merritt, E. A. (2006). Acta Cryst. D62, 439–450. [DOI] [PubMed]
- Redfield, C. (2004). Methods Mol. Biol. 278, 233–254. [DOI] [PubMed]
- Roddy, P., Howard, N., Van Kerkhove, M. D., Lutwama, J., Wamala, J., Yoti, Z., Colebunders, R., Palma, P. P., Sterk, E., Jeffs, B., Van Herp, M. & Borchert, M. (2012). PLoS One, 7, e52986. [DOI] [PMC free article] [PubMed]
- Sheffield, P., Garrard, S. & Derewenda, Z. (1999). Protein Expr. Purif. 15, 34–39. [DOI] [PubMed]
- Shen, Y. & Bax, A. (2013). J. Biomol. NMR, 56, 227–241. [DOI] [PMC free article] [PubMed]
- Sherwood, L. J. & Hayhurst, A. (2013). PLoS One, 8, e61232. [DOI] [PMC free article] [PubMed]
- Shi, W., Huang, Y., Sutton-Smith, M., Tissot, B., Panico, M., Morris, H. R., Dell, A., Haslam, S. M., Boyington, J., Graham, B. S., Yang, Z.-Y. & Nabel, G. J. (2008). J. Virol. 82, 6190–6199. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vranken, W. F., Boucher, W., Stevens, T. J., Fogh, R. H., Pajon, A., Llinas, P., Ulrich, E. L., Markley, J. L., Ionides, J. & Laue, E. D. (2005). Proteins, 59, 687–696. [DOI] [PubMed]
- Walczak, M. J., Samatanga, B., van Drogen, F., Peter, M., Jelesarov, I. & Wider, G. (2014). Angew. Chem. Int. Ed. 53, 1320–1323. [DOI] [PubMed]
- Watanabe, S., Noda, T. & Kawaoka, Y. (2006). J. Virol. 80, 3743–3751. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wukovitz, S. W. & Yeates, T. O. (1995). Nature Struct. Mol. Biol. 2, 1062–1067. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: C-terminal domain of Ebola (Bundibugyo) nucleoprotein, 5dsd
PDB reference: C-terminal domain of Ebola (Taï Forest) nucleoprotein, 5e2x
Supporting Information.. DOI: 10.1107/S2059798315021439/gm5041sup1.pdf