Abstract
The order in which genes are organized within a genome is generally not conserved between distantly related species. However, within virus orders and families, strong conservation of gene order is observed. The factors that constrain or promote gene-order diversity are largely unknown, although the regulation of gene expression is one important constraint for viruses. Here we investigate why gene order is conserved for a positive-strand RNA virus encoding a single polyprotein in the context of its authentic multicellular host. Initially, we identified the most plausible trajectory by which alternative gene orders could evolve. Subsequently, we studied the accessibility of key steps along this evolutionary trajectory by constructing two virus intermediates: (1) duplication of a gene followed by (2) loss of the ancestral gene. We identified five barriers to the evolution of alternative gene orders. First, the number of viable positions for reordering is limited. Second, the within-host fitness of viruses with gene duplications is low compared to the wild-type virus. Third, after duplication, the ancestral gene copy is always maintained and never the duplicated one. Fourth, viruses with an alternative gene order have even lower fitness than viruses with gene duplications. Fifth, after more than half a year of evolution in isolation, viruses with an alternative gene order are still vastly inferior to the wild-type virus. Our results show that all steps along plausible evolutionary trajectories to alternative gene orders are highly unlikely. Hence, the inaccessibility of these trajectories probably contributes to the conservation of gene order in present-day viruses.
Keywords: gene order, genome evolution, virus evolution, experimental evolution
THE organization of genes within a genome can vary greatly between phylogenetically distant species. Several comparative studies of bacterial, archaeal, and eukaryotic genomes have concluded that, in general, gene order is not conserved (Himmelreich et al. 1997; Kolstø 1997; Koonin and Galperin 1997; Siefert et al. 1997; Watanabe et al. 1997; Dandekar et al. 1998; Rocha 2008). In stark contrast, gene order within virus orders and families is often conserved. Viral genomes tend to be smaller, with minimal intergenic sequences and in some cases overlapping genes (Lynch 2006; Belshaw et al. 2007; Koonin 2009). The reasons why a particular gene order supports the required patterns of virus gene expression and virus replication have in many cases also been elucidated. For example, different expression levels for viral gene products can arise through the generation of subgenomic RNAs (de Haan et al. 2003), frameshifts (Chung et al. 2008), stuttering of the RNA polymerase in intergenic regions (Wertz et al. 1998), having multiple genome segments with different regulatory elements (Sullivan and Ahlquist 1997), or varying the frequency of different genome segments (Sicard et al. 2013). Altering gene order in viral genomes therefore can be associated with great fitness costs (Novella et al. 2004; Springman et al. 2005), and rearrangement of essential genes is not always reversible (Wertz et al. 1998). Nevertheless, it is not always obvious why gene order has been so well conserved in viruses.
Phylogenetic approaches have helped to unveil interesting patterns in gene-order evolution. An intriguing example is the endornaviruses found in plants, fungi, and protists (Valverde et al. 1990; Wakarchuk and Hamilton 1990; Fukuhara et al. 2006), which have acquired domains with similar functions from these different hosts (Song et al. 2013). Despite their distinct origins, these domains have a strict functional order within the Endornavirus genus (Roossinck et al. 2011; Song et al. 2013), even though they highly vary as to presence or absence. Whereas phylogenetic approaches can identify patterns in gene-order evolution, experimental evolution potentially could shed light on the short-term dynamics and underlying mechanisms. The evolution of gene order has been explored experimentally for phage T7 (Springman et al. 2005) and Vesicular stomatitis virus (VSV) (Pesko et al. 2015).
T7 has a double-stranded DNA genome of ∼40 kb. The T7 genome contains three promoters for the Escherichia coli RNA polymerase, and host-mediated transcription draws the first part of the T7 genome into the cell. Once the T7 RNA polymerase protein in this early region is expressed, it initiates transcription for the rest of the genome from its associated promoters, internalizing the remaining part of the T7 genome and achieving a high level of transcription of the late genes. The artificial repositioning of the T7 RNA polymerase downstream of its normal location resulted in a delay of the phage life cycle and had severe impacts on viral fitness (Endy et al. 2000; Springman et al. 2005). Subsequent experimental evolution led to only a modest recovery in fitness (Springman et al. 2005). In one evolved line, the RNA polymerase was restored to the wild-type position, but at the same time, other genes in T7 genome were relocated, and a full regain of fitness was not observed. This study demonstrates that gene order is important for fitness and that the wild-type levels of fitness are not rapidly re-evolved after reorganizing the genome.
VSV is a nonsegmented negative-strand RNA virus with a genome size of ∼11.2 kb that encodes five proteins. Transcription of VSV is regulated by a single promoter located at the 3′ end of the genome. Stuttering of the VSV RNA polymerase causes greater messenger RNA (mRNA) production in upstream genes, which is a strategy to regulate gene expression. Gene order in VSV was altered by moving the nucleocapsid (N) gene, located at the 3′ end, sequentially downstream in the genome (Wertz et al. 1998). This led to a stepwise decrease in N mRNA production and protein expression (Wertz et al. 1998). The initial fitness of the VSV variants was low (Novella et al. 2004), but fitness gains were observed in evolutionary time, and fitness improved the most for the variant with the lowest initial fitness (Pesko et al. 2015). Nevertheless, the variant with the wild-type gene order still grew better than the other variants.
T7 and VSV are different in nature, gene content, and structure, and both use different replication strategies. Despite these differences, gene order is important for the regulation of gene expression in both viruses. Moreover, the different constraints on gene-order evolution observed in these two studies raise the question of their general applicability. Do most viruses and viral genome architectures suffer from the major constraints, as has been observed for T7? What about viruses that do not use promoters for the transcription of mRNA, such as the positive-strand RNA viruses? Many emerging viruses with large societal impacts, as well as viral model systems with great relevance to fundamental research, are positive-strand RNA viruses, making it relevant to address these questions.
The positive-strand RNA viruses represent the largest group of viruses (Francki et al. 1991) and are classified into three tribes: picorna-, alpha-, and flavi-like (Koonin and Dolja 1993; Fauquet et al. 2005). These viruses are characterized by conserved gene clusters and especially the helicase-polymerase arrangement, where the helicase gene is typically located upstream of the polymerase gene (Koonin and Dolja 1993). In particular, the picorna-like tribe is identified by the partial conservation of core genes that consist of the RNA-dependent RNA polymerase (RdRp), a chymotrypsin-like protease (3CPro), a superfamily 3 helicase (S3H), and a genome-linked protein (VPg) (Koonin and Dolja 1993; Fauquet et al. 2005; Koonin et al. 2008). Moreover, core genes tend to form ordered arrays, whereas noncore genes are responsible for genome reorganization and recombination between distant groups of viruses from all three tribes (Koonin and Dolja 1993; Fauquet et al. 2005).
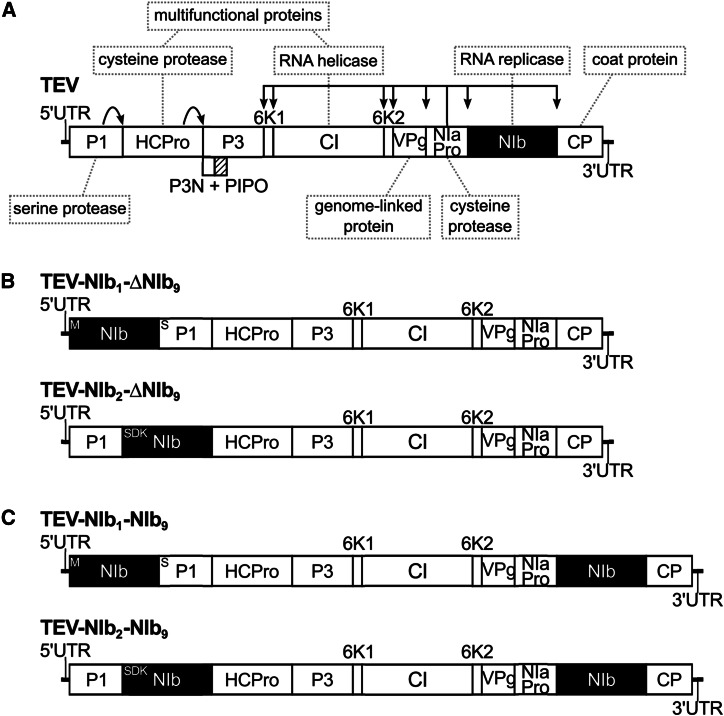
To study the evolution of alternative gene orders in positive-strand RNA viruses in the context of a real multicellular-host infection, we used the picorna-like Tobacco etch virus (TEV; genus Potyvirus, family Potyviridae). TEV has a 9.5 kb genome that codes for a single polyprotein that is further processed into 11 mature peptides (Figure 1A). Because it is composed of positive-strand RNA, the TEV genome can be immediately translated on entering a cell. Unlike the bacteriophage T7 and VSV, replication of TEV is not regulated by a promoter but by the VPg protein linked to the 5′ end of the genome, which helps to initiate RNA replication (Puustinen and Mäkinen 2004). Then the viral NIa-Pro protease is responsible for processing the polyprotein at most of its proteolytic cleavage sites (Revers and García 2015), except for the processing of the first two proteins, P1 serine protease and HC-Pro cysteine protease, which are self-cleaving. Therefore, the rate of synthesis of the mature proteins depends on three factors: the amount of positive-strand RNA accessible to ribosomes, the rate and effectiveness of translation into the polyprotein, and the efficiency of its proteolysis by the viral proteases. Within the Potyviridae family, gene order has been strictly conserved, including the Bymovirus genus, which has evolved a segmented bipartite genome (Revers and García 2015).
Figure 1.
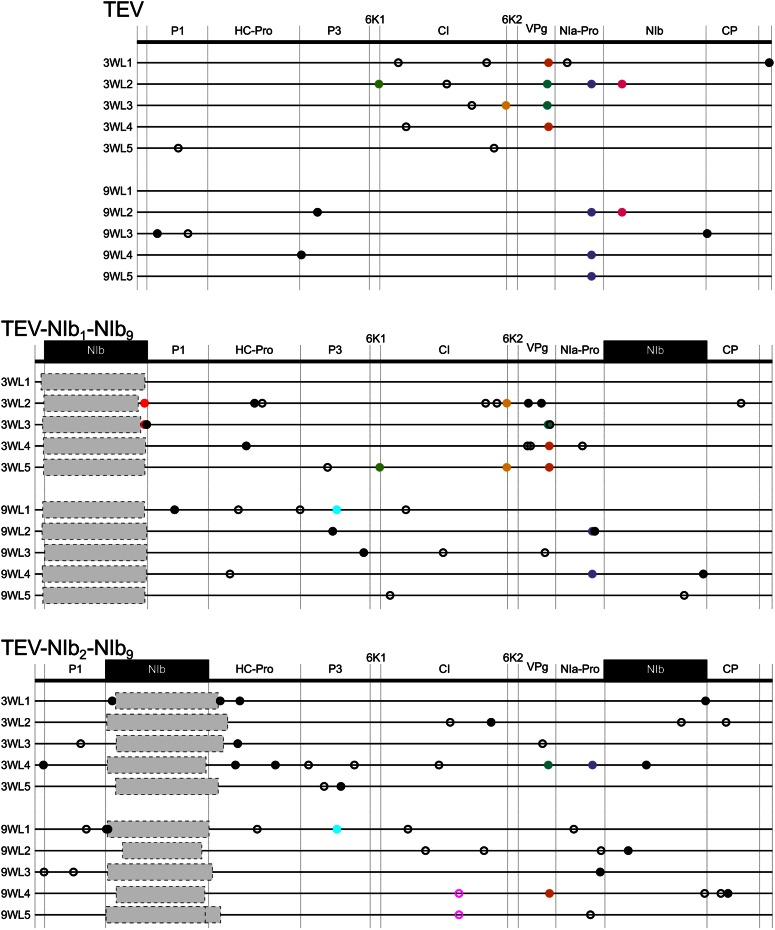
Schematic representations of the different Tobacco etch virus genotypes used in this study. The wild-type TEV (A) codes for 11 mature peptides, including P3N-PIPO embedded within the P3 protein at a +1 frameshift. Relevant names of the proteins products are indicated in the gray dashed boxes. For more information on the features and proposed functions of the proteins, consult Revers and García (2015). For constructing the viral genotypes with the NIb replicase in an alternative position, a copy of NIb was moved to the first and second positions within the TEV genome, removing NIb from its original position (B). For constructing the viral genotypes with a duplication of NIb, a second copy of NIb was introduced into the first and second positions within the TEV genome, preserving the original NIb and thereby generating a duplication event (C). For simplification, P3N-PIPO is drawn only at the wild-type TEV.
