Abstract
Polymorphic chromosomal rearrangements can bind hundreds of genes into single genetic loci with diverse effects. Rearrangements are often associated with local adaptation and speciation and may also be an important component of genetic variation within populations. We genetically and phenotypically characterize a segregating inversion (inv6) in the Iron Mountain (IM) population of Mimulus guttatus (yellow monkeyflower). We initially mapped inv6 as a region of recombination suppression in three F2 populations resulting from crosses among IM plants. In each case, the F1 parent was heterozygous for a derived haplotype, homogenous across markers spanning over 5 Mb of chromsome 6. In the three F2 populations, inv6 reduced male and female fitness components. In addition, inv6 carriers suffered an ∼30% loss of pollen viability in the field. Despite these costs, inv6 exists at moderate frequency (∼8%) in the natural population, suggesting counterbalancing fitness benefits that maintain the polymorphism. Across 4 years of monitoring in the field, inv6 had an overall significant positive effect on seed production (lifetime female fitness) of carriers. This benefit was particularly strong in harsh years and may be mediated (in part) by strong positive effects on flower production. These data suggest that opposing fitness effects maintain an intermediate frequency, and as a consequence, inv6 generates inbreeding depression and high genetic variance. We discuss these findings in relation to the theory of inbreeding depression and the maintenance of fitness variation.
Keywords: inbreeding depression, polymorphism, variation, chromosomal rearrangement, Mimulus guttatus
POLYMORPHIC chromosomal inversions are an important component of genetic variability (Sturtevant and Mather 1938; Hoffmann and Rieseberg 2008). They are associated with species differentiation in both plants (Rieseberg et al. 1999; Fishman et al. 2013; Hermann et al. 2013) and animals (Noor et al. 2001). Within species, inversions often exhibit clines suggestive of adaptation to latitudinal environmental variables (Balanyà et al. 2003; Hoffmann and Rieseberg 2008; Cheng et al. 2012; Fang et al. 2012). Similarly, putatively adaptive trait differences cosegregate with inversions within many species, including Drosophila (Krimbas and Powell 1992), Anopheles (Coluzzi et al. 2002), Rhagoletis (Feder et al. 2003), seaweed flies (Gilburn and Day 1999), monkeyflowers (Lowry and Willis 2010), and sticklebacks (Jones et al. 2012). These patterns support the idea that inversions contribute to local adaptation and speciation because they suppress recombination among multiple genetic variants with context-dependent effects on fitness (Kirkpatrick and Barton 2006).
The same evolutionary processes that generate differences among geographical populations can also maintain chromosomal polymorphisms within populations. Environmental fluctuations or frequency dependence could allow the persistence of alternative arrangements containing sets of co-adapted alleles, each set being optimal under different conditions (Dobzhansky 1970). This hypothesis may account for segregating “supergenes” such as those determining mating types in heterostylous plants or wing mimicry polymorphisms in Heliconius butterflies [reviewed in Schwander et al. (2014) and Thompson and Jiggins (2014)]. An alternative scenario for polymorphism is that a newly arising chromosomal rearrangement may happen to contain one or more intrinsically detrimental alleles along with an advantageous allele (Kirkpatrick and Barton 2006). Recessive or partially recessive deleterious alleles should be individually rare, but they are common as a class of variant (Muller 1918; Kondrashov 1988) and likely the major cause of inbreeding depression (Charlesworth and Willis 2009). A stable polymorphism may result if the beneficial variants caught by a new inversion increase fitness as heterozygotes (i.e., are at least partially dominant), whereas the detrimental alleles are at least partially recessive. The inversion will increase in frequency initially, as its deleterious recessive alleles will be rare and thus hidden in heterokaryotypes. However, as the inversion frequency rises, recessive costs are expressed more often. Eventually, the benefits and costs balance, resulting in an equilibrium frequency. Thus, rather than promoting adaptation, the inversion brakes the spread of a beneficial allele and leads to elevated frequencies of deleterious alleles.
Here, we describe a polymorphic inversion (hereafter inv6) with strong but conflicting effects on different fitness components. We initially mapped this feature in 2003. The results of that study, presented for the first time in this article, were paradoxical. inv6 segregated in each of three independent QTL mapping crosses, suggesting an intermediate population frequency (it was present in at least three of six plants sampled). However, the fitness effects of inv6 were estimated to be entirely negative in the greenhouse. The Iron Mountain (IM) population, in which inv6 segregates, is a large, stable population and it is extremely unlikely that a genetic variant with uniformly detrimental fitness effects would rise to high frequency. To address this mystery, we conducted five subsequent experiments using a combination of genetic, genomic, and ecological methods.
The original experiment mapped major QTL for multiple fitness components to inv6 in each of three F2 populations, demonstrating substantial negative effects on both male and female fertility under greenhouse conditions. We then generated crosses within inv6 karyotypes to demonstrate that the recombination-suppressed region exhibits normal levels of recombination in crosses with collinear genomes. We then genotyped a random sample of plants from the natural population at markers diagnostic of inv6, which provided an initial estimate of frequency within IM. This experiment also revealed a distinctive haplotype structure for inv6, suggesting that it is recently derived. Next, we conducted a genomic study of eight IM lines with the standard karyotype in combination with two that are homozygous for inv6. These data confirm the recent origin of inv6 and identify several genic regions of elevated sequence divergence. Finally, we performed two different field studies, a one-generation study of male fitness (conducted in 2007) and a four-generation study of female fitness variation (2010–2013). The first demonstrated that the negative inv6 effect on pollen viability, first observed in the greenhouse, is reiterated in the field. Surprisingly, however, the second field study revealed a net positive effect of inv6 on lifetime female fitness, mediated in part by effects on flower production. These estimates are the first compelling evidence of a fitness advantage associated with inv6 and a possible resolution to its unexpected abundance in nature.
When an allele (or karyotype) has both positive and negative effects, gene action (dominance or recessivity) is a critical evolutionary factor. A limitation of our field studies is that they are based on observed genotype frequencies in nature, where inv6 occurs almost entirely as a heterozygote. However, key corroborative evidence comes from two other studies (Scoville et al. 2009; Bodbyl Roels and Kelly 2011) showing that inv6 is strongly deleterious in homozygotes because it is rapidly “purged” when a population is experimentally inbred. inv6 increased in frequency within a third study (Kelly 2008; Kelly et al. 2013), but evolving populations of this experiment were prevented from inbreeding, and homozygous fitness costs were directly ameliorated. Given the aggregate of results from these many experiments, we argue that the positive heterozygous effects on female fecundity have allowed inv6 to increase from a rarity to a substantial population frequency within IM (≈8%). However, its strongly negative homozygous effects likely prevent this variant from fixing in the population. We discuss the possibility that inversions under balancing selection may be common and potentially important contributors to both genetic variation and inbreeding depression in natural populations.
Materials and Methods
Study system
Mimulus guttatus (2n = 28, Phrymaceae) is a self-compatible wildflower that grows throughout western North America. It is the most common member of an eponymous species complex composed of highly polytypic, partially interfertile subspecies (Vickery 1978; Wu et al. 2007). We focus on a population located on Iron Mountain in the Western Cascades of Oregon. The site is an alpine meadow composed of a steep north-facing slope at an elevation of 1470 m over an area of ∼600 m2. The population usually comprises hundreds of thousands of flowering individuals each year, is bee-pollinated, and has a mixed mating system with an estimated selfing rate of 0–25% (Willis 1993; Sweigart et al. 1999). The population shows no evidence of spatial genetic structure (Sweigart et al. 1999) or biparental inbreeding (Kelly and Willis 2002).