Given the need for correct polyprotein processing, a polyprotein-mediated gene-expression strategy is likely to impose constraints on gene-order evolution. Rearranged viral genomes must conserve proteolytic cleavage sites, and as a consequence, most recombination events are likely to disrupt polyprotein processing. However, even if they do not, would the resulting viruses be viable? The RdRp of TEV is coded by the NIb gene, and deletion thereof leads to virus variants that cannot replicate on their own (Li and Carrington 1995). In a previous study, we considered whether virus infectivity was maintained when the NIb gene was relocated to all possible intergenic positions in the TEV genome without maintaining the original NIb copy (Majer et al. 2014). Only two of nine viruses with reordered genomes were viable: the genotypes with NIb relocated to the first two intergenic positions (Figure 1B). A variant with NIb relocated to the third intergenic position was not infectious in wild-type plants, while it could cause infection in transgenic plants expressing NIb in trans, albeit at a lower frequency (Majer et al. 2014). Moreover, in these cases, we always found an exact deletion of the NIb gene, and therefore, we do not consider this a viable virus for reordering.
In this study, we used experimental evolution to better understand the dynamics of genome architecture evolution and gene-order conservation. Why has gene order been conserved within positive-strand RNA virus orders and families? Are there accessible evolutionary trajectories to alternative orders, or is a lack of accessible trajectories an important impediment in present-day viruses? Here we use the two viable reordered TEV variants to address these issues. We first explore whether there are accessible evolutionary trajectories that result in these two viable reordered viruses. To consider whether such natural trajectories exist, genomes containing duplications of the NIb gene were generated, tested for viability, and evolved in plants. We consistently found that the NIb gene at the alternative position was rapidly lost owing to the occurrence of large genomic deletions. Finally, we explored the evolutionary potential of viruses with reordered genomes by evolving viruses with a single NIb gene in an alternative position. We then measured virus accumulation and fitness and used next-generation sequencing to identify genomic changes. Although we found evidence for adaptation of these reordered viruses in terms of increasing virus accumulation, they were still less fit than the wild-type virus. This study therefore revealed multiple barriers to the evolution of alternative gene orders.
Materials and Methods
Viral constructs, virus stocks, and plant infections
TEV-NIb1-ΔNIb9, TEV-NIb2-ΔNIb9, TEV-NIb1-NIb9, and TEV-NIb2-NIb9 were generated from complementary DNA (cDNA) clones constructed using plasmid pGTEVa, which consists of a TEV infectious cDNA (accession no. DQ986288, including two silent mutations, G273A and A1119G) flanked by Cauliflower mosaic virus (CaMV) 35S promoter and terminator in a binary vector derived from pCLEAN-G181 (Thole et al. 2007). Clones were constructed using standard molecular biology techniques, including PCR amplification of cDNAs with the high-fidelity Phusion DNA Polymerase (Thermo Scientific), DNA digestion with Eco31I (Thermo Scientific) for assembly of DNA fragments (Engler et al. 2009), DNA ligation with T4 DNA Ligase (Thermo Scientific), and transformation of E. coli DH5α by electroporation. Sanger sequencing confirmed the sequences of the resulting plasmids. The ancestral and resulting binary plasmids were transformed in Agrobacterium tumefaciens C58C1 harboring helper plasmid pCLEAN-S48 (Thole et al. 2007). Nicotiana tabacum L cv. Xanthi (NN) plants were agroinoculated with A. tumefaciens cultures (Bedoya and Daròs 2010), and symptomatic tissue was collected 7 days postinoculation (dpi). To generate large virus stocks, the collected tissue was homogenized, ground into fine powder using liquid nitrogen and a mortar, and resuspended 1:1 in phosphate buffer (50 mM KH2PO4, pH 7.0, and 3% polyethylene glycol 6000). The third true leaf of 4-week-old N. tabacum plants was mechanically inoculated with 50 μl of the TEV genotypes. N. tabacum plants were kept in a BSL-2 greenhouse at 24° with 16 hr of light. All systemically infected tissues were harvested 7 dpi and stored at −80°.
For the serial passage experiments, 500 mg of homogenized stock tissue was ground into fine powder and diluted in 500 μl of phosphate buffer. From this mixture, 20 μl was then mechanically inoculated on the third true leaf using carborundum. Depending on the virus variant and the passage duration, seven or five independent replicates were used. At the end of the designated passage duration (3 or 9 weeks), all leaves above the inoculated leaf were collected and stored at −80°. For subsequent passages, the frozen tissue was homogenized, and a sample of the homogenized tissue was ground and resuspended with an equal amount of phosphate buffer (Zwart et al. 2014). Then 20 μl was rub-inoculated on the third true leaf. All the experiments involving plant infections have been performed following the Spanish National Guidelines for Plant Research.
Reverse transcription polymerase chain reaction (RT-PCR)
To determine whether deletions had occurred at the NIb locus, RNA was extracted from 100 mg of homogenized infected tissue using the InviTrap Spin Plant RNA Mini Kit (Stratec Molecular). Reverse transcription (RT) was performed using M-MuLV Reverse Transcriptase (Thermo Scientific) and the reverse primer 5′-CGCACTACATAGGAGAATTAG-3′ located in the 3′ UTR of the TEV genome. PCR was then performed with Taq DNA Polymerase (Roche) and primers flanking the NIb gene inserted at the first and second positions—forward 5′-GCAATCAAGCATTCTACTTC-3′ and reverse 5′-CCTGATATGTTTCCTGATAAC-3′—as well as primers flanking the NIb gene at the original position—forward 5′-TCATTACAAACAAGCACTTG-3′ and reverse 5′-GCAAACTGCTCATGTGTGG-3′. PCR products were electrophoresed using 1% agarose gel. Based on the amplicon size and the genome size of the ancestral viruses, genome size was estimated for the corresponding evolved viruses, assuming that deletions occurred only within the amplified region.
Accumulation and within-host competitive fitness assays
Prior to performing assays, the viral copy numbers per 100 mg of tissue of the ancestral virus stocks and all evolved lineages were determined for subsequent assays. The InviTrap Spin Plant RNA Mini Kit (Stratec Molecular) was used to isolate the total RNA of 100 mg of homogenized infected tissue. Real-time quantitative RT-PCR (RT-qPCR) was then performed using the One-Step SYBR PrimeScript RT-PCR Kit II (Takara) in accordance with the manufacturer’s instructions in a StepOnePlus Real-Time PCR System (Applied Biosystems). Specific primers for the CP (coat protein) gene were used—forward 5′-TTGGTCTTGATGGCAACGTG-3′ and reverse 5′-TGTGCCGTTCAGTGTCTTCCT-3′. StepOne Software v.2.2.2 (Applied Biosystems) was used to analyze the data. The concentration of genome equivalents per 100 mg of tissue was then normalized to that of the sample with the lowest concentration using phosphate buffer.
For the accumulation assays, 4-week-old N. tabacum plants were inoculated with 50 μl of these dilutions of ground tissue. For each ancestral and evolved lineage, three independent plant replicates were used. Plant height was measured (data not shown) and leaf tissue was harvested 7 dpi. Virus accumulation then was determined by means of RT-qPCR for the CP gene for TEV, TEV-NIb1-ΔNIb9, TEV-NIb2-ΔNIb9, TEV-NIb1-NIb9, TEV-NIb2-NIb9, and the evolved lineages of these viruses.
To measure within-host competitive fitness, we used TEV carrying an enhanced green fluorescent protein (TEV-eGFP) (Bedoya and Daròs 2010) as a common competitor. TEV-eGFP has proven to be stable for up to 6 weeks (using 1- and 3-week serial passages) in N. tabacum (Zwart et al. 2014) and is therefore not subjected to eGFP loss in our 1-week-long competition experiments. All ancestral and evolved viral lineages were again normalized to the sample with the lowest concentration, and 1:1 mixtures of TEV-NIb1-NIb9 and TEV-NIb2-NIb9 genome equivalents were made with TEV-eGFP. For both TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9, the ratio was 10 genome equivalents set to 1 of TEV-eGFP. The mixture was mechanically inoculated on N. tabacum plants using three independent plant replicates per viral lineage. The plant leaves were collected at 7 dpi and stored at −80°. RNA was extracted from 100 mg of homogenized tissue as described earlier. RT-qPCR for the CP gene was used to determine total viral accumulation, and independent RT-qPCR reactions also were performed for the eGFP sequence using specific primers—forward 5′-CGACAACCACTACCTGAGCA-3′ and reverse 5′-GAACTCCAGCAGGACCATGT-3′. The ratio R of the evolved and ancestral lineages to TEV-eGFP is then , where and are the RT-qPCR measured copy numbers of CP and eGFP, respectively. Then we can estimate the within-host competitive fitness as , where R0 is the ratio at the start of the experiment, and Rt is the ratio after t days of competition (Carrasco et al. 2007).
Sanger sequencing
To determine the exact positions of the deletions detected by RT-PCR in the evolved TEV-NIb1-NIb9 and TEV-NIb2-NIb9 lineages, the genomes were partly sequenced by Sanger’s method. RT was performed using AccuScript Hi-Fi (Agilent Technologies) reverse transcriptase and the reverse primer 5′-TTGCACCTTGTGTGACCAC-3′ located in the P3 gene of the TEV genome. PCR was then performed with Phusion DNA Polymerase (Thermo Scientific) and primers flanking the deletions of NIb at the first and second positions—forward 5′-GCAATCAAGCATTCTACTTC-3′ and reverse 5′-CCTGATATGTTTCCTGATAAC-3′. Sanger sequencing was performed at GenoScreen (Lille, France, www.genoscreen.com) with an ABI3730XL DNA analyzer. Six sequencing reactions were done per lineage using the same two outer primers as used for PCR amplification plus four inner primers—forward 5′-CAATTGTTCGCAAGTGTGC-3′ and 5′-ACACGTACTGGCTGTCAGCG-3′ and reverse 5′-GCTCTTCTTGCTAATGAT-3′ and 5′-ATGGTATGAAGAATGCCTC-3′. Sequences were assembled using Geneious v.8.0.3 (www.geneious.com), and the start and end positions of the deletions were determined. Based on the original TEV-NIb1-NIb9 and TEV-NIb2-NIb9 reference sequences, new reference sequences were constructed for each of the evolved lineages.
Illumina next-generation sequencing (NGS), variants, and SNP calling
For Illumina NGS of the evolved and ancestral lineages, the viral genomes were amplified by RT-PCR using AccuScript Hi-Fi Reverse Transcriptase (Agilent Technologies) and Phusion DNA Polymerase (Thermo Scientific), with six independent replicates that were pooled. TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9 were amplified using three primer sets (set 1: 5′-GCAATCAAGCATTCTACTTCTATTGCAGC-3′ and 5′-CCTGATATGTTTCCTGATAAC-3′; set 2: 5′-ACACGTACTGGCTGTCAGCG-3′ and 5′-CATCAATGTCAATGGTTACAC-3′; set 3: 5′-CCCGTGAAACTCAAGATAG-3′ and 5′-CGCACTACATAGGAGAATTAG-3′). TEV-NIb1-NIb9 and TEV-NIb2-NIb9 were amplified using three different primer sets (set 1: 5′-GCAATCAAGCATTCTACTTCTATTGCAGC-3′ and 5′-TATGGAAGTCCTGTGGATTTTCCAGATCC-3′; set 2: 5′-TTGACGCTGAGCGGAGTGATGG-3′ and 5′-AATGCTTCCAGAATATGCC-3′; set 3: 5′-TCATTACAAACAAGCACTTG-3′ and 5′-CGCACTACATAGGAGAATTAG-3′). Equimolar mixtures of the three PCR products were made. We did not have any Illumina data for the TEV-NIb1-NIb9 3WL1 (3-week passages, lineage 1) because this sample was lost during the preprocessing steps for sequencing. Sequencing was performed at GenoScreen. Illumina HiSeq2500 2 × 100-bp paired-end libraries with dual-index adaptors were prepared along with an internal PhiX control. Libraries were prepared using the Nextera XT DNA Library Preparation Kit (Illumina). Sequencing quality control was performed by GenoScreen based on PhiX error rate and Q30 values. Read artifact filtering and quality trimming (3′ minimum Q28 and minimum read length of 50 bp) were done using FASTX-Toolkit v.0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Dereplication of the reads and 5′ quality trimming requiring a minimum of Q28 were done using PRINSEQ-lite v.0.20.4 (Schmieder and Edwards 2011). Reads containing undefined nucleotides (N) were discarded. As an initial mapping step, the evolved sequences were mapped using Bowtie v.2.2.4 (Langmead and Salzberg 2012) against their corresponding ancestral sequence: TEV (GenBank accession number DQ986288, including two silent mutations: G273A and A1119G), TEV-NIb1-ΔNIb9 (GenBank accession number KT203714), TEV-NIb2-ΔNIb9 (GenBank accession number KT203715), TEV-NIb1-NIb9 ancestral (GenBank accession number KT203712), TEV-NIb2-NIb9 ancestral (GenBank accession number KT203713), and against the evolved TEV-NIb1-NIb9 and TEV-NIb2-NIb9 references including the corresponding deletions. Subsequently, mutations were detected using SAMtools’ mpileup (Li et al. 2009) in the evolved lineages compared with their ancestral lineages. We were only interested in mutations at a frequency of >10%. Therefore, we present frequencies as reported by SAMtools, which has a low sensitivity for detecting low-frequency variants (Spencer et al. 2014).
After the initial premapping step, error correction was done using Polisher v2.0.8 (available for academic use from the Joint Genome Institute), and consensus sequences were defined for every lineage. Subsequently, the cleaned reads were remapped using Bowtie v.2.2.4 against the corresponding consensus sequence for every lineage. The remapping was efficient, with about 84–89% and 89–92% of the paired reads mapping exactly one time for the TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9 lineages, respectively. For the TEV-NIb1-NIb9 and TEV-NIb2-NIb9 lineages, this was 86–89% and 85–89%, respectively. For each new consensus, SNPs within each virus population were identified using SAMtools’ mpileup and VarScan v.2.3.7 (Koboldt et al. 2012). For SNP calling, maximum coverage was set to 40,000, and SNPs with a frequency of <1% were discarded.
Data availability
The sequences of the ancestral viral stocks were submitted to GenBank (GenBank accession numbers KT203711—KT203715). Supplemental Material, Figure S1 and Figure S2 show the distribution of the SNP frequencies in the evolved virus lineages. Table S1 shows the results of a nonlinear regression analysis of the viral genome sizes over time. Table S2 shows the high-frequency mutations of the evolved TEV-NIb1-NIb9 and TEV-NIb2-NIb9 lineages compared with their ancestral sequence. Table S3 shows the within-population sequence variation of the evolved and ancestral TEV-NIb1-NIb9 lineages after the remapping step. Table S4 shows the within-population sequence variation of the evolved and ancestral TEV-NIb2-NIb9 lineages after the remapping step. Table S5 shows the high-frequency mutations of the evolved TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9 lineages compared with their ancestral sequence. Table S6 shows the within-population sequence variation of the evolved and ancestral TEV-NIb1-ΔNIb9 lineages after the remapping step. Table S7 shows the within-population sequence variation of the evolved and ancestral TEV-NIb2-ΔNIb9 lineages after the remapping step.
Results
Study framework: plausible evolutionary trajectories to alternative gene orders
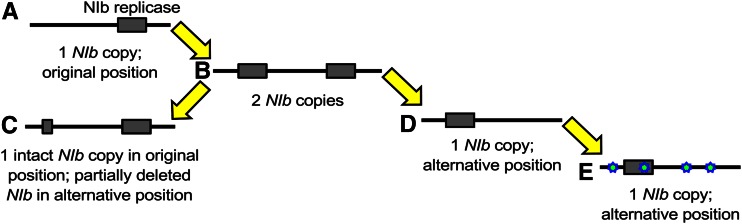
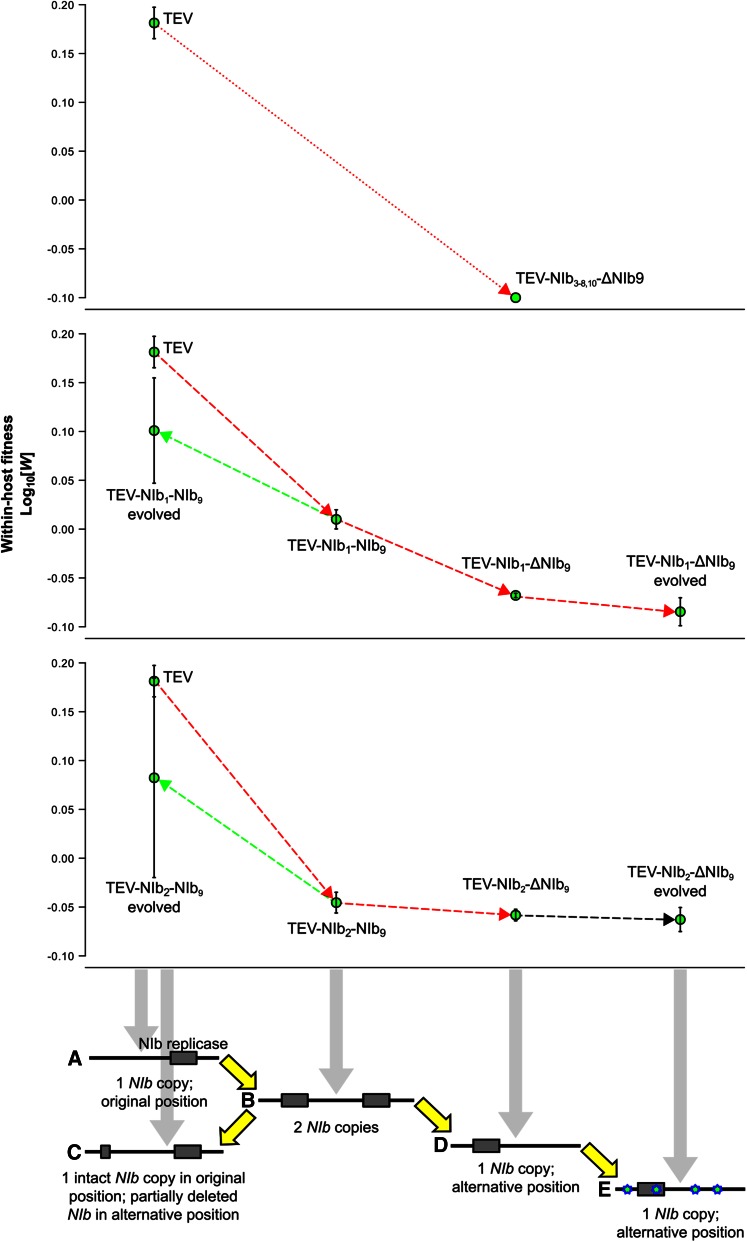
For the rearrangement of genes in a viral genome, many potential evolutionary trajectories can be envisioned. Here we use “plausible” to denote trajectories that could be traversed by the virus in terms of consistently maintaining replication (i.e., virological perspective), whereas we use the term “accessible” to denote trajectories that could be traversed if the fitness and stability of intermediate steps are considered (i.e., evolutionary perspective). In the potyvirus model we are considering, a number of constraints conspire to effectively limit the number of plausible trajectories to one. First, we are considering repositioning of the essential NIb replicase, which must be present in every cell that will contribute to infection and between-host transmission. When plants are inoculated with multiple potyvirus genotypes, the observed rate of cellular co-infection is typically very low, with the main exception being early infection prior to systemic movement (Dietrich and Maiss 2003; Zwart et al. 2011; Tromas et al. 2014a; Gutiérrez et al. 2015). It is therefore not surprising that while TEV missing the NIb gene (TEV-ΔNIb) can autonomously infect plants expressing NIb (Li and Carrington 1995), it cannot co-infect wild-type tobacco plants when co-inoculated with a wild-type virus (Tromas et al. 2014b). Since each intermediate along a reordering trajectory must be capable of autonomous replication, a plausible trajectory for an essential gene—such as NIb—will necessarily involve a gene-duplication event. Second, although higher gene expression as a consequence of gene duplication may have benefits, the increase in genome size and the possible disruption of the expression of other genes could significantly reduce viral fitness. Therefore, the most plausible trajectory to a new gene order is gene duplication, followed by deletion of the ancestral copy (Figure 2). A complication in this process is that variants with a deletion of the new gene copy may very well be favored owing to the fine-tuning of polyprotein processing and expression levels at this position. A deletion of the new gene copy would, however, be a cul-de-sac along this evolutionary trajectory (Figure 2, variant c). Alternatively, viruses with only the new copy may be fixed (Figure 2, variant d), successively followed by the evolutionary fine-tuning (e.g., adaptive mutations) of the relocated gene (Figure 2, variant e).
Figure 2.
Plausible evolutionary trajectory for reordering of an essential gene in a potyvirus genome. First, the essential gene must be duplicated (variant a → variant b) to ensure that the virus can replicate autonomously at all times. Subsequently, we anticipate that one copy of the duplicate gene will be lost: either the second new copy is deleted (variant c) or the alternative gene copy is deleted (variant d). In the latter event, further evolution will take place to accommodate this reordered genome (variant e).
Given that biological constraints strongly suggest that an evolutionary trajectory through gene duplication is the most plausible route to gene reordering, we decided to study this route in detail for the repositioning of NIb in the TEV genome. To get a complete picture of the likelihood that a new gene order can evolve by means of this route, we decided to consider the following five key steps: (1) the fitness of viruses with gene duplications, rendering an indication of how long such variants can persist, (2) the evolutionary potential of viruses with gene duplications, focusing on the stability of the new gene copy because this will show whether the viruses can act as a bridge to the evolution of a new gene order, (3) the fitness of viruses with a single NIb copy in an alternative position to determine the likely fate of such viruses in the background of their direct ancestor, the corresponding double NIb genotype, and (4) the evolutionary potential of viruses with a single NIb copy in an alternative position because should such a virus occur and have low fitness, we can infer whether—following a period of reproductive isolation—it could eventually be competitive with wild-type viruses.
From the outset, however, we are faced with a barrier to the evolution of alternative gene orders for TEV: there are only two alternative positions to place the NIb replicase for which viruses appear to be viable (Majer et al. 2014): (1) before the P1 serine protease gene and (2) between P1 and the HC-Pro cysteine protease genes (Figure 1B). A first barrier to the evolution of alternative gene order is therefore the number of potentially viable intergenic sites, which is limited to only two of nine for TEV. Recombination events leading to the movement of a gene, as well as conservation of the reading frame and polyprotein processing, will be rare, and in addition, all other things being equal, seven of nine of these events will lead down trajectories that are ultimately cul-de-sacs. The number of viable alternative positions, and therefore the effective supply of first-step recombinants leading to alternative gene order, is almost 10-fold smaller than that suggested by the mutational supply alone.
We therefore focus on those evolutionary pathways to reach the two viable alternative gene orders, and consequently, four different TEV genotypes were constructed. The NIb gene was inserted at the first and second positions in the TEV genome while preserving NIb at the original position (Figure 1C). Henceforth, we refer to these viruses with a duplication of NIb as TEV-NIb1-NIb9 and TEV-NIb2-NIb9 (see Table 1), with subscripts denoting the intergenic positions of NIb. Note that NIb9 is referring to NIb at its original position. We also generated viruses in which NIb was moved to alternative positions and the original gene was deleted (Figure 1B). We refer to these viruses with a single NIb copy at alternative positions as TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9 (Table 1). For all viruses generated, the termini of the relocated NIb were adjusted such that this protein is properly translated and sites for cleavage from the viral polyprotein are provided (Figure 1), similar to the original proteolytic cleavage sites at the corresponding positions. For these four key intermediate viruses along an evolutionary trajectory to altered gene order, we could then measure virulence, viral accumulation, and competitive fitness and study their evolutionary potential.