Mapping populations
As part of our study to investigate the genetic basis of complex trait variation and inbreeding depression within the IM population, we crossed six plants to produce three mapping populations (hereafter, the replicated F2 experiment). The grandparents of the replicated F2 were sampled from a selection experiment on flower size (Kelly 2008; Kelly et al. 2013). Selection was sustained for six generations (bidirectional on corolla width) within populations that were maintained at large size. After a generation of random mating without selection within each population, three Low parents (sampled from the low-selected population) were selected from the Low population, and each was randomly paired to a distinct High parent. We crossed the plants within each High–Low pair and randomly selected a single F1 offspring. Three mapping populations, each consisting of 378–384 F2 individuals, were derived from selfing the three F1 progenitors. These are called the c2, c3, and c4 mapping populations, and each F2 was scored as High–High, High–Low, or Low–Low at each locus according to the grandparental origin of a marker allele.
Following discovery of the inversion on chromosome 6 (see below), we generated an additional cross between two inbred lines from Iron Mountain (IM179 and IM767) with the same orientation of markers on chromosome 6. We selfed a single F1 from this cross to produce 86 F2 plants that we grew and then genotyped at 18 marker loci.
Genotypes and phenotypes in the replicated F2 experiment
As part of our original QTL mapping design, we measured days to flower, pollen viability, pollen number, and supplemented seed set on each F2 individual in the University of Kansas greenhouses in the Spring of 2003. On each plant, the first and second flower were sampled for pollen while the third and fourth were hand-pollinated. We later harvested and counted seed to estimate supplemented seedset. We used a common pollen donor (IM62, a standard inbred line derived from Iron Mountain) for all flowers. Pollen number and viability were measured using a Coulter Counter following the protocol of Kelly et al. (2002). We collected bud meristem tissue from each of 378, 384, and 384 individuals of mapping population c2, c3, and c4, respectively, and extracted DNA using a CTAB procedure (Kelly and Willis 1998).
Nearly all of the markers used for this study (prefixed with MgSTS for M. guttatus sequence tagged site or simply “e” for expressed sequence tag) are exon-primed markers spanning introns (MgSTS markers are available from http://www.mimulusevolution.org). To identify informative markers in each cross, the High and Low outbred parents and progenitor individuals were screened at 748 MgSTS markers that had been successfully amplified in IM62. The forward primer was tagged in the 5′ end with fluorescent dye, and the resulting labeled PCR products were run on ABI 3730 or 3700 Genetic Analyzers (Applied Biosystems, Foster City, CA). PCR amplification followed a touchdown protocol (see Fishman et al. 2001) with multiplexing and pooling based on expected allele sizes. We scored genotypes using Genemarker software (Softgenetics, State College, PA) based on the segregation of length-variable alleles. These markers are usually codominant and single copy, allowing genetic maps from different experiments to be tied together through markers in orthologous genes. We also genotyped several microsatellite markers [prefixed by “aat” (Fishman et al. 2001)] and custom-designed markers (prefixed by “yw”) in each mapping population to fill large gaps in the maps (>30 cM).
Linkage map construction
We built the linkage maps following three iterative rounds of data quality control. We used JOINMAP 4.0 (Stam 1993) to construct preliminary maps by regression mapping and examined each marker for amount of missing data and for incidence of double crossovers. Markers with significant missing data (20% or more) were re-amplified depending on whether the marker was critical to fill a gap; otherwise, it was discarded. We discarded questionable markers. Special effort was made to identify and place markers that filled in large gaps and markers that were shared between crosses. The final missing data proportions were 4.7, 2.8, and 3.3% for c2, c3, and c4, respectively.
We finalized linkage maps for each mapping population with the Kosambi mapping function using the maximum-likelihood algorithm in JOINMAP 4.0 run with default settings. We made two versions of linkage group 6 for each map as this chromosome exhibited extensive recombination suppression involving 20–30 markers in each map. One version was made using the maximum-likelihood algorithm as above, with all but one representative marker in the suppressed recombination region deleted. Deleting excess markers in the region of suppressed recombination was necessary for permutation tests of significance. The second version of chromosome 6 was made to illustrate recombination suppression. We included all genotyped markers on chromosome 6 and performed regression mapping. Taking the ends of linkage groups into account, total map length was calculated by adding 2s to the estimated length of each linkage group, where “s” equals the average intermarker distance for that linkage group. Assuming random distribution of markers, estimated genome coverage was calculated as c = 1 − (e − 2dn/L), the proportion of the genome within distance “d” of a marker, where “n” is the number of markers and “L” is total map length.
QTL mapping and inv6 phenotypic effects
For genome-wide mapping of fertility traits in the three F2 populations, we used composite interval mapping in Windows QTL Cartographer 2.5 (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm) to set priors for Bayesian mapping as implemented in Rqtlbim (Yandell et al. 2007; Yi et al. 2007). Details of the QTL mapping methods are given in File S1. To estimate the effects of inv6 on fertility traits in the mapping populations, we used ANOVA with the inv6 genotype inferred from diagnostic markers with ambiguous genotypes treated as missing data.
Identification of the inversion haplotype
We grew and extracted DNA from a sample of 96 outbred individuals from the Zia-1 base population, the source population for the artificial selection experiment (Kelly 2008). We genotyped each sample for 18 markers spanning chromosome 6, in both recombination-suppressed and freely recombining regions.
Sequencing of IM664 and analysis of IM genomic data
Flagel et al. (2014) fully genome-sequenced nine inbred lines from a large collection of homozygous lines formed from randomly sampled IM plants (Willis 1999b). Genotyping of the lines at diagnostic markers by Scoville et al. (2009) indicates that eight of the nine lines sequenced by Flagel et al. (2014) have the standard karyotype for chromosome 6 (lines IM109, IM1145, IM320, IM479, IM62, IM624, IM693, IM767), while one is inv6 (IM835). To examine patterns of sequence variation within inv6, we resequenced an additional inv6 line (IM664). We extracted genomic DNA from a single plant of IM664 using a CTAB extraction protocol (Kelly and Willis 1998) followed by column purification (MoBio PowerClean DNA kit, MoBio Laboratories, Carlsbad CA). We prepared a genomic DNA library with the Illumina Nextera kit (Illumina Inc., San Diego), following standard protocols. The IM664 library was sequenced (along with 19 other libraries unrelated to this project) in one 150-bp PE run of Illumina NextSeq.
Sequence data from IM664 was combined with the data from the nine previously sequenced IM lines (reads downloaded from the JGI Short Read Archive). We processed reads from each line with Scythe (https://github.com/vsbuffalo/scythe/) to remove adaptor contamination and then with Sickle (https://github.com/najoshi/sickle/) to trim low-quality sequence. Using BWA (http://bio-bwa.sourceforge.net/) with default parameters, we mapped read pairs to the v2.0 draft of the M. guttatus genome (http://www.phytozome.net/) after masking repetitive sequence. We used Picard tools (http://broadinstitute.github.io/picard/) to add ReadGroups to the resulting bam files and then indexed these files using SAMtools (http://samtools.sourceforge.net/). Finally, we called SNPs across the 10 lines simultaneously using the UnifiedGenotyper of GATK v2.5 (https://www.broadinstitute.org/gatk/). We analyzed the resulting VCF file using custom python scripts (Supplemental material Materials). We filtered SNPs to include only sites within coding regions of the 14 main chromosomes (32,579,125 bp of the genome) with a minimum mapping quality score of ≥30. We suppressed all sites that exhibited heterozygosity in any line and required at least five reads at a site to call a plant as homozygous for either the reference or alternative base. With these filters, we found a total of 791,416 SNPs. We calculated pairwise differences (π) among all lines across the genome within 50-kb windows.
Field measures of male and female fitness components
In 2007, we collected the pollen from all four anthers of newly opened flowers (one per plant) at IM and then harvested plants for later DNA extraction (Fishman and Saunders 2008). Pollen was stained with aniline blue, and a subset of fertile and sterile grains was counted using a hemocytometer. Individuals (n = 187) were genotyped at e423 and e723, markers diagnostic for inv6. Genotypic effects on male fertility traits were analyzed with t-tests in JMP. In 2010–2014, we collected entire senescing M. guttatus plants from the Iron Mountain population and then recovered and counted their fruits, seeds, and (in 2012 and 2013) flowers (Fishman and Kelly 2015). We then extracted genomic DNA from the remaining tissue, and genotyped individuals at the inv6 diagnostic marker e423 (n = 1248 over the 4 years). Because we collected only plants that survived to mature at least one fruit, our measures of female fitness do not include survival-to-reproduction.