Table 1. TEV genotypes constructed for this study.
| Viral genotype | Original NIb gene present? | Alternative position of NIb gene |
|---|---|---|
| TEV-NIb1-ΔNIb9 | No | 1 |
| TEV-NIb2-ΔNIb9 | No | 2 |
| TEV-NIb1-NIb9 | Yes | 1 |
| TEV-NIb2-NIb9 | Yes | 2 |
Viruses with a duplication of NIb have reduced fitness and accumulation
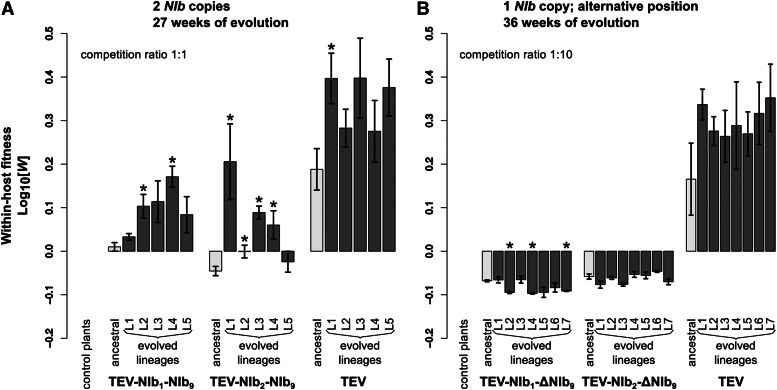
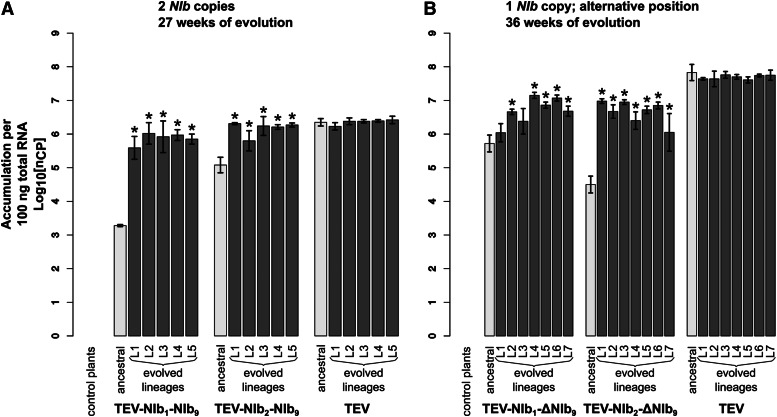
TEV-NIb1-NIb9 and TEV-NIb2-NIb9 were reconstituted from infectious clones and inoculated in N. tabacum L. cv. Xanthi-nc (NN) plants. The collected tissue from these plants served as the starting material for all succeeding experiments. Subsequently, we measured within-host competitive fitness (W) by individually competing the two viruses with NIb duplications against the wild-type virus carrying a GFP marker (TEV-eGFP) and viral accumulation by measuring the number of virions (genome equivalents) present in the host plant in the absence of a competing virus. After normalizing the number of virions for each viral genotype to the same concentration, both competition and accumulation experiments were performed for a total duration of 1 week. For both ancestral viruses TEV-NIb1-NIb9 and TEV-NIb2-NIb9, we observed statistically significant decreases—compared to the ancestral wild-type virus—in competitive fitness (Figure 3A; TEV-NIb1-NIb9: t4 = −6.379, P =0.003; TEV-NIb2-NIb9: t4 = −8.348, P = 0.001) and accumulation (Figure 4A; TEV-NIb1-NIb9: t4 = −45.097, P < 0.001; TEV-NIb2-NIb9: t4 = −8.650, P < 0.001). The magnitude of change in competitive fitness for both genotypes is similar (∼0.6), but the magnitude of change in accumulation is much larger for TEV-NIb2-NIb9 (0.054) than for TEV-NIb1-NIb9 (0.001). Compare the light gray bars labeled “ancestral” in panel A of both Figure 3 and Figure 4 to see how the duplicated viruses performed compared to the wild-type virus. Because of the increase in genome size, we were not surprised that duplication of NIb leads to a virus with reductions in these fitness components. Nevertheless, this observation has an important implication; the first step that must be taken along the plausible evolutionary route we have suggested is already unlikely. Viruses with NIb duplications cannot be maintained in virus populations for long periods of time. These viruses therefore must establish a bridgehead in viral populations by means of genetic drift, from whence they can continue along the evolutionary trajectory to alternative gene order. Consequently, the low fitness of viruses with NIb duplications constitutes a second barrier to reordering.
Figure 3.
Within-host competitive fitness of the evolved and ancestral lineages. Fitness W, as determined by competition experiments and RT-qPCR, of the different viral genotypes with a duplication of NIb (A) and with NIb moved to an alternative position (B) with wild-type TEV and healthy control plants with respect to a common competitor, TEV-eGFP. In A, the competition experiment was started with an inoculum containing a 1:1 mixture of both competitors, while a 1:10 mixture (common competitor and virus of interest) was used in B. The ancestral lineages are indicated with light-gray bars and the evolved lineages with dark-gray bars. The viruses in A were evolved using five replicate lineages each (L1–L5) for a total of 27 weeks, and viruses in B were evolved using seven replicate lineages each (L1–L7) for a total of 36 weeks. Evolved lineages that tested significantly different compared to their ancestral lineage are indicated with an asterisk (t-test with Holm-Bonferroni correction for multiple tests).
Figure 4.
Virus accumulation of the evolved and ancestral lineages. Virus accumulation, as measured by RT-qPCR, of the different viral genotypes with a duplication of NIb (A) and with NIb moved to an alternative position (B) with the wild-type TEV and healthy control plants. The ancestral lineages are indicated with light-gray bars and the evolved lineages with dark-gray bars. The viruses in A were evolved using five replicate lineages each (L1–L5) for a total of 27 weeks, and the viruses in B were evolved using seven replicate lineages each (L1–L7) for a total of 36 weeks. Evolved lineages that tested significantly different compared to their ancestral lineage are indicated with an asterisk (t-test with Holm-Bonferroni correction for multiple tests).
An evolutionary cul-de-sac: after duplication, NIb is pervasively deleted from an alternative position
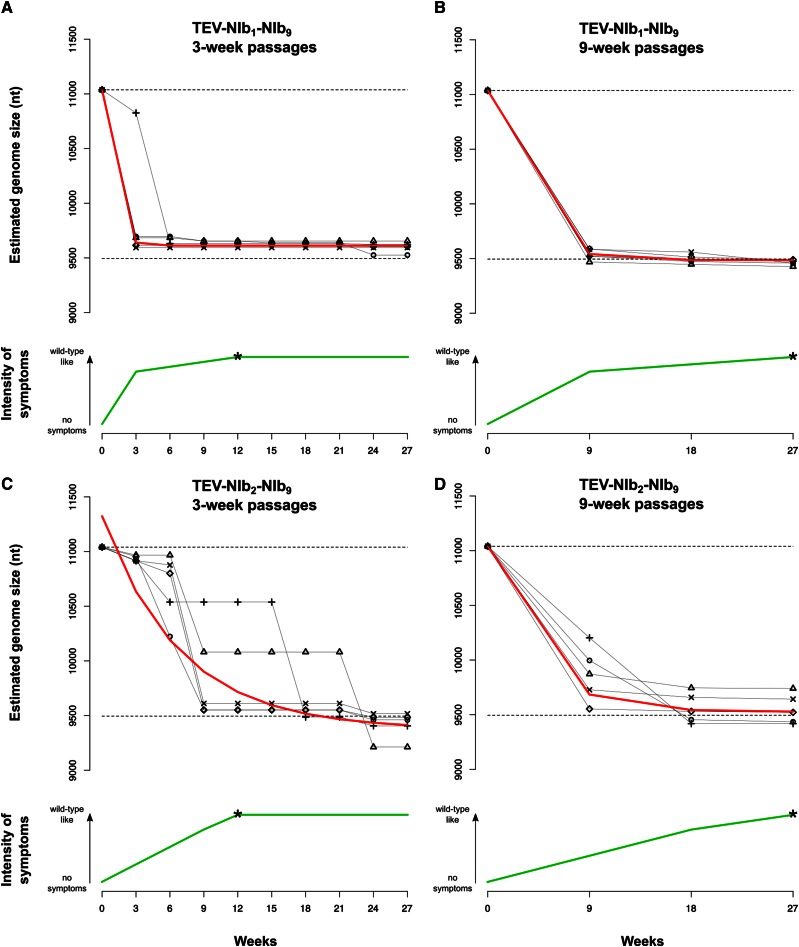
The next key stage in the evolution of alternative gene order is to determine the evolutionary potential of viruses with gene duplications TEV-NIb1-NIb9 and TEV-NIb2-NIb9. The genotypes containing two NIb copies (Figure 1C) were therefore evolved in N. tabacum plants for a total of 27 weeks using both nine 3-week passages and three 9-week passages. Our choice for passage duration was based on a previous study in which we showed that selection appears to act more strongly for longer-duration passages, while nonfunctional sequences are more stable during shorter passages (Zwart et al. 2014). Hence, these conditions may fulfill requirements for further evolution of duplicated viruses by (1) retaining the duplicated gene copy while (2) still allowing for selection—and not mainly genetic drift—to act on these virus populations. At the start of the evolution experiment, the TEV-NIb1-NIb9 and TEV-NIb2-NIb9 viruses had reduced infectivity and showed little or no symptoms, consisting of fewer than seven sparse small chlorotic spots per leaf. These symptoms are much weaker than typical symptoms observed for wild-type TEV, which consist of vein clearing, mosaic mottling, chlorosis, and stunting of the leaves together with reduced plant growth (Velasquez et al. 2014; Revers and García 2015). However, during the evolution experiment, these symptoms changed to wild-type-like symptoms, indicated by the green lines in Figure 5. In the first 3- and 9-week passages, the reduced symptoms turned into mild wild-type-like symptoms for the TEV-NIb1-NIb9 lineages (Figure 5). For the TEV-NIb2-NIb9 lineages, mild wild-type-like symptoms appeared later: for the 3-week lineages in passage 3 (9 weeks on the x-axis in Figure 5) and for the 9-week lineages in passage 2 (18 weeks on the x-axis in Figure 5). At the end of the evolution experiment (27 weeks), all lineages of both viruses showed wild-type-like symptoms in tobacco plants.
Figure 5.
Symptomatology and genome size over time of genotypes with a duplication of NIb. The different panels display how the genome size and symptomatology of the TEV-NIb1-NIb9 (A and B) and TEV-NIb2-NIb9 (C and D) genotypes change along the evolution experiments performing 3-week (A and C) and 9-week (C and D) serial passages. The green lines in each panel plot the observed severity of symptoms. The arrows on the left sides of the plots indicate the gradient from no symptoms to wild-type-like symptoms. The asterisk on each green line indicates the passage where we first observed wild-type-like symptoms (as described in the text). Note that for all genotypes we observed few or no symptoms at the beginning and wild-type-like symptoms at the end of the evolution experiment. The black symbols connected by continuous black lines represent the estimated genome size of the different viral lineages along the evolution experiment. The upper dashed lines represent the genome size of the ancestral viruses, and the lower dashed lines represent the genome size of the wild-type virus. The red line plots the median estimates of a nonlinear regression model. Note that the reduction in genome size appears to be negatively correlated with the severity of symptoms developed.
Through RT-PCR, deletions were detected in the second alternative copy of NIb but never in the original one. Genome size for all the evolved lineages was estimated at every passage (black symbols and continuous lines in Figure 5). As deletions occur, the viral genome reduces in size to one that is similar to that of wild-type TEV. An exponential decay model [] was fitted to the estimated genome size S over time t for every lineage (red line in Figure 5 and Table S1), and the rates of size change b were compared by means of a generalized linear model (GLM) using a gamma probability distribution. Whereas PASSAGE DURATION did not have a significant effect on genome size, both virus GENOTYPE and the interaction GENOTYPE × PASSAGE DURATION were significant (Table 2). The alternative NIb copy was deleted more quickly in the TEV-NIb1-NIb9 lineages than in the TEV-NIb2-NIb9 lineages (Figure 5; compare A and B to C and D), whereas the interaction term suggests that the effect of genotype is particularly strong for the 3-week passages (Table 2). Additionally, in the TEV-NIb2-NIb9 lineages, there appears to be more variation in the time points at which the second NIb copy was deleted in the evolution experiment (Figure 5, C and D). The alternative NIb copy from TEV-NIb2-NIb9 is therefore more stable, suggesting that the second position in the TEV genome is a more accessible trajectory for duplication of a gene and subsequent reorganization within the genome. The decrease in genome size also appears to correlate with the appearance of stronger symptoms (Figure 5). At the end of the evolution experiment, all lineages had a genome size that was very similar to that of the wild-type virus. All TEV-NIb1-NIb9 3-week lineages and four of five 9-week lineages even evolved to a genome size smaller than the wild-type virus by deleting a part of the 5′ UTR. For TEV-NIb2-NIb9, four of five 3-week lineages and three of five 9-week lineages evolved to a smaller genome size by deleting part of the HC-Pro cysteine protease.
Table 2. GLM analysis of the genome size data.
| Term | LRT | df | P |
|---|---|---|---|
| Intercept | 28.420 | 1 | <0.001 |
| GENOTYPE | 10.280 | 1 | 0.001 |
| PASSAGE DURATION | 0.005 | 1 | 0.943 |
| GENOTYPE × PASSAGE DURATION | 4.707 | 1 | 0.030 |
For all the evolved lineages, we then measured within-host competitive fitness and accumulation (Figure 3A and Figure 4A). Compare bars labeled “ancestral” and “evolved lineages” in Figure 3A. When pairwise comparisons were made between the ancestral virus and the evolved lineages (t-test with Holm-Bonferroni correction), significant increases in within-host competitive fitness were found for two of five TEV-NIb1-NIb9 lineages and for four of five TEV-NIb2-NIb9 lineages (Figure 3A and Table 3). However, fitness never reached the fitness of the wild-type virus, and therefore, we did find a significant effect of treatment [ANOVA with post hoc Tukey honestly significant difference (HSD)] comparing the evolved TEV-NIb1-NIb9 and TEV-NIb2-NIb9 lineages to the evolved TEV lineages (Table 4). Now compare bars labeled “ancestral” and “evolved lineages” in Figure 4A. Whereas no significant increases in accumulation were found comparing the ancestral virus and the evolved lineages of the wild-type TEV, we did find significant increases in viral accumulation for all the evolved lineages of both TEV-NIb1-NIb9 and TEV-NIb2-NIb9. And these lineages reached similar accumulation levels as the evolved wild-type lineages. However, we did find a significant effect of treatment on accumulation comparing the evolved lineages of the viral genotypes owing to the differences between the evolved TEV-NIb1-NIb9 and the wild-type TEV lineages (Table 4).
Table 3. Two-sample t-test (4 df) with Holm-Bonferroni correction comparing ancestral against evolved lineages.
| Accumulation | Within-host fitness | |||||
|---|---|---|---|---|---|---|
| Genotype | Lineage | t | P | t | P | |
| 2 NIb copies | TEV-NIb1-NIb9 | 9WL1 | −11.744 | <0.001a | −3.101 | 0.036 |
| 9WL2 | −14.697 | <0.001a | −5.610 | 0.005a | ||
| 9WL3 | −9.690 | 0.001a | −3.663 | 0.022 | ||
| 9WL4 | −27.962 | <0.001a | −10.713 | <0.001a | ||
| 9WL5 | −29.966 | <0.001a | −2.991 | 0.040 | ||
| TEV-NIb2-NIb9 | 9WL1 | −9.305 | 0.001a | −4.849 | 0.003a | |
| 9WL2 | −3.284 | 0.030a | −4.358 | 0.012a | ||
| 9WL3 | −5.600 | 0.005a | −12.814 | <0.001a | ||
| 9WL4 | −8.222 | 0.001a | −5.348 | 0.006a | ||
| 9WL5 | −8.832 | 0.001a | −1.405 | 0.233 | ||
| TEV | 9WL1 | 1.3475 | 0.249 | −4.836 | 0.008a | |
| 9WL2 | −0.2692 | 0.801 | −2.549 | 0.063 | ||
| 9WL3 | −0.4693 | 0.663 | −3.542 | 0.024 | ||
| 9WL4 | −0.5268 | 0.626 | −1.782 | 0.149 | ||
| 9WL5 | −0.7859 | 0.476 | −4.028 | 0.016 | ||
| 1 NIb copy, alternative position | TEV-NIb1-ΔNIb9 | 9WL1 | −1.521 | 0.203 | −0.395 | 0.713 |
| 9WL2 | −6.133 | 0.004a | 15.041 | <0.001a | ||
| 9WL3 | −2.534 | 0.064 | −0.515 | 0.634 | ||
| 9WL4 | −9.322 | 0.001a | 19.454 | <0.001a | ||
| 9WL5 | −7.363 | 0.002a | 3.986 | 0.016 | ||
| 9WL6 | −8.738 | 0.001a | 2.652 | 0.057 | ||
| 9WL7 | −5.670 | 0.005a | 17.220 | <0.001a | ||
| TEV-NIb2-ΔNIb9 | 9WL1 | −16.224 | <0.001a | 3.139 | 0.035 | |
| 9WL2 | −11.531 | <0.001a | 0.805 | 0.466 | ||
| 9WL3 | −16.134 | <0.001a | 4.564 | 0.010 | ||
| 9WL4 | −9.068 | 0.001a | −0.989 | 0.379 | ||
| 9WL5 | −13.961 | <0.001a | −0.502 | 0.642 | ||
| 9WL6 | −14.943 | <0.001a | −3.544 | 0.024 | ||
| 9WL7 | −4.320 | 0.012a | 2.374 | 0.077 | ||
| TEV | 9WL1 | 1.377 | 0.241 | −3.311 | 0.030 | |
| 9WL2 | 0.986 | 0.380 | −2.152 | 0.098 | ||
| 9WL3 | 0.476 | 0.659 | −1.674 | 0.170 | ||
| 9WL4 | 0.908 | 0.415 | −1.642 | 0.176 | ||
| 9WL5 | 1.464 | 0.217 | −1.867 | 0.135 | ||
| 9WL6 | 0.621 | 0.569 | −2.388 | 0.075 | ||
| 9WL7 | 0.497 | 0.646 | −2.865 | 0.046 | ||
Significant after Holm-Bonferroni correction.
Table 4. Nested ANOVAs on accumulation and within-host fitness of evolved lineages with a post hoc Tukey HSD test.
| Trait | Source of variation | df | SS | F | P | Treatments compared | P | ||
|---|---|---|---|---|---|---|---|---|---|
| 2 NIb copies | Accumulation | Treatment | 2 | 1.818 | 19.476 | <0.001 | TEV-NIb1-NIb9 | TEV-NIb2-NIb9 | 0.002 |
| Lineage within treatment | 12 | 0.922 | 1.646 | 0.131 | TEV-NIb1-NIb9 | TEV | <0.001 | ||
| Error | 30 | 1.400 | TEV-NIb2-NIb9 | TEV | 0.053 | ||||
| Within-host fitness | Treatment | 2 | 0.665 | 117.296 | <0.001 | TEV-NIb1-NIb9 | TEV-NIb2-NIb9 | 0.593 | |
| Lineage within treatment | 12 | 0.208 | 6.124 | <0.001 | TEV-NIb1-NIb9 | TEV | <0.001 | ||
| Error | 32 | 0.091 | TEV-NIb2-NIb9 | TEV | <0.001 | ||||
| 1 NIb copy, alternative position | Accumulation | Treatment | 2 | 14.451 | 18.236 | <0.001 | TEV-NIb1-ΔNIb9 | TEV-NIb2-ΔNIb9 | 0.832 |
| Lineage within treatment | 18 | 4.778 | 6.699 | <0.001 | TEV-NIb1-ΔNIb9 | TEV | <0.001 | ||
| Error | 42 | 1.664 | TEV-NIb2-ΔNIb9 | TEV | <0.001 | ||||
| Within-host fitness | Treatment | 2 | 1.966 | 686.952 | <0.001 | TEV-NIb1-ΔNIb9 | TEV-NIb2-ΔNIb9 | 0.161 | |
| Lineage within treatment | 18 | 0.028 | 1.078 | 0.405 | TEV-NIb1-ΔNIb9 | TEV | <0.001 | ||
| Error | 42 | 0.060 | TEV-NIb2-ΔNIb9 | TEV | <0.001 | ||||
The evolution of viruses with two NIb copies results in an increase in fitness related to the reduction in genome size. However, we consistently observed the deletion of NIb at an alternative position, leading back to the ancestral wild-type virus, and we never observed the deletion of NIb at its ancestral position. This evolutionary cul-de-sac therefore represents a third barrier to reordering.
Whole-genome sequences of evolved lineages of viruses with a NIb duplication
All evolved and ancestral lineages described in this study have been fully sequenced using the Illumina technology. The sequences of the ancestral lineages were used as an initial reference for the evolved lineages. Furthermore, for the genotypes that originally had two NIb copies, parts of the genome were sequenced by Sanger to determine the exact deletion sites, which have been detected previously by RT-PCR (Figure 5). Most deletion variants were used to construct new reference sequences for each of the evolved TEV-NIb1-NIb9 and TEV-NIb2-NIb9 lineages. After the initial mapping step, mutations were detected in the evolved lineages compared to their corresponding ancestor.
At the sequence level, the main changes in TEV-NIb1-NIb9 and TEV-NIb2-NIb9 were large genomic deletions in the second NIb copy. In other words, for both viruses, we consistently observed pseudogenization of the duplicated NIb copy, in accordance with RT-PCR results. In TEV-NIb1-NIb9, none of these deletions included the N-terminus of P1, while for nine of ten lineages these deletions included the 3′ end of the 5′ UTR (Figure 6). Only in half of the TEV-NIb1-NIb9 lineages is the reading frame maintained after pseudogenization. Because the deletion occurs at the start of the genome, it is not necessary to maintain the reading frame as long as the original second methionine in P1 is preserved. In TEV-NIb2-NIb9, none of the deletions included the C-terminus of P1, but for seven of ten lineages the deletions included the N-terminal region of the HC-Pro cysteine protease (Figure 6), similar to results obtained by previous studies (Dolja et al. 1993; Zwart et al. 2014). HC-Pro cysteine protease is a multifunctional protein, and the N-terminal region of HC-Pro cysteine protease is implicated in transmission by aphids (Thornbury et al. 1990; Atreya et al. 1992) and is not essential for replication and movement (Dolja et al. 1993; Cronin et al. 1995). In the other genotype, TEV-NIb2-NIb9, the reading frame was maintained in the sequence of all lineages after pseudogenization. This could be explained simply by the fact that these lineages depend on only one methionine codon at the beginning of the polyprotein, at the start of the coding region in P1.
Figure 6.
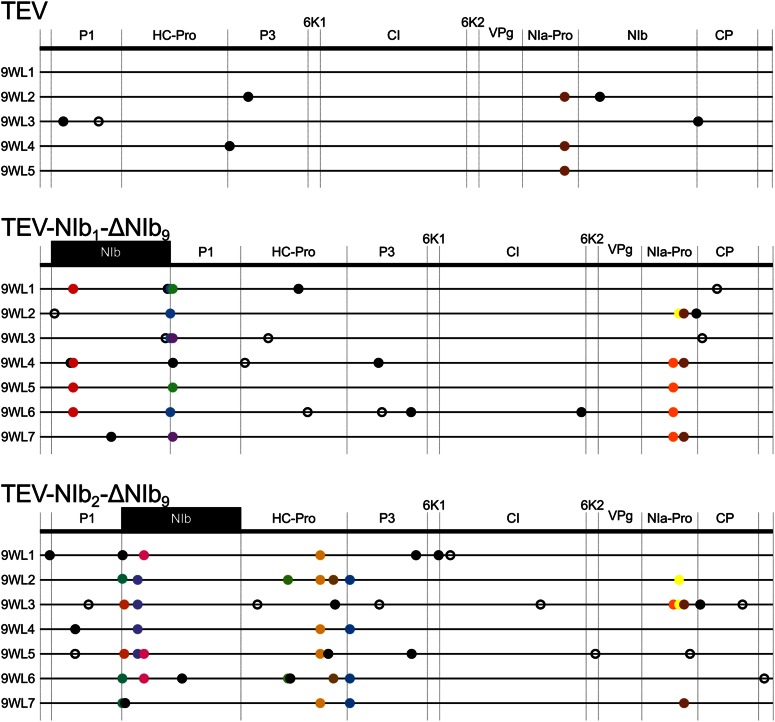
Genomes of the evolved lineages with a duplication of NIb. Mutations were detected using NGS data of the evolved TEV-NIb1-NIb9 and TEV-NIb2-NIb9 lineages compared to their ancestral lineages. The wild-type TEV is given for comparative purposes. The names on the left identify lineages (e.g., 3WL1 is the final population of 3-week passages, lineage 1). No NGS data were available for TEV-NIb1-NIb9 3WL1. Filled circles and open circles represent nonsynonymous and synonymous substitutions, respectively. Black substitutions occur in only one lineage, whereas color-coded substitutions are repeated in two or more lineages. Note that the mutations are present at different frequencies, as reported by SAMtools (>10%). Gray boxes indicate genomic deletions in the majority variant, as detected by Sanger sequencing technology.