There were too few inv6 homozygotes per year to include all three genotypes in the analyses. Thus, we coded individuals as inv6 carriers if they were either heterozygotes or inv6 homozygotes. This allows us to evaluate dominant and/or additive effects of the inversion, but not recessive ones. We examined female reproductive trait variation (flowers, fruits, seeds/fruit, and total seeds) using generalized linear models in JMP 11 (SAS Institute, Cary NC) with year and inv6 as main effects. For the first three traits, we used an overdispersed Poisson distribution (log-link function), whereas we analyzed log-transformed seed number (+1) using a normal distribution and identity link (as in Fishman and Kelly 2015).
The plants genotyped for inv6 only partially overlapped those genotyped at the D female meiotic drive locus, which affected several female fitness traits over the same seed collections (Fishman and Kelly 2015). To maximize sample size per year, we did not include D genotype in these analyses, but did verify that there was no statistical association between the two polymorphisms (Pearson χ2, P = 0.61). There were never significant year × genotype interactions [lowest P = 0.09 for log(total seeds)], so we present analyses without interaction effects. However, our study period spanned two fairly standard growth years (2010 and 2012: one to two fruits, 40–60 seeds per plant) and two relatively benign years (2011 and 2013). Using mean seedset as a proxy for year quality, we then asked whether the inversion had different effects in good (mean seedset 100–250) and poor (mean seedset 40–55) years in an analysis with quality, year (quality), inv6, and inv6 × quality interaction.
Data availability
The genetic, phenotypic, and fitness data specific to inv6 are contained in File S2. The SNP data (within genes) for the 10 resequenced IM lines is reported as Table S3. The raw sequence data is available from the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra).
Results
Discovery of inv6 region by recombination suppression
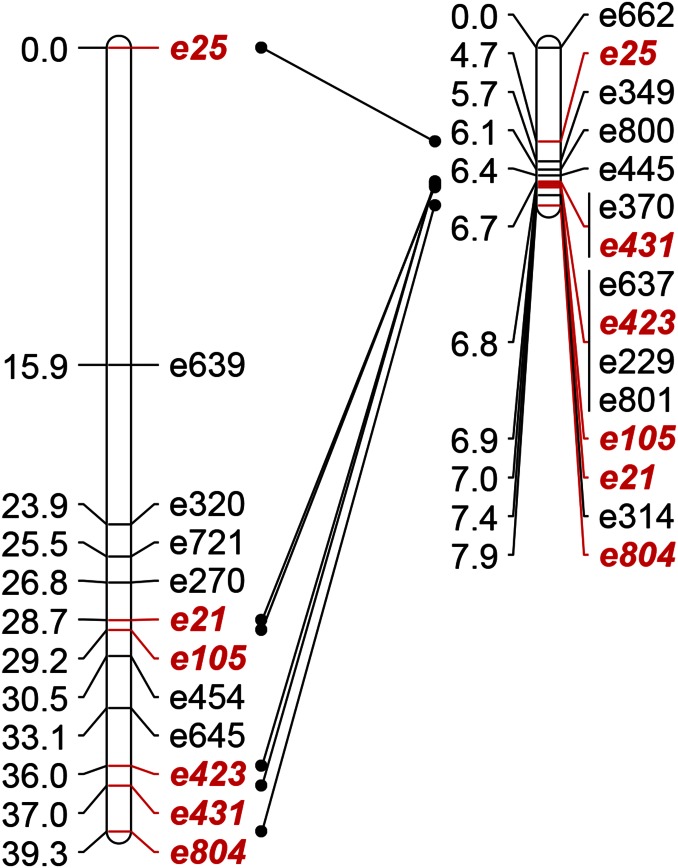
All three of the replicate High × Low F2 genetic maps (Supplemental Material, Figure S1) revealed a large cluster of markers on chromosome 6 that appeared to be completely linked. This nonrecombining region, spanning marker e25 to e804 on the consensus map (Figure 1), corresponds to ∼4.2 Mb of the v2.0 M. guttatus physical map (http://phytozome.jgi.doe.gov/). In contrast, the same set of markers corresponds to 40 cM in the IM79 × IM767 cross (Figure 1), as well as in other M. guttatus linkage maps (Lowry and Willis 2010; Fishman et al. 2014; Holeski et al. 2014). Each High × Low F2 segregated for a particular shared haplotype (i.e., a specific set of alleles) at contiguous MgSTS loci across the recombination-suppressed region. inv6 was contributed by the Low grandparent (small flower size) in c2, but from the High grandparents in c3 and c4.
Figure 1.
Comparative linkage mapping of the upper end of chromosome 6 in Iron Mountain M. guttatus hybrids. In a freely recombining cross (left: IM179 × IM767 F2; n = 86), this region spans ∼40 cM, whereas recombination is highly suppressed in all three F2 mapping populations segregating for the inv6 haplotype (right; c3 map shown). Marker names are to the right, and centimorgans are to the left of bar. Shared markers are highlighted in italic red type.
QTL for fitness traits map to Inv6
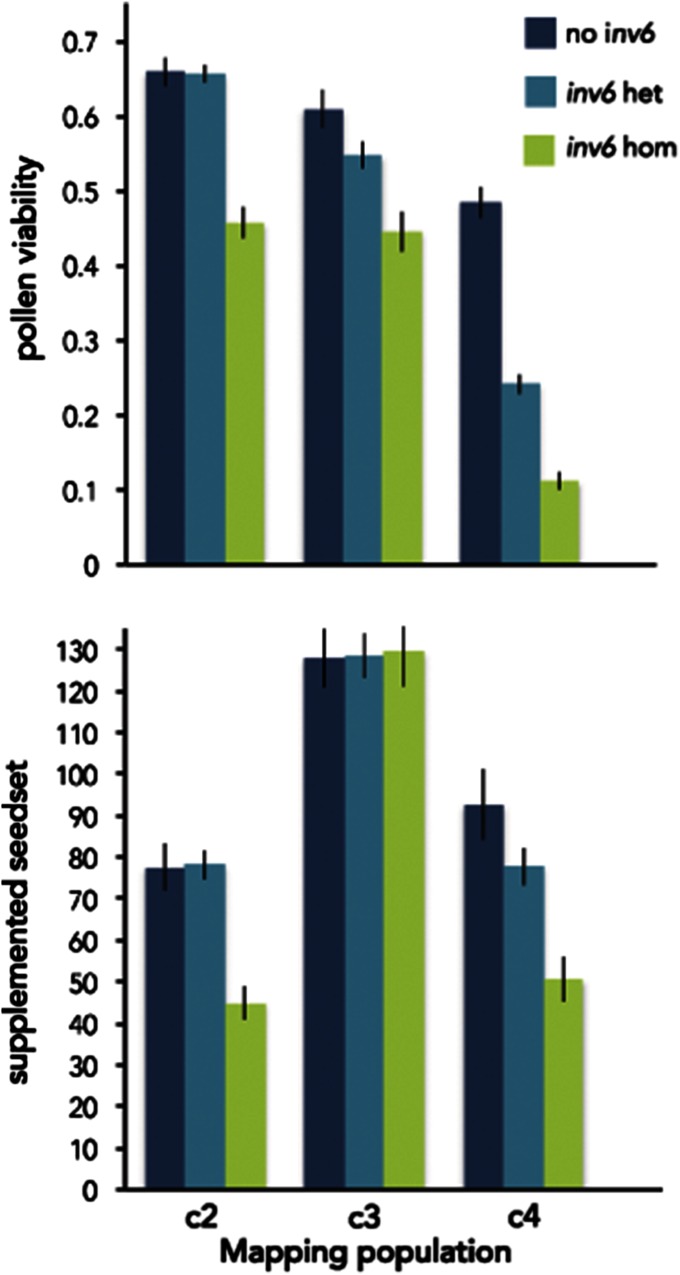
inv6 affected trait means differently in each of the three mapping populations, but all significant effects were negative (Figure 2). In c2, the inversion reduced total pollen (F2,369 = 11.26, P < 0.001), pollen viability (proportion of grains viable; F2,369 = 40.32, P < 0.001), and supplemented seedset (female reproductive capacity; F2,364 = 14.87, P < 0.001) per flower. For all three measurements, gene action was recessive with the negative effect limited to inv6 homozygotes (Figure 2). In c3, there were no effects on female fertility, but a significant negative effect on pollen viability (F2,379 = 9.43, P < 0.001). Gene action is more nearly additive in c3 (Figure 2). In c4, Inv6 had strongly deleterious effects on female fertility (F2,343 = 10.40, P < 0.001), total pollen (F2,345 = 10.04, P < 0.001), and pollen viability (F2,345 = 92.67, P < 0.001). Heterozygotes were intermediate for all three measurements in c4; inv6 was partially recessive its effects on female fertility and total pollen but slightly dominant for pollen viability.