However, for these lineages, little evidence for adaptive evolution was found at the level of single-nucleotide mutations (Figure 6, Table S2, Table S3, and Table S4). We determined mutations present in the evolved lineages with respect to their corresponding ancestral sequences. In the TEV-NIb1-NIb9 3-week lineages, we detected three high-frequency (>10%) convergent nonsynonymous mutations occurring in two of five lineages located in the pseudogenized alternative NIb copy (A1643U) at the 3′ end of CI (U7066C) and in VPg (U7703A). For the TEV-NIb1-NIb9 9-week lineages, we observed one convergent nonsynonymous mutation that occurred in two of five lineages in NIa-Pro (A8347G). The same mutation also was found in TEV wild-type 3- and 9-week lineages. For the TEV-NIb2-NIb9 3-week lineages, no repeated mutations were found, and in the 9-week lineages, one convergent synonymous mutation was found in CI (C6351U) for two of five lineages. For more information of the mutations found in both genotypes, see Table S2. After remapping the cleaned reads against a new defined consensus sequence for each lineage, we looked at the variation within each lineage. SNPs were detected at a frequency as low as 1%. In the evolved TEV-NIb1-NIb9 lineages, a total of 301 SNPs were detected, with a median of 36 (27–45) per lineage. In the evolved TEV-NIb2-NIb9 lineages, a total of 220 SNPs were detected, with a median of 23.5 (4–44) per lineage. In both virus genotypes, most of the SNPs were present at low frequency, with a higher percentage of synonymous (TEV-NIb1-NIb9, 66.4%; TEV-NIb2-NIb9, 64.5%) vs. nonsynonymous changes (Figure S1). However, the difference in the distribution of synonymous vs. nonsynonymous SNP frequency was not significant (Kolmogorov-Smirnov test; TEV-NIb1-NIb9: D = 0.146, P = 0.073; TEV-NIb2-NIb9: D = 0.144, P = 0.217). For more details on the frequency of the SNPs within every lineage, see Table S3 and Table S4.
The results from the whole-genome data are congruent with the RT-PCR results and phenotypic assays: the main change in the evolved lineages of the viruses with NIb duplications is the deletion of the second copy, which turns into a virus that is in all respects similar to the ancestral wild-type virus. Although there are some convergent single-nucleotide mutations, these occur only in a small fraction of lineages and, moreover, are often shared between the TEV, TEV-NIb1-NIb9, and TEV-NIb2-NIb9 lineages. These mutations therefore appear to represent general adaptations, without a strong link to the transient presence of the second NIb copy.
Viruses with NIb moved to an alternative position have further reductions in fitness and viral accumulation
The viruses for which NIb was moved to an alternative position without conserving the original NIb copy, TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9, were reconstituted from infectious clones. Subsequently, we measured their within-host competitive fitness and viral accumulation. Compare bars labeled “ancestral” in both Figure 3B and Figure 4B. For both TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9, we observed significant decreases—compared to the wild-type virus—in competitive fitness (Figure 3B; TEV-NIb1-ΔNIb9: t4 = −4.897, P = 0.008; TEV-NIb2-ΔNIb9: t4 = −4.692, P = 0.009) and accumulation (Figure 4B; TEV-NIb1-ΔNIb9: t4 = −10.463, P < 0.001; TEV-NIb2-ΔNIb9: t4 = −16.453, P < 0.001). Deletion of the original NIb copy therefore leads to further reductions in viral fitness, suggesting that TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9 will be rapidly outcompeted by both their direct ancestors (TEV-NIb1-NIb9 and TEV-NIb2-NIb9, respectively) and viruses with deletions in the new copy of NIb (fitness similar to wild-type virus). Therefore, we are confronted with a fourth evolutionary barrier: viruses with a single copy of NIb in an alternative position cannot outcompete the duplicated virus, meaning that they must be maintained, or probably fixed, by genetic drift to have the opportunity to undergo further evolution.
Limited short-term evolutionary potential of viruses with NIb moved to an alternative position
In the evolutionary trajectory we have postulated, the final step is the evolution of a virus with NIb only in an alternative position. If these viruses managed to occur through a series of chance events and could exist in isolation from their ancestral viruses for a period of time, what would their evolutionary fate be? Could these viral populations readily converge on a fitness peak that allowed them to be comparable or superior to the ancestral TEV?
To address these questions, TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9 were evolved in tobacco plants for a total of 36 weeks using four 9-week serial passages. We did not perform 3-week passages because we expected these genomes to be stable and therefore considered only a condition with maximal selection and minimal drift by intermittent bottlenecks. Both reordered viruses have very low infectivity, and serial passage was completed successfully for only seven of ten lineages. For both TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9, tobacco plants had very weak or no symptoms of infection, and these symptoms did not become more severe over time. Therefore, infection had to be confirmed by RT-PCR. Two different regions of the reordered genomes were amplified using primers flanking the region of the new NIb position as well as primers flanking the original position of the removed NIb. No evidence was found of any insertions or deletions within these amplified sites, indicating that reordered viruses were stable over time and that there was no presence of wild-type-like viruses.
For all the evolved lineages, we measured within-host competitive fitness and accumulation (Figure 3B and Figure 4B). Compare bars labeled “ancestral” and “evolved lineages” in Figure 3B. Pairwise comparisons between the ancestral and evolved lineages showed there were no significant increases in within-host competitive fitness (Figure 3B and Table 3), while in three of five TEV-NIb1-ΔNIb9 lineages we even found significant decreases. Now compare bars labeled “ancestral” and “evolved lineages” in Figure 4B. Accumulation levels of the wild-type virus did not change significantly compared to the ancestral TEV, while for five of seven of the evolved TEV-NIb1-ΔNIb9 lineages and all the TEV-NIb2-ΔNIb9 lineages, accumulation increased significantly (Figure 4B and Table 3). However, these accumulation levels never reached the same levels as the wild-type virus. Comparing only the evolved lineages, there was a significant effect of treatment (ANOVA with post hoc Tukey HSD; Table 4) on viral accumulation and within-host competitive fitness, indicating that the wild-type TEV outperforms the two reordered viruses for both fitness components.
Whole-genome sequences of evolved lineages of viruses with a single NIb copy at an alternative position
We found evidence of adaptive convergent evolution comparing the evolved and ancestral lineages containing one reordered NIb copy (Figure 7, Table S5, Table S6, and Table S7). Mutations in the TEV-NIb1-ΔNIb9 lineages were found in (1) the reordered NIb gene at the first position (U428C), (2) in P1 around the proteolytic cleavage site of NIb and P1 (U1688C, U1697C, and U1697A), and (3) in NIa-Pro (U8210C and A8347G). Mutation U1688C modifies the start codon of P1 (M563T). This is explained by the introduction of an additional start codon at the first position of the reordered genome that makes the original methionine redundant. Mutations in the TEV-NIb2-ΔNIb9 lineages were found (1) in the reordered NIb gene at the second position (G1066A, G1090A, G1264A, and U1346C), (2) in HC-Pro (G3213U, A3632G, and U3803C), (3) in P3 (A4016G), and (4) in NIa-Pro (U8285C and A8350G). The former mutation in NIa-Pro (U8285C) also was found in one lineage of TEV-NIb1-ΔNIb9, and the latter mutation (A8350G) also was found in the evolved lineages of TEV-NIb1-ΔNIb9 and the wild-type TEV. Not a single mutation was detected in VPg, which is putatively involved in translation and replication. For more information of the mutations found in both genotypes, see Table S5.
Figure 7.
Genomes of the evolved lineages with NIb moved to an alternative position. Mutations were detected using NGS data of the evolved TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9 lineages compared to their ancestral lineages. The wild-type TEV is given for comparative purposes. The names on the left identify lineages (e.g., 9WL1 is the final population of 9-week passages, lineage 1). Filled circles and open circles represent nonsynonymous and synonymous substitutions, respectively. Black substitutions occur in only one lineage, whereas color-coded substitutions are repeated in two or more lineages. Note that the mutations are present at different frequencies, as reported by SAMtools (>10%).
As for the within-population variation, in the evolved TEV-NIb1-ΔNIb9 lineages, we detected 137 SNPs, with a median of 22 (7–37) per lineage. In the evolved TEV-NIb2-ΔNIb9 lineages, 155 SNPs were detected, with a median of 22 (18–48) per lineage. In both virus genotypes, most of the SNPs were low-frequency SNPs, with a higher percentage of synonymous changes (58.4%) in the TEV-NIb1-ΔNIb9 lineages, while in the TEV-NIb2-ΔNIb9 lineages, the percentage of nonsynonymous changes was higher (63.9%) (see Figure S2). However, no significant difference was found in the distribution of synonymous vs. nonsynonymous SNP frequencies (Kolmogorov-Smirnov test; TEV-NIb1-ΔNIb9: D = 0.203, P = 0.078; TEV-NIb2-ΔNIb9: D = 0.152, P = 0.318). For more information on the frequency of the SNPs within each lineage, see Table S6 and Table S7.
The mutations appeared to be contingent on the ancestral genotype; most of the convergent mutations that were found in the lineages of one reordered genotype were not found in the other genotype. All convergent mutations found were nonsynonymous. Note that the convergent mutations are present at different frequencies and that none of these convergent mutations were fixed in all the replicate lineages (Table S5).
For TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9, we therefore see markedly different patterns of genome evolution than for TEV-NIb1-NIb9 and TEV-NIb2-NIb9. In agreement with RT-PCR results, TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9 show no signs of major genomic rearrangements, such as the large deletions seen in TEV-NIb1-NIb9 and TEV-NIb2-NIb9. Given the strong selection, which undoubtedly would occur for a variant with wild-type gene order, these results suggest that the mutation supply and nonviability of viruses without NIb are major limiting factors. This result provides support for our conjecture that gene-order evolution involving essential genes must occur through gene duplications in potyviruses. However, the observed convergent single-nucleotide mutations are congruent with the observed improvement in virus accumulation, suggesting that selection is acting on these virus populations.
Studying the evolutionary potential of the reordered viruses TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9, although demonstrating clear signatures of adaptive evolution, therefore also shows further barriers to the evolution of alternative gene orders. First, increased accumulation and sequence-level convergent evolution illustrate that adaptive evolution occurred, meaning that the reordered viruses, despite initially having low levels of accumulation, are still somewhat evolvable and show marked improvement in key fitness components. Conversely, accumulation was still significantly lower than that of the wild-type virus. Furthermore, within-host competitive fitness remained similar to that of the ancestral TEV-NIb1-ΔNIb9 or TEV-NIb2-ΔNIb9 or, strikingly, even decreased in some lineages. These observations suggest that even after 36 weeks of evolution under conditions that optimize selection, these reordered viruses remain grossly inferior competitors to the wild-type virus. Therefore, even if a virus population should overcome all four of these barriers, a final evolutionary barrier to the reordering of potyvirus genomes remains.