Figure 2.
Effect of inv6 genotype on pollen viability (mean ± 1 SE) and supplemented seedset within each of the F2 mapping populations.
The heterogeneity of inv6 effects among crosses could be due to variation in the non-inv6 alleles and/or differences in the remainder of the genetic backgrounds. Trait means differed substantially among the three mapping populations (Table S1). Consistent with a highly polygenic basis for fertility variation, we mapped 30 QTL for pollen number or pollen viability in other parts of the genome, as well as two QTL for supplemented seed set (Table S2). The chromosome 11 meiotic driver D (Fishman and Saunders 2008) also segregated in each cross and, as expected, the drive allele reduced male fitness. Most fertility QTL exhibit intermediate dominance, but we did map one under-dominant QTL (for viable pollen number) and three overdominant QTL (two for viable pollen number and one for supplemented seedset). However, none of these were present in more than one mapping population. Excluding the two major QTL (inv6 and D) as well as the over/underdominant QTL, low alleles were moderate in effect (average s = 0.31) and partially recessive (average h = 0.18). These estimates, combined with the fact that these QTL were always segregating in only one of the three crosses, suggests that they may be deleterious alleles segregating at low frequencies in the natural population.
Frequency of inv6 in nature
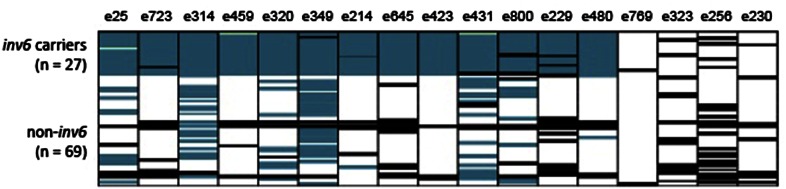
In the Zia-1 base population, which was derived via two generations of hand-outcrossing from a wild IM sample of >1000 plants, the inv6 haplotype had an overall frequency of 15% (Figure 3). In the flowering plants sampled from the field, the estimated frequency of inv6 was slightly lower (7–8%; see Genome sequence variation within and between inv6 carriers). Allelic variation in this population suggests that inv6 is a derived haplotype, as it shares alleles with the alternative arrangement at many loci. However, in this sample, inv6 has private alleles at a few apparently diagnostic loci (e.g., e723 and e423). This pattern is confirmed with enormously increased replication of polymorphism in the genomic survey.
Figure 3.
Delineation of inv6 haplotype block in M. guttatus individuals derived from wild IM plants by one generation of outbreeding (n = 96). Each cell represents the genotype of an individual genotyped at 17 markers across chromosome 6. Genotypes carrying at least one inv6-associated allele are shown in blue (for the 13 inv6-spanning markers) and non-inv6 genotypes are shown in white. Black indicates missing data, and three non-inv6 genotypes (likely genotyping errors or double crossovers) within the inv6 block are shown in green.
Genome sequence variation within and between inv6 carriers
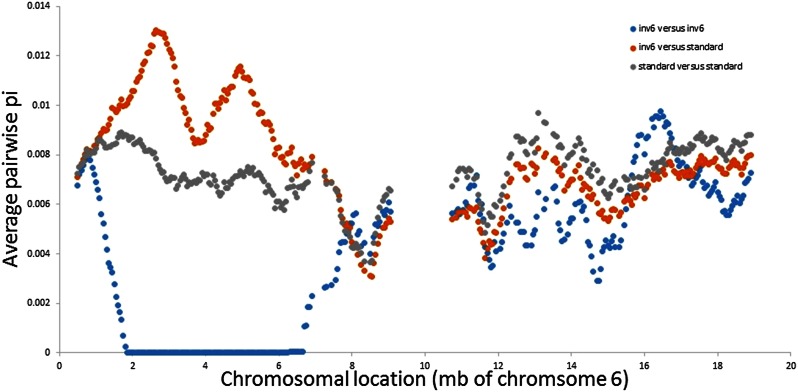
The two inv6 lines (IM664 and IM835) are nearly identical over 5.4 Mb of chromosome 6 (Figure 4). They differ at only six nucleotide positions within the interval from location 1,336,275 to 6,751,183. This is not generally a region of low variation. Average divergence among standard lines (gray points in Figure 4) is greater than the genome-wide average π = 0.0063 for all lines and over a thousand times the divergence between IM664 and IM835 across this region (π = 0.0000051). Importantly, these two lines are not especially similar outside of the inv6 region. The divergence between IM664 and IM835 across the other 13 chromosomes is typical of the distance between any two random lines from IM (π = 0.0058 for IM664 vs. IM835, the overall nonchromosome 6 average π = 0.0061 between lines). At each of the six sites differing between IM664 and IM835 within inv6, one of these two lines harbors a “singleton.” In other words, all lines have the reference base except IM664 (which has the alternative base at two SNPs) or IM835 (which has the alternative at the other four SNPs).
Figure 4.
The average pairwise sequence divergence (π / bp) is reported across chromosome 6 for three distinct contrasts: blue indicates inv6/inv6, orange indicates inv6/standard, and gray indicates standard/standard. Points are based on a 1-Mb moving average (each is calculated by averaging π/bp estimates from contiguous 50-kb windows). The absence of data from 9 to 11 Mb corresponds to the putative centromere.
At SNPs within the inverted region, the base exhibited by inv6 lines is usually, but not always, segregating within the other eight lines (reiterating pattern of Figure 3). About 15% of the SNPs in the inverted region represent “fixed differences” (the two inv6 lines exhibit one base and all other lines are homozygous for the alternative), and these SNPs exhibit some clustering (note peaks of orange trajectories in Figure 4). This divergence (15%) is elevated relative to the genome-wide average; outside of chromosome 6, IM664 and/or IM835 differ from all other genotyped lines at 4.3% of SNPs. However, given that the sample includes only eight standard lines, a fixed difference in the sample does not imply that the inv6 base is absent from the larger population of standard karyotypes. The full set of genotype calls (10 lines scored at 791,416 SNPs) is reported in Table S3.
Inv6 fitness effects in the field
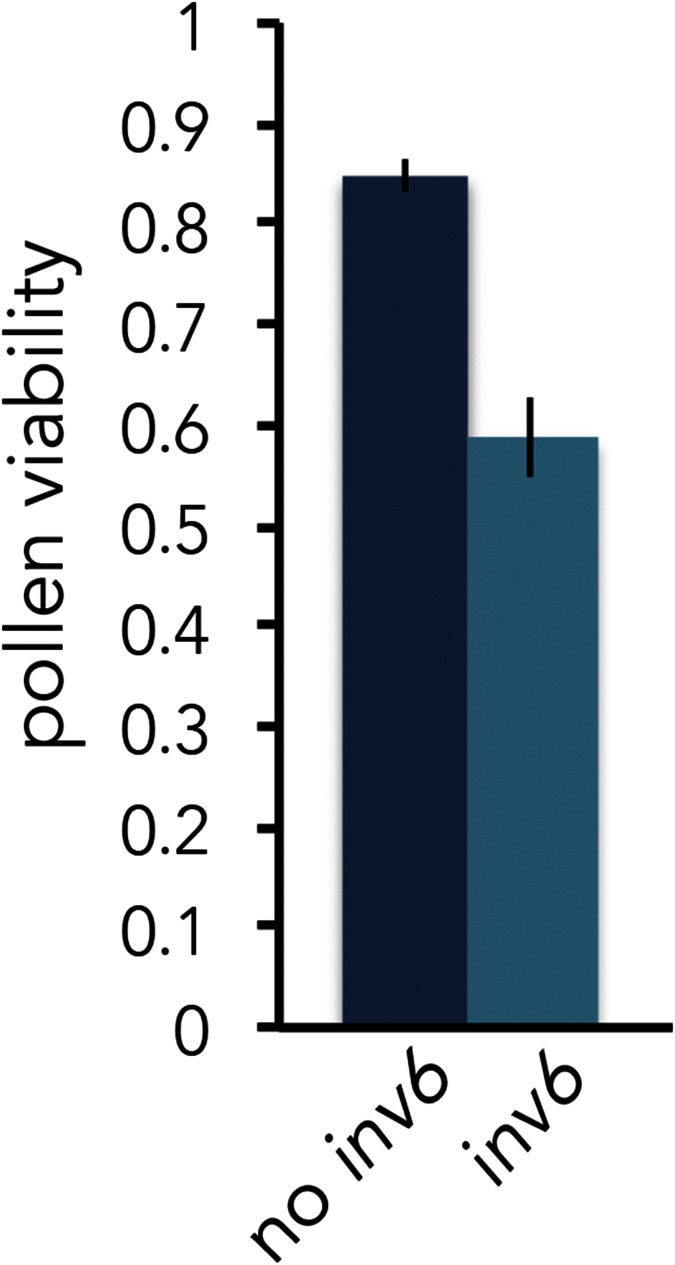
Given strong negative effects on fertility traits in the greenhouse, the nontrivial frequency of inv6 in the IM population is surprising. To evaluate both the frequency and the fitness effects of inv6 more directly, we capitalized on two existing samples of wild IM plants. First, we examined pollen viability and pollen number in field samples collected in 2007. As in the greenhouse experiments (particularly c4, where it was additively deleterious), carriers of inv6 had 30% lower pollen viability than noncarriers (P < 0.0001, n = 187; Figure 5). Homozygotes are too rare in the field to estimate dominance effects. These deleterious effects in heterozygotes make the observed frequency of inv6 (8% in the 2007 sample of wild plants) extremely unlikely without counterbalancing benefits. Second, we examined effects of the inv6 genotype on female fertility traits (flower number, fruit number, seeds/fruit, and total seed number) from 2010 through 2013. There was strong year-to-year variation (all P < 0.0001) in all these traits, indicating that this sample captures the breadth of natural environmental variation in female fertility. Similar to 2007, the frequency of inv6 was ∼7% (∼14% heterozygous plants) and was fairly constant across years in the 2010–2013 samples.
Figure 5.
Effect of inv6 genotype on pollen viability (mean ± 1 SE) of wild Iron Mountain M. guttatus plants (2007; n = 177). Individuals were assigned to genotypic categories (inv6, no inv6) based on their alleles at the diagnostic markers e423 and e723. An inv6 assignment indicates a heterozygous individual, as the two inv6 homozygotes in the dataset were excluded.
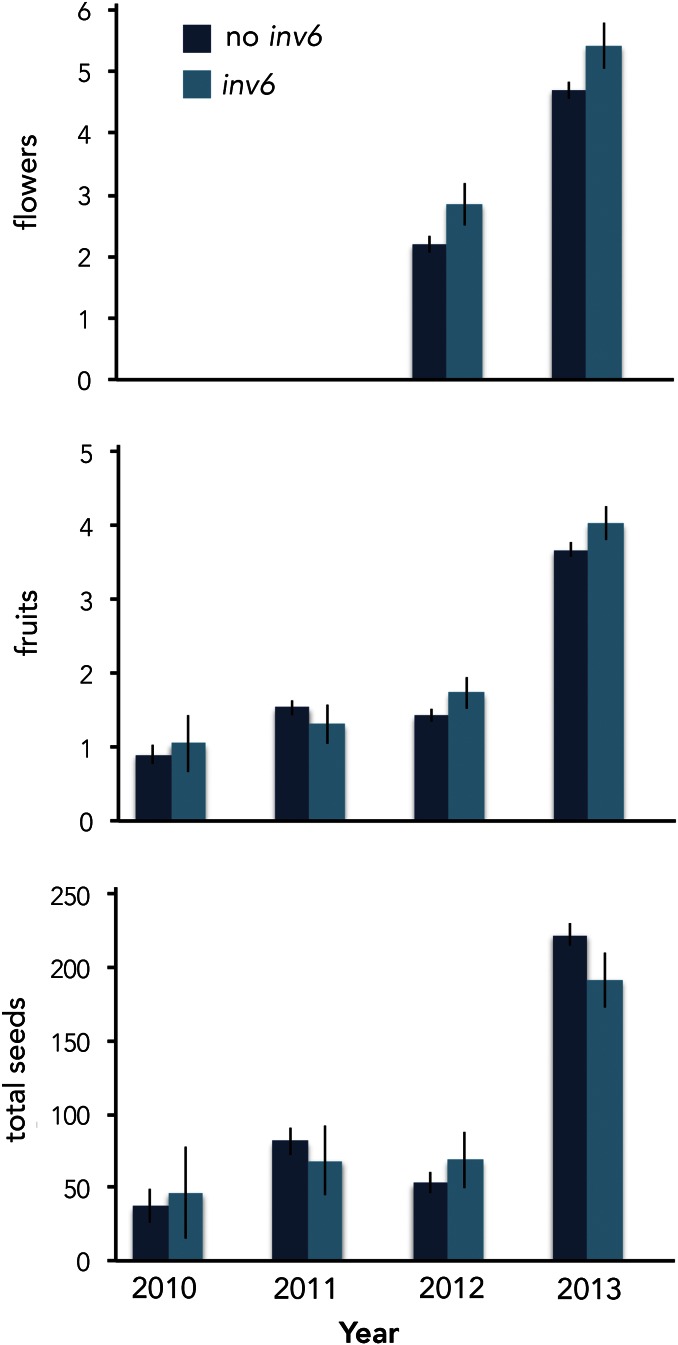
In contrast to inv6’s negative effects on seedset per flower in the greenhouse and on male fertility in both environments, we detected only positive effects of inv6 on female reproductive traits (flower, fruit, and total seed number) in the field (Figure 6). Most strikingly, for the 2 years in which we had flower counts (2012 and 2013), inv6 carriers produced significantly more flowers than alternative genotypes (GLM, LR χ2 = 7.63; P = 0.006, n = 889). In those 2 years, increased flower production translated into increased fruit set (P = 0.04); however, the effect on fruit number was marginally nonsignificant across all 4 years (P = 0.12, n = 1242). In contrast to the greenhouse experiments, we saw no inv6 effect on seeds per fruit (P = 0.78). However, across all 4 years, there was a significant positive effect of inv6 on log(total seeds) (P = 0.047). Although the year × inv6 interaction was not significant for seed number (P = 0.13), there did seem to be a pattern. In 2010 and 2012, which had relatively low fecundity (population mean = 43 and 53 seeds, respectively), inv6 appeared beneficial, whereas it had slightly negative effects in 2011 and 2013 (population mean = 110 and 227, respectively) (Figure 6). Grouping years by quality (2011 and 2013 good, 2010 and 2012 bad), there was a significant quality × inv6 interaction (P = 0.02), as well as a significant effect of inv6 (P = 0.03), in the full model.