Discussion
In this study, we have explored whether the most plausible evolutionary trajectory for the rearrangement of gene order in a positive-strand RNA potyvirus is accessible. Overall, we have identified five barriers to the evolution of viruses with the essential NIb replicase gene moved to an alternative position. First, only two of nine viruses with NIb moved to an alternative position are viable. Second, the fitness of viruses with NIb duplications was low, meaning that such viruses would be quickly displaced from populations if they arose (Figure 8 shows a summary of fitness data). Third, for viruses with gene duplications, the new NIb copy was lost in all lineages, whereas the original copy was maintained. This propensity represents a second cul-de-sac because loss of the new NIb copy entails a trajectory that leads back to the ancestral gene order. Fourth, viruses with only a single NIb copy at an alternative position had low fitness and accumulation, notably lower than viruses with duplications (Figure 8). Therefore, to reach a virus with a single NIb copy in an alternative position, two rare recombination events must occur within a small time window because the intermediate step is unstable. Moreover, the low-fitness recombinants would need to be maintained or fixed by genetic drift because they would be outcompeted otherwise. Fifth, even if this unlikely sequence of events occurs and the resulting virus becomes reproductively isolated, after more than half a year of evolution under optimal conditions, this virus still would not stand a chance in head-to-head competition with its ancestor (Figure 8). We therefore conclude that—under the conditions we have considered—the evolution of alternative gene orders for TEV is highly unlikely because the evolutionary trajectory to alternative gene order we have studied is not accessible. These results suggest that one reason gene order has been conserved in potyviruses is therefore the lack of accessible trajectories to alternative gene orders.
Figure 8.
Within-host competitive fitness of all trajectories explored. For all the trajectories proposed (x-axis), the fitness (W at the y-axis) was plotted. The viruses with the NIb gene moved to intergenic positions 3–8 and 10 are nonviable genotypes (upper panel; W < 0). The trajectories of viruses with NIb moved to intergenic positions 1 and 2 are indicated in the middle and lower panels, respectively. Red lines indicate a decrease in fitness, black lines indicate no significant change in fitness, and green lines indicate an increase in fitness. Note how the ancestral and evolved TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9 lineages have an inferior within-host competitive fitness compared to their ancestral TEV-NIb1-NIb9 and TEV-NIb2-NIb9 viruses (respectively) and the wild-type TEV.
The observation that the evolved virus lineages with an alternative gene order have improved accumulation while having unchanged or deteriorated within-host fitness is noteworthy. Our serial passage experiment was conducted in single-host plants, and we would therefore expect within-host competitive fitness to improve. In other words, the virus variant that is present at highest frequency in the final population has the highest probability of carrying over to the next round of infection, irrespective of the level of accumulation. We hypothesize that unchanged or lowered competitive fitness is probably due to low infection levels during serial passaging of TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9. These low infection levels would result in limited direct competition for space within the host, with adaptation occurring by means of more prudent use of cellular resources and improved repression of host immune responses, for example. Evolution during low-level infections therefore may not improve performance in direct competition and may even lower it as a result of antagonistic pleiotropy, given that accumulation and competitive fitness do not correlate for TEV (Zwart et al. 2014). Since the common competitor used in the fitness assays is a strain derived from TEV, high-level infections will occur in these assays. For these reasons, fitness improvements in TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9 lineages may very well not be conspicuous in direct competitions with the ancestral TEV and can only be detected systemically by measuring virus accumulation.
The first step in our proposed model, gene duplication, is a rare event in the recent history of RNA viruses (Simon-Loriere and Holmes 2013). The few cases describing gene duplication in RNA viruses seem to occur through either homologous or nonhomologous recombination (Forss and Schaller 1982; Tristem et al. 1990; Boyko et al. 1992; Walker et al. 1992; Wang and Walker 1993; Karasev et al. 1995; LaPierre et al. 1999; Peng et al. 2001; Valli et al. 2007; Simon-Loriere and Holmes 2013). In our experiments, the gene duplications are engineered artificially, while loss of the duplicated gene generates shorter genomes that are efficiently selected for, probably by means of slippage of the RdRp. We also observed deletions in the N-terminal region of the HC-Pro cysteine protease because there is no selection to maintain the intact HC-Pro. However, these specific deletions probably would not perform well in nature because the resulting truncated protein would preclude aphid-mediated transmission.
We speculate that the rearrangement of genes within the TEV genome could lead to differences in efficiency of cleavage at the proteolytic sites. Given the lower conservation of gene order and sequence homology at the 5′ end in potyviruses, repositioning of genes to this side of the genome would least disturb polyprotein processing. Therefore, it is not surprising that the two positions available for relocation of the NIb replicase gene are located at the N-terminus, before and between the P1 and HC-Pro genes. Within the Potyviridae family, both P1 and the multifunctional HC-Pro have the lowest sequence conservation (Adams et al. 2005; Revers and García 2015). P1 is not an essential gene (Verchot and Carrington 1995a), but the cleavage that separates P1 and HC-Pro is required (Verchot and Carrington 1995b). The introduction of NIb before or between P1 and HC-Pro could result in a delay in this separation or even make P1 inactive, which, in turn, would result in a low accumulation of the virus (Verchot and Carrington 1995a). We do observe that the viral genomes with an alternative gene order have low accumulation levels, but the accumulation levels are able to improve over time with the same gene order. This suggests that the adaptive mutations alone are responsible for the increase in accumulation levels. Nevertheless, these adaptive mutations could not compensate for the rearrangements done because within-host competitive fitness remains low throughout evolutionary time.
It can be postulated that the early expression of the NIb gene would have advantages for viral replication. However, the position of NIb in potyviruses’ polyprotein seems to be optimized for its interaction with VPg for initiation of replication. Furthermore, it is thought that the interaction with both VPg and NIa-Pro targets NIb to the membranous structures where viral RNA replication takes place (Dufresne et al. 2008). Thus, the movement of NIb away from these interacting proteins might result in a delay in the interaction, replication, and gene expression. This is a difficult, maybe even impossible, burden to overcome for a virus.
The relocation of NIb in our model system did lead to the evolution of adaptive or compensatory mutations that allowed for an increase in viral accumulation. Interestingly, convergent mutations occur in both the relocated NIb and two regions that are responsible for proteolytic activity: the C-terminal region of HC-Pro and the main viral protease NIa-Pro. When relocating the NIb replicase to the first position in the genome, and thereby introducing a second methionine codon before NIb in the viral genome, we observe a mutation that changes the original methionine (in P1) to a threonine. Therefore, a reversion of NIb to its original position is not a subsequent step we would expect to see in this viral genotype.
Comparing our study to the experimental evolution done on the repositioning of the T7 RNA polymerase (Springman et al. 2005), similar results were obtained despite T7 having a different genome composition, architecture, and replication and gene expression strategies. In T7 studies, fitness is measured as the rate of population growth, which is comparable to our accumulation measurements. As in our study, the fitness of the rearranged phages improved but never reached the wild-type level. Additionally, as in our study, the reordered T7 genomes are stable over a long period of time. Concerning these similarities, the lack of accessible evolutionary trajectories to alternative gene orders cannot be entirely explained by the most obvious impediment: that potyviruses with reordered genomes most likely would be nonviable owing to improper autocatalytic processing of the polyprotein. In our study, experimental evolution of the reordered viruses illustrates other important barriers to gene order evolution.
In VSV, the evolution of alternative gene orders is more plausible than for TEV because the movement of the N gene does not affect viability of the virus (Wertz et al. 1998). The evolvability of variants of VSV with the N in alternative positions was higher in a cell line from an alternative host (Pesko et al. 2015). Our results for the evolution of TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9 are comparable, although we considered evolution in a permissive host and, as such, expected to see little adaptation of the ancestral TEV. Pesko et al. (2015) did not consider the evolutionary trajectory leading up to the formation of a virus with a rearranged genome. However, at least in the cell-culture environment where MOI can be high for VSV, this may not be as big an impediment as for TEV. A computational study suggests that point mutations in VSV intergenic regions preceded or coevolved with fixation of the wild-type gene order, resulting in a suboptimal genome organization (Lim and Yin 2009).
For our experiments, we have considered only one present-day potyvirus as well as variants with rearranged gene orders derived from it. Whether our results will extend to other present-day potyviruses is a valid question, but it is equally important to consider that these results may have only a limited bearing on the potential for gene-order evolution in ancestral potyviruses. As a result of epistasis, adaptive evolution can limit accessible evolutionary trajectories (Salverda et al. 2011), while purifying selection can result in entrenchment (Shah et al. 2015). It is therefore plausible that evolutionary trajectories to alternative gene orders may have been more accessible to ancestral viruses. We therefore do not rule out that other factors may have been important in conserving potyvirus gene order, in particular, at early time points in their evolution.
What conditions could make the evolutionary trajectory we have studied accessible for present-day viruses or could open up alternative evolutionary trajectories? We have explored evolvability of a single potyvirus genotype at different points along the trajectory to alternative gene orders. It is possible that certain genotypes could be less constrained, although given the low fitness of the intermediates considered and the instability of the new NIb copy, this potentiating variation probably would have to preexist prior to the first recombination step leading to gene duplication. Prime candidates for mutations that may mitigate such constraints are the convergent mutations found in the evolved lineages of TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9. An alternative host species in which high MOI occurs also could open up new evolutionary trajectories by allowing complementation between viral genomes, thereby widening the set of plausible trajectories beyond only those involving gene duplication. We think that it is unlikely that such hosts exist, however, given that (1) in general, MOI estimates for plant RNA viruses tend to be low (Zwart et al. 2013), and (2) alternative hosts would tend to be semipermissive, and we therefore would intuitively expect lower infection levels and MOI in such a host. We think that the most promising avenue for further research on alternative gene orders is to consider the impact of potentiating mutations.
By showing these different barriers to alternative gene orders in viruses, we expect to drive further research on the diversity of gene order over different organisms. Our results serve as a roadmap for testing which factors constrain or promote gene-order conservation across different viruses and could be compared to the great diversity of gene order in other taxa.
Acknowledgments
We thank Francisca de la Iglesia and Paula Agudo for excellent technical assistance. This work was supported by the John Templeton Foundation (grant 22371), by the European Commission 7th Framework Program EvoEvo Project (grant ICT610427), and by the Spanish Ministerio de Economía y Competitividad (MINECO) (grants BFU2012-30805 to S.F.E. and BIO2011-26741 and BIO2014-54269-R to J.A.D). A.W. was supported by a John Templeton Foundation grant and the EvoEvo Project. M.P.Z. was supported by a Juan de la Cierva postdoctoral contract from MINECO. E.M. was the recipient of a predoctoral fellowship (AP2012-3751) from the Spanish Ministerio de Educación, Cultura y Deporte. The opinions expressed in this publication are those of the authors and do not necessarily reflect the views of the John Templeton Foundation. The authors declare that they have no competing interests.
Author contributions: A.W., M.P.Z., and S.F.E. designed the study. A.W., M.P.Z., and N.T. performed the experiments. E.M. and J.A.D. prepared the viral constructs. A.W., M.P.Z., and S.F.E. analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Footnotes
Communicating editor: J. J. Bull
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.185017/-/DC1.
Literature Cited
- Adams M. J., Antoniw J. F., Fauquet C. M., 2005. Molecular criteria for genus and species discrimination within the family Potyviridae. Arch. Virol. 150: 459–479. [DOI] [PubMed] [Google Scholar]
- Atreya C. D., Atreya P. L., Thornbury D. W., Pirone T. P., 1992. Site-directed mutations in the potyvirus HC-Pro gene affect helper component activity, virus accumulation, and symptom expression in infected tobacco plants. Virology 191: 106–111. [DOI] [PubMed] [Google Scholar]
- Bedoya L. C., Daròs J.-A., 2010. Stability of Tobacco etch virus infectious clones in plasmid vectors. Virus Res. 149: 234–240. [DOI] [PubMed] [Google Scholar]
- Belshaw R., Pybus O. G., Rambaut A., 2007. The evolution of genome compression and genomic novelty in RNA viruses. Genome Res. 17: 1496–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko V. P., Karasev A. V., Agranovsky A. A., Koonin E. V., Dolja V. V., 1992. Coat protein gene duplication in a filamentous RNA virus of plants. Proc. Natl. Acad. Sci. USA 89: 9156–9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco P., Daròs J.-A., Agudelo-Romero P., Elena S. F., 2007. A real-time RT-PCR assay for quantifying the fitness of tobacco etch virus in competition experiments. J. Virol. Methods 139: 181–188. [DOI] [PubMed] [Google Scholar]
- Chung B. Y., Miller W. A., Atkins J. F., Firth A. E., 2008. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 105: 5897–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin S., Verchot J., Haldeman-Cahill R., Schaad M. C., Carrington J. C., 1995. Long-distance movement factor: a transport function of the potyvirus helper component proteinase. Plant Cell 7: 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar T., Snel B., Huynen M., Bork P., 1998. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 23: 324–328. [DOI] [PubMed] [Google Scholar]
- de Haan C. A. M., van Genne L., Stoop J. N., Volders H., Rottier P. J. M., 2003. Coronaviruses as vectors: position dependence of foreign gene expression. J. Virol. 77: 11312–11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C., Maiss E., 2003. Fluorescent labelling reveals spatial separation of potyvirus populations in mixed infected Nicotiana benthamiana plants. J. Gen. Virol. 84: 2871–2876. [DOI] [PubMed] [Google Scholar]
- Dolja V. V., Herndon K. L., Pirone T. P., Carrington J. C., 1993. Spontaneous mutagenesis of a plant potyvirus genome after insertion of a foreign gene. J. Virol. 67: 5968–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne P. J., Thivierge K., Cotton S., Beauchemin C., Ide C., et al. , 2008. Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 374: 217–227. [DOI] [PubMed] [Google Scholar]
- Endy D., You L., Yin J., Molineux I. J., 2000. Computation, prediction, and experimental tests of fitness for bacteriophage T7 mutants with permuted genomes. Proc. Natl. Acad. Sci. USA 97: 5375–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C., Gruetzner R., Kandzia R., Marillonnet S., 2009. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A., 2005. Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego. [Google Scholar]
- Forss S., Schaller H., 1982. A tandem repeat gene in a picornavirus. Nucleic Acids Res. 10: 6441–6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francki R. I. B., Fauquet C. M., Knudson D. L., Brown F., 1991. Classification and nomenclature of viruses: fifth report of the International Committee on Taxonomy of Viruses. Arch. Virol. 17: 1–405. [Google Scholar]
- Fukuhara T., Koga R., Aoki N., Yuki C., Yamamoto N., et al. , 2006. The wide distribution of endornaviruses, large double-stranded RNA replicons with plasmid-like properties. Arch. Virol. 151: 995–1002. [DOI] [PubMed] [Google Scholar]
- Gutiérrez S., Pirolles E., Yvon M., Baecker V., Michalakis Y., et al. , 2015. The multiplicity of cellular infection changes depending on the route of cell infection in a plant virus. J. Virol. 89: 9665–9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelreich R., Plagens H., Hilbert H., Reiner B., Herrmann R., 1997. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 25: 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasev A. V., Boyko V. P., Gowda S., Nikolaeva O. V., Hilf M. E., et al. , 1995. Complete sequence of the citrus tristeza virus RNA genome. Virology 208: 511–520. [DOI] [PubMed] [Google Scholar]
- Koboldt D. C., Zhang Q., Larson D. E., Shen D., McLellan M. D., et al. , 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22: 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolstø A. B., 1997. Dynamic bacterial genome organization. Mol. Microbiol. 24: 241–248. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., 2009. Evolution of genome architecture. Int. J. Biochem. Cell Biol. 41: 298–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V., Dolja V. V., 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28: 375–430. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Galperin M. Y., 1997. Prokaryotic genomes: the emerging paradigm of genome-based microbiology. Curr. Opin. Genet. Dev. 7: 757–763. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Wolf Y. I., Nagasaki K., Dolja V. V., 2008. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6: 925–939. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPierre L. A., Holzschu D. L., Bowser P. R., Casey J. W., 1999. Sequence and transcriptional analyses of the fish retroviruses walleye epidermal hyperplasia virus types 1 and 2: evidence for a gene duplication. J. Virol. 73: 9393–9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. H., Carrington J. C., 1995. Complementation of tobacco etch potyvirus mutants by active RNA polymerase expressed in transgenic cells. Proc. Natl. Acad. Sci. USA 92: 457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K., Yin J., 2009. Computational fitness landscape for all gene-order permutations of an RNA virus. PLOS Comput. Biol. 5: e1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2006. Streamlining and simplification of microbial genome architecture. Annu. Rev. Microbiol. 60: 327–349. [DOI] [PubMed] [Google Scholar]
- Majer E., Salvador Z., Zwart M. P., Willemsen A., Elena S. F., et al. , 2014. Relocation of the NIb gene in the tobacco etch potyvirus genome. J. Virol. 88: 4586–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novella I. S., Ball L. A., Wertz G. W., 2004. Fitness analyses of vesicular stomatitis strains with rearranged genomes reveal replicative disadvantages. J. Virol. 78: 9837–9841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C. W., Peremyslov V. V., Mushegian A. R., Dawson W. O., Dolja V. V., 2001. Functional specialization and evolution of leader proteinases in the family Closteroviridae. J. Virol. 75: 12153–12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesko K., Voigt E. A., Swick A., Morley V. J., Timm C., et al. , 2015. Genome rearrangement affects RNA virus adaptability on prostate cancer cells. Front. Genet. 6: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puustinen P., Mäkinen K., 2004. Uridylylation of the potyvirus VPg by viral replicase NIb correlates with the nucleotide binding capacity of VPg. J. Biol. Chem. 279: 38103–38110. [DOI] [PubMed] [Google Scholar]
- Revers F., García J. A., 2015. Molecular biology of potyviruses. Adv. Virus Res. 92: 101–199. [DOI] [PubMed] [Google Scholar]
- Rocha E. P. C., 2008. The organization of the bacterial genome. Annu. Rev. Genet. 42: 211–233. [DOI] [PubMed] [Google Scholar]
- Roossinck M. J., Sabanadzovic S., Okada R., Valverde R. A., 2011. The remarkable evolutionary history of endornaviruses. J. Gen. Virol. 92: 2674–2678. [DOI] [PubMed] [Google Scholar]
- Salverda M. L. M., Dellus E., Gorter F. A., Debets A. J. M., van der Oost J., et al. , 2011. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 7: e1001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R., Edwards R., 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P., McCandlish D. M., Plotkin J. B., 2015. Contingency and entrenchment in protein evolution under purifying selection. Proc. Natl. Acad. Sci. USA 112: E3226–E3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard A., Yvon M., Timchenko T., Gronenborn B., Michalakis Y., et al. , 2013. Gene copy number is differentially regulated in a multipartite virus. Nat. Commun. 4: 2248. [DOI] [PubMed] [Google Scholar]
- Siefert J. L., Martin K. A., Abdi F., Widger W. R., Fox G. E., 1997. Conserved gene clusters in bacterial genomes provide further support for the primacy of RNA. J. Mol. Evol. 45: 467–472. [DOI] [PubMed] [Google Scholar]
- Simon-Loriere E., Holmes E. C., 2013. Gene duplication is infrequent in the recent evolutionary history of RNA viruses. Mol. Biol. Evol. 30: 1263–1269. [DOI] [PubMed] [Google Scholar]
- Song D., Cho W. K., Park S.-H., Jo Y., Kim K.-H., 2013. Evolution of and horizontal gene transfer in the Endornavirus genus. PLoS One 8: e64270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer D. H., Tyagi M., Vallania F., Bredemeyer A. J., Pfeifer J. D., et al. , 2014. Performance of common analysis methods for detecting low-frequency single nucleotide variants in targeted next-generation sequence data. J. Mol. Diagn. 16: 75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springman R., Badgett M. R., Molineux I. J., Bull J. J., 2005. Gene order constrains adaptation in bacteriophage T7. Virology 341: 141–152. [DOI] [PubMed] [Google Scholar]
- Sullivan M. L., Ahlquist P., 1997. cis-Acting signals in bromovirus RNA replication and gene expression: networking with viral proteins and host factors. Semin. Virol. 8: 221–230. [Google Scholar]
- Thole V., Worland B., Snape J. W., Vain P., 2007. The pCLEAN dual binary vector system for Agrobacterium-mediated plant transformation. Plant Physiol. 145: 1211–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornbury D. W., Patterson C. A., Dessens J. T., Pirone T. P., 1990. Comparative sequence of the helper component (HC) region of Potato virus Y and a HC-defective strain, Potato virus C. Virology 178: 573–578. [DOI] [PubMed] [Google Scholar]
- Tristem M., Marshall C., Karpas A., Petrik J., Hill F., 1990. Origin of vpx in lentiviruses. Nature 347: 341–342. [DOI] [PubMed] [Google Scholar]
- Tromas N., Zwart M. P., Lafforgue G., Elena S. F., 2014a Within-host spatiotemporal dynamics of plant virus infection at the cellular level. PLoS Genet. 10: e1004186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas N., Zwart M. P., Forment J., Elena S. F., 2014b Shrinkage of genome size in a plant RNA virus upon transfer of an essential viral gene into the host genome. Genome Biol. Evol. 6: 538–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli A., López-Moya J. J., García J. A., 2007. Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae. J. Gen. Virol. 88: 1016–1028. [DOI] [PubMed] [Google Scholar]
- Valverde R. A., Nameth S., Abdallha O., Al-Musa O., Desjardins P., et al. , 1990. Indigenous double-stranded RNA from pepper (Capsicum annuum). Plant Sci. 67: 195–201. [Google Scholar]
- Velasquez N., Hossain M. J., Murphy J. F., 2014. Differential disease symptoms and full-length genome sequence analysis for three strains of Tobacco etch virus. Virus Genes 50: 442–449. [DOI] [PubMed] [Google Scholar]
- Verchot J., Carrington J. C., 1995a Evidence that the potyvirus P1 proteinase functions in trans as an accessory factor for genome amplification. J. Virol. 69: 3668–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot J., Carrington J. C., 1995b Debilitation of plant potyvirus infectivity by P1 proteinase-inactivating mutations and restoration by second-site modifications. J. Virol. 69: 1582–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakarchuk D. A., Hamilton R. I., 1990. Partial nucleotide sequence from enigmatic dsRNAs in Phaseolus vulgaris. Plant Mol. Biol. 14: 637–639. [DOI] [PubMed] [Google Scholar]
- Walker P. J., Byrne K. A., Riding G. A., Cowley J. A., Wang Y., et al. , 1992. The genome of bovine ephemeral fever rhabdovirus contains two related glycoprotein genes. Virology 191: 49–61. [DOI] [PubMed] [Google Scholar]
- Wang Y., Walker P. J., 1993. Adelaide river rhabdovirus expresses consecutive glycoprotein genes as polycistronic mRNAs: new evidence of gene duplication as an evolutionary process. Virology 195: 719–731. [DOI] [PubMed] [Google Scholar]
- Watanabe H., Mori H., Itoh T., Gojobori T., 1997. Genome plasticity as a paradigm of eubacteria evolution. J. Mol. Evol. 44(Suppl. 1): S57–S64. [DOI] [PubMed] [Google Scholar]
- Wertz G. W., Perepelitsa V. P., Ball L. A., 1998. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc. Natl. Acad. Sci. USA 95: 3501–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart M. P., Daròs J.-A., Elena S. F., 2011. One is enough: in vivo effective population size is dose-dependent for a plant RNA virus. PLoS Pathog. 7: e1002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart M. P., Tromas N., Elena S. F., 2013. Model-selection-based approach for calculating cellular multiplicity of infection during virus colonization of multi-cellular hosts. PLoS One 8: e64657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart M. P., Willemsen A., Daròs J. A., Elena S. F., 2014. Experimental evolution of pseudogenization and gene loss in a plant RNA virus. Mol. Biol. Evol. 31: 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences of the ancestral viral stocks were submitted to GenBank (GenBank accession numbers KT203711—KT203715). Supplemental Material, Figure S1 and Figure S2 show the distribution of the SNP frequencies in the evolved virus lineages. Table S1 shows the results of a nonlinear regression analysis of the viral genome sizes over time. Table S2 shows the high-frequency mutations of the evolved TEV-NIb1-NIb9 and TEV-NIb2-NIb9 lineages compared with their ancestral sequence. Table S3 shows the within-population sequence variation of the evolved and ancestral TEV-NIb1-NIb9 lineages after the remapping step. Table S4 shows the within-population sequence variation of the evolved and ancestral TEV-NIb2-NIb9 lineages after the remapping step. Table S5 shows the high-frequency mutations of the evolved TEV-NIb1-ΔNIb9 and TEV-NIb2-ΔNIb9 lineages compared with their ancestral sequence. Table S6 shows the within-population sequence variation of the evolved and ancestral TEV-NIb1-ΔNIb9 lineages after the remapping step. Table S7 shows the within-population sequence variation of the evolved and ancestral TEV-NIb2-ΔNIb9 lineages after the remapping step.