Figure 6.
Effects of inv6 genotype on female fitness components (mean ± 1 SE) of wild Iron Mountain M. guttatus plants (n = 1248). Individuals were assigned to genotypic categories (inv6, no inv6) based on their alleles at the diagnostic marker e423. There were only a few inv6 homozygotes in the entire 4-year dataset (not enough to include in the statistical analyses), so an inv6 assignment indicates a heterozygous individual. We show raw means and standard errors here, but the statistical tests in the text were done in a GLM framework (Poisson, log link for fruits and flowers) or with log-transformed values (normal, identity link for seeds).
Discussion
Chromosomal rearrangements such as inversions are increasingly recognized as contributors to local adaptation and speciation. Inversion polymorphisms may also be an important component of complex trait variation within populations, but have been little explored beyond a few supergenes and selfish elements (Ford 1971; Thomas et al. 2008; Thompson and Jiggins 2014). Here, we genetically and phenotypically characterize an intermediate-frequency inversion polymorphism in the Iron Mountain population of M. guttatus (yellow monkeyflower). We identified inv6 on the basis of no recombination among markers on chromosome 6 spanning over 40 cM in a freely recombining cross (Figure 1). Genomic data confirms it to be a distinct and apparently recently derived haplotype. We demonstrate that inv6 has strong and partially recessive deleterious effects on male and female fertility traits in the greenhouse and negative effects on pollen fertility in wild plants, but positive effects on lifetime female fitness likely mediated through increases in flower production. These results contribute to the evidence that inversion polymorphisms may commonly segregate within populations because they bind deleterious and beneficial variants into genetic loci with balanced fitness effects.
Why is inv6 polymorphic?
The mapping experiment that first identified inv6 revealed only strongly negative fitness effects (Figure 2). Despite this, extensive field sampling indicates that inv6 is surprisingly common in the largely outbred IM population. The estimated frequency of 7–10% translates to ∼50,000 copies of inv6 among the ≈300,000 flowering adults in a typical year at IM. IM exhibits very high levels of nucleotide variation (∼2% genome wide), and there is no evidence of a population bottleneck that could have recently inflated the frequency of an unconditionally deleterious mutation (J. Puzey, J. Willis, J. Kelly, unpublished results). Although we do not yet know the full phenotypic effects of genes within inv6, the synthesis of data from this and several other experiments provides a partial explanation for this paradoxical abundance. Hardy–Weinberg proportions are immediately relevant to this explanation. In the field, there are 10–20× as many inv6 heterozygotes as homozygotes. As a consequence, a slight advantage of inv6 in heterozygotes may allow it to increase when rare even if it is strongly deleterious when homozygous.
The balance of evidence suggests variable, but on average, partially recessive deleterious effects for inv6. We see no evidence for the underdominance expected when sterility is caused by chromosomal differences per se, i.e., gametes with duplications/deletions resulting from crossovers in inversion loops (White 1969). In the replicated F2, one of three crosses (c2) revealed recessive gene action, while the other two were more nearly additive (Figure 2). This variability is not surprising, given that the alternative to inv6 is not a single allele but many distinct haplotypes (Figure 3 and Figure 4). Previous experiments conducted on the IM population corroborate average recessive deleterious effects for inv6. Willis (1999b) initiated >1000 independent inbred lines, starting each from an outbred IM plant. Scoville et al. (2009) genotyped 138 of these lines after six or more generations of self-fertilization (predicted inbreeding coefficient >0.98). Only 4 of 138 (≈3%) carried inv6, which is much reduced from the frequency in the initial sample. In a distinct experiment, Bodbyl Roels and Kelly (2011) synthesized large synthetic populations by intercrossing the F2 populations described here for subsequent experimental evolution. Two replicate populations were maintained at large size but compelled to self-fertilize. The initial frequency of inv6 was 37% in each replicate, but declined to 1 and 16%, respectively, after five generations. In two other populations that were supplied with bumblebee pollinators, inv6 declined to a lesser extent (14 and 21%, respectively).
The field data for inv6 (Figure 5 and Figure 6) paint a different picture, but these estimates are based almost entirely on the heterozygous effects of the inversion (very few inv6 homozygotes were sampled). The single year that male fitness was estimated (2007) suggests a 30% pollen viability cost in wild heterozygotes (Figure 5). This estimate is intermediate to that obtained from c3 and c4 of the replicated F2 experiment (Figure 2). In contrast, field data on female fitness show that, particularly in poor years, inv6 carriers set significantly more seeds than noncarriers. This appears to be primarily mediated through increases in flower and fruit number.
inv6 would not occur primarily in heterozygotes if its natural population frequency was near 50% as initially suspected. However, the estimated 7–10% frequency suggests it unlikely that inv6 would have segregated in each of the three F2 mapping populations if the parents were directly sampled from IM. Instead, the parents of the QTL study were sampled from the diverging populations of an artificial selection experiment (Kelly 2008). Large experimental populations were founded from IM and maintained with enforced outcrossing for 10 generations (the parents were sampled from generation 6). Over 10 generations, inv6 rose from an initial frequency of 15 to ∼65% in each of three independent populations, one that experienced selection for larger flowers, one for smaller flowers, as well as the unselected control (Kelly et al. 2013). Even though inv6 would routinely occur in homozygotes once at high frequency, this would not translate into a fitness disadvantage given the methods of propagation within that experiment. All adult plants were randomly paired to another survivor from the same population, one assigned as sire and the other as dam. We repeatedly hand-pollinated dam from sire to ensure sufficient seedset. Even plants with 30–40% reduced pollen viability would still sire far more seed than required to found a family of the next generation. Because we equalized the contribution of each parent pair to the next generation (each contributed ∼10 progeny), any intrinsic difference in female fecundity would also be inconsequential. The parallel increase of inv6 within up, down, and unselected populations suggests that inv6 conferred some unmeasured fitness benefits common to all populations in that experiment. One possibility is germination requirements/timing, which could both respond to inadvertent greenhouse selection and underlie the genotypic differences in flower number and seedset seen in the wild. Seed germination was one of the few life stages in the selection experiment with opportunity for uncontrolled selection.
The internal homogeneity of inv6 is relevant to its origin and frequency. Figure 3 indicates genotype matching of 27 inv6 carriers at 13 markers. Figure 4 shows near identity of two inv6 homozygotes across millions of bases. Thus, inv6 appears recently derived and exhibits the molecular pattern of a partial sweep, similar to the pattern evident at the D locus on chromosome 11, which is strongly indicated as a balanced polymorphism (Fishman and Saunders 2008; Fishman and Kelly 2015). Importantly, inv6 alleles/bases are usually, but not always, present in non-inv6 haplotypes. The latter exhibit many different combinations of alleles, indicative of free recombination in the majority of the population. Together, these observations indicate that inv6 is the derived arrangement, but its absolute age is difficult to infer. Under the neutral infinite sites model, the expected π between two lines is 2μ Tmrca (Tajima and Nei 1983), where μ is the mutation rate per generation and Tmrca is the number of generations since the common ancestor of the two lines. The strict validity of this model for the present data is certainly questionable, but it provides a useful guide regarding the relative age of the feature. The estimated average Tmrca for standard karyotypes in the region is ∼1500 times the Tmrca of the two inv6 sequences. This may be due to the fact that the inversion occurred relatively recently within IM, allowing minimal time to accumulate new “internal” variation via mutation. Alternatively, a selective sweep may have occurred within inv6 (but not the larger population), eliminating any internal variation previously accumulated.
The observation that inv6 increased from rarity to a population frequency of nearly 10% does not imply that it is a balanced polymorphism. The evidence for balance is the low fitness estimates for inv6 homozygotes in Figure 2, as well as the negative effect of inbreeding evident in previous experiments (Scoville et al. 2009; Bodbyl Roels and Kelly 2011). The evidence is not sufficient to determine if the IM population is currently at, or close to, an equilibrium frequency for inv6, given conflicting positive and negative effects. Moreover, a balanced polymorphism does not even necessarily predict a fixed allele frequency. The fluctuating selection suggested by Figure 6 does not predict a fixed equilibrium.
Genetic basis of inbreeding depression
Inbreeding depression is the decrease in fitness that occurs with increased homozygosity caused by mating between relatives or self-fertilization. It has been a focus of research in genetics for >150 years (Darwin 1876; East 1908; Crow 1993). Two models have dominated thinking about inbreeding depression: dominance and overdominance. The dominance model posits that inbreeding depression is caused by recessive or partially recessive alleles with deleterious effects on fitness. Recessivity of segregating deleterious mutations is predicted because selection more rapidly eliminates additive and dominant mutations from a population. In contrast, the overdominance model states that heterozygotes have superior fitness compared to either alternative homozygote, such that fitness declines as homozygosity increases with inbreeding. At present, a preponderance of data favors deleterious, partially recessive mutations rather than overdominance to explain the bulk of inbreeding depression (Charlesworth and Willis 2009).
inv6 is the second major chromosomal polymorphism to have been mapped within IM, following the drive locus on chromosome 11 (D). The latter is a structural variant of the centromeric region of chromosome 11 that exhibits centromere-associated drive over the alternative chromosomal type. D gains an ∼60:40 transmission advantage in heterozygote individuals by driving through female meiosis (Fishman and Saunders 2008). Consistent with this selective advantage, patterns of nucleotide diversity suggest a recent and rapid spread of the D variant at Iron Mountain. However, D is prevented from reaching fixation because it exhibits recessive negative effects on both male (pollen viability) and female (seed set) fitness components in nature. Together, these linked recessive costs maintain the D chromosomal variant at intermediate frequency (30–40%) near the predicted equilibrium (Fishman and Kelly 2015). Although we do not know as much about inv6 yet, its shared features with D suggest a general alternative model for inbreeding depression.
inv6 and D each generate substantial inbreeding depression, but neither polymorphism conforms to either the dominance or overdominance model. These loci exhibit partially recessive deleterious effects (like the dominance model) but intermediate allele frequencies (like the overdominance model). Because of the latter feature, these polymorphisms generate considerable inbreeding depression and also genetic variance for fitness. For both D and inv6, the existence of such intermediate frequency deleterious variation depends on structural variants that prevent (at least in the short-term) recombination from breaking up the association between alleles with positive and negative effects. Otherwise, the alleles with deleterious effects (particularly if not entirely recessive) should be driven to low frequency by selection. Both deleterious recessive mutations and structural mutations are common; in combination with rare beneficial variants or driving selfish elements, they may often contribute to balanced polymorphism.
Apart from D and inv6, the data from the replicated F2 experiment are consistent mainly with the dominance model; inbreeding depression maintained by mutation-selection balance. We mapped >20 additional QTL for male fertility (Table S2). Nearly all were mapped in only one of the three crosses. This is consistent with the prediction that deleterious alleles should be rare in the population, and as a consequence, each such allele was sampled into only one founding parent of the six used to generate the mapping populations. A few QTL did exhibit apparent overdominance. However, these QTL were also unique to a mapping population, contrary to expectations for alleles maintained at intermediate population frequencies by balancing selection. The level of mapping resolution in the replicated F2 experiment cannot distinguish true overdominance from associative overdominance (deleterious alleles linked in repulsion phase, also known as “pseudo-overdominance”). The remaining QTL exhibit average partially recessive gene action of the low allele. The average dominance coefficient (excluding the chromosomal rearrangements) of h = 0.18 is very close to the previous estimate of h ∼0.15 (Willis 1999a). It is likely that many additional deleterious mutations are segregating in IM, but are yet unmapped because the individual effects are below our detection limit.
Conclusion
Modern tools of genetic mapping, combined with population sampling, provide a direct means to investigate the maintenance of genetic variation in fitness. This work contributes to a larger effort to identify genetic components of fitness variation within the Iron Mountain population of M. guttatus and reveals their individual (and apparently complex) histories. Balancing selection facilitated by recombination suppression in inversions may be an unexpectedly significant and general factor in the maintenance of fitness variation within populations, in keeping with the prominent role of inversions in speciation and divergence.
A great deal remains unknown about the fitness costs and benefits of inv6. It is reasonable to hypothesize that the negative effects of inv6 are due to partially recessive deleterious mutations captured within a novel inversion. This inversion has reached an unexpectedly high frequency owing to (perfectly) linked alleles with beneficial heterozygous effects. This interpretation is generally congruent with theory (Sturtevant and Mather 1938; Kirkpatrick and Barton 2006). An inversion polymorphism that has captured both advantageous and deleterious alleles is maintained if the advantageous effect of the inversion is (i) greater than its disadvantageous effects in the heterozygote and (ii) smaller than the disadvantageous effects in the homozygote. Of course, these are conditions for polymorphism under almost any diploid model, inversion or not. However, inversions might greatly increase the likelihood that the conditions are met; advantageous and deleterious effects are bundled together by recombination suppression in heterokaryotypes.
The genetic basis and the selective factors underlying the positive heterozygous effect of inv6 are presently unknown. The inverted genomic region contains >1200 annotated genes. The advantageous effect could owe to one mutation (producing associative overdominance in combination with linked deleterious alleles) or to multiple genetic changes. In the latter case, it is plausible that inv6 has captured a constellation of alleles that are locally advantageous within IM, as postulated by Kirkpatrick and Barton (2006). However, it is also possible that co-adaptation among alleles within inv6 underlie its beneficial effects. Linked with deleterious recessive alleles, inv6 could represent a “supergene with baggage.” Epistasis for fitness-related traits is common in M. guttatus (Kelly 2005; Monnahan and Kelly 2015); if positively interacting alleles are located on the same chromosome, inversions may often be favored because they suppress recombination among them. The environmental dependence of inv6 fitness effects, evident from differences between greenhouse and field and between years at the field site, is also likely important. Finally, the inversion itself (e.g., mutations caused by the breakpoints rather than genic variants within the rearranged region) may cause either positive or negative fitness effects (e.g., Küpper et al. 2016). Much remains to be learned.
Acknowledgments
We thank Arpiar Saunders, Tyler Huggins, Angela Stathos, Dan Crowser, Becky Fletcher, Katie Zarn, and Mariah McIntosh for assistance with field collections, counting of pollen and seeds, and genotyping of markers. This research was supported by National Institute of Health grant GM073990 (to J.K.K. and J.H.W.) and by National Science Foundation grant DEB-0918902 (to L.F.).
Footnotes
Communicating editor: L. C. Moyle
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.183566/-/DC1.
Literature Cited
- Balanyà J., Serra L., Gilchrist G. W., Huey R. B., 2003. Evolutionary pace of chromosomal polymorphism in colonizing populations of Drosophila subobscura: an evolutionary time series. Evolution 57: 1837–1845. [DOI] [PubMed] [Google Scholar]
- Bodbyl Roels S. A., Kelly J. K., 2011. Rapid evolution caused by pollinator loss in Mimulus guttatus. Evolution 65: 2541–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D., Willis J. H., 2009. The genetics of inbreeding depression. Nat. Rev. Genet. 10: 783–796. [DOI] [PubMed] [Google Scholar]
- Cheng C., White B. J., Kamdem C., Mockaitis K., Costantini C., et al. , 2012. Ecological genomics of Anopheles gambiae along a latitudinal cline: a population-resequencing approach. Genetics 190: 1417–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluzzi M., Sabatini A., della Torre A., Di Deco M. A., Petrarca V., 2002. A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298: 1415–1418. [DOI] [PubMed] [Google Scholar]
- Crow, J. F. 1993 Mutation, mean fitness, and genetic load. Oxf. Surv. Evol. Biol. 9: 3–42.
- Darwin C. R., 1876. The Effects of Cross- and Self-Fertilisation in the Vegetable Kingdom. John Murray, London. [Google Scholar]
- Dobzhansky T., 1970. Genetics of the Evolutionary Process. Columbia University Press, New York. [Google Scholar]
- East E. M., 1908. Inbreeding in Corn. Rep. Conn. Agric. Exp. Sta; 419–428. [Google Scholar]
- Fang Z., Pyhäjärvi T., Weber A. L., Dawe R., Glaubitz J. C., et al. , 2012. Megabase-scale inversion polymorphism in the wild ancestor of maize. Genetics 191: 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder J. L., Roethele J. B., Filchak K., Niedbalski J., Romero-Severson J., 2003. Evidence for inversion polymorphism related to sympatric host race formation in the apple maggot fly, Rhagoletis pomonella. Genetics 163: 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L., Kelly J. K., 2015. Centromere-associated meiotic drive and female fitness variation in Mimulus. Evolution 69: 1208–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L., Saunders A., 2008. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322: 1559–1562. [DOI] [PubMed] [Google Scholar]
- Fishman L., Kelly A. J., Morgan E., Willis J. H., 2001. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159: 1701–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L., Stathos A., Beardsley P., Williams C. F., Hill J. P., 2013. Chromosomal rearrangements and the genetics of reproductive barriers in Mimulus (monkey flowers). Evolution 67: 2547–2560. [DOI] [PubMed] [Google Scholar]
- Fishman L., Willis J. H., Wu C. A., Lee Y. W., 2014. Comparative linkage maps suggest that fission, not polyploidy, underlies near-doubling of chromosome number within monkeyflowers (Mimulus; Phrymaceae). Heredity 112: 562–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L. E., Willis J. H., Vision T. J., 2014. The standing pool of genomic structural variation in a natural population of Mimulus guttatus. Genome Biol. Evol. 6(1): 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford E. B., 1971. Ecological Genetics. Chapman and Hall, London. [Google Scholar]
- Gilburn A. S., Day T. H., 1999. Female mating behaviour, sexual selection and chromosome I inversion karyotype in the seaweed fly, Coelopa frigida. Heredity 82: 276–281. [DOI] [PubMed] [Google Scholar]
- Hermann K., Klahre U., Moser M., Sheehan H., Mandel T., et al. , 2013. Tight genetic linkage of prezygotic barrier loci creates a multifunctional speciation island in Petunia. Curr. Biol. 23: 873–877. [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A., Rieseberg L. H., 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation. Annu. Rev. Ecol. Evol. Syst. 39: 21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holeski, L., P. Monnahan, B. Koseva, N. McCool, R. L. Lindroth, and J. K. Kelly, 2014 A high-resolution genetic map of yellow monkeyflower identifies chemical defense QTLs and recombination rate variation. G3 (Bethesda) 4: 813–821. [DOI] [PMC free article] [PubMed]
- Jones F. C., Grabherr M. G., Chan Y. F., Russell P., Mauceli E., et al. , 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A. J., Willis J. H., 1998. Polymorphic microsatellite loci in Mimulus guttatus and related species. Mol. Ecol. 7: 769–774. [Google Scholar]
- Kelly J. K., 2005. Epistasis in monkeyflowers. Genetics 171: 1917–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. K., 2008. Testing the rare alleles model of quantitative variation by artificial selection. Genetica 132: 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. K., Willis J. H., 2002. A manipulative experiment to estimate bi-parental inbreeding in monkeyflowers. Int. J. Plant Sci. 163: 575–579. [Google Scholar]
- Kelly J. K., Rasch A., Kalisz S., 2002. A method to estimate pollen viability from pollen size variation. Am. J. Bot. 89: 1021–1023. [DOI] [PubMed] [Google Scholar]
- Kelly J. K., Koseva B., Mojica J. P., 2013. The genomic signal of partial sweeps in Mimulus guttatus. Genome Biol. Evol. 5: 1457–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., Barton N., 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov A. S., 1988. Deleterious mutations and the evolution of sexual reproduction. Nature 336: 435–440. [DOI] [PubMed] [Google Scholar]
- Krimbas C. B., Powell J. R., 1992. Drosophila Inversion Polymorphism. CRC Press, Boca Raton, FL. [Google Scholar]
- Küpper C., Stocks M., Risse J. E., dos Remedios N., Farrell L. L., et al. , 2016. A supergene determines highly divergent male reproductive morphs in the ruff. Nat. Genet. 48: 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry D. B., Willis J. H., 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8: e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnahan P. J., Kelly J. K., 2015. Naturally segregating loci exhibit epistasis for fitness. Biol. Lett. 11: pii: 20150498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1918. Genetic variability, twin hybrids and constant hybrids, in a case of balanced lethal factors. Genetics 3: 422–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor M. A. F., Grams K. L., Bertucci L. A., Reiland J., 2001. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 98: 12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg L. H., Whitton J., Gardner K., 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152: 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander T., Libbrecht R., Keller L., 2014. Supergenes and complex phenotypes. Curr. Biol. 24: R288–R294. [DOI] [PubMed] [Google Scholar]
- Scoville A., Lee Y. W., Willis J. H., Kelly J. K., 2009. Contribution of chromosomal polymorphisms to the G-matrix of Mimulus guttatus. New Phytol. 183: 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam P., 1993. Construction of integrated genetic linkage maps by means of a new computer package: Join Map. Plant J. 3: 739–744. [Google Scholar]
- Sturtevant A. H., Mather K., 1938. The interrelations of inversions, heterosis and recombination. Am. Nat. 72: 447–452. [Google Scholar]
- Sweigart A., Karoly K., Jones A., Willis J. H., 1999. The distribution of individual inbreeding coefficients and pairwise relatedness in a population of Mimulus guttatus. Heredity 83: 625–632. [DOI] [PubMed] [Google Scholar]
- Tajima F., Nei M., 1983. Estimation of evolutionary distance between nucleotide-sequences. Jpn. J. Genet. 58: 684–685. [DOI] [PubMed] [Google Scholar]
- Thomas J. W., Cáceres M., Lowman J. J., Morehouse C. B., Short M. E., et al. , 2008. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics 179: 1455–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M. J., Jiggins C. D., 2014. Supergenes and their role in evolution. Heredity 113: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery R. K., 1978. Case studies in the evolution of species complexes in Mimulus. Evol. Biol. 11: 405–507. [Google Scholar]
- White M. J. D., 1969. Chromosomal rearrangements and speciation in animals. Annu. Rev. Genet. 3: 75–98. [Google Scholar]
- Willis J. H., 1993. Partial self fertilization and inbreeding depression in two populations of Mimulus guttatus. Heredity 71: 145–154. [Google Scholar]
- Willis J. H., 1999a Inbreeding load, average dominance, and the mutation rate for mildly deleterious alleles in Mimulus guttatus. Genetics 153: 1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis J. H., 1999b The role of genes of large effect on inbreeding depression in Mimulus guttatus. Evolution 53: 1678–1691. [DOI] [PubMed] [Google Scholar]
- Wu C. A., Lowry D. B., Cooley A. M., Wright K. M., Lee Y. W., et al. , 2007. Mimulus is an emerging model system for the integration of ecological and genomic studies. Heredity 100: 220–230. [DOI] [PubMed] [Google Scholar]
- Yandell B. S., Mehta T., Banerjee S., Shriner D., Venkataraman R., et al. , 2007. R/qtlbim: QTL with Bayesian Interval Mapping in experimental crosses. Bioinformatics 23: 641–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi N., Banerjee S., Pomp D., Yandell B. S., 2007. Bayesian mapping of genomewide interacting quantitative trait loci for ordinal traits. Genetics 176: 1855–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genetic, phenotypic, and fitness data specific to inv6 are contained in File S2. The SNP data (within genes) for the 10 resequenced IM lines is reported as Table S3. The raw sequence data is available from the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra).