Abstract
Deciphering the genetic basis of human disease requires a comprehensive knowledge of genetic variants irrespective of their class or frequency. Although an impressive number of human genetic variants have been catalogued, a large fraction of the genetic difference that distinguishes two human genomes is still not understood at the base-pair level. This is because the emphasis has been on single-nucleotide variation as opposed to less tractable and more complex genetic variants, including indels and structural variants. The latter, we propose, will have a large impact on human phenotypes but require a more systematic assessment of genomes at deeper coverage and alternate sequencing and mapping technologies.
Keywords: human genetic variation, indels, structural variation, copy number variation, segmental duplication
UNCOVERING the genetic basis of human disease and phenotype requires an understanding of the nature and pattern of human genetic variation. This includes not only variant discovery and accurate genotyping but a resolution of the haplotype structure and the mutational properties that have shaped our genome. The completion of phase 3 of the 1000 Genomes Project (Auton et al. 2015) was an important landmark in this regard. More than 2500 “normal genomes” were sequenced from 26 different human populations, revealing an impressive 84.7 million single-nucleotide variants (SNVs), 3.6 million insertion/deletion (indel) variants, and >60,000 structural variants (SVs). The latter are distinguished from indels based on event sizes greater than or equal to 50 bp in length (Sudmant et al. 2015b). While there are other similar population-based genome sequencing projects that have been recently completed (Genome of the Netherlands Consortium 2014; Sudmant et al. 2015a) or are underway (e.g., UK10K Consortium et al. 2015), most are smaller in scale and/or have more restrictions with respect to data access and use. As a result, the 1000 Genomes Project variants serve as one of the most powerful resources for understanding the normal pattern of human genetic variation.
There are two current limitations with this catalog of human genetic variation. First, it is derived from relatively sparse genome sequence data (six- to sevenfold sequence coverage). The decision to sequence genomes at this level of coverage was only partially an economic one. It was driven largely by population genetic theory where most of the common genetic variation (>1% allele frequency) could be resolved by imputation as a result of linkage disequilibrium (1000 Genomes Project Consortium 2010). As a result, more genomes were strategically sequenced rather than sequencing fewer genomes more deeply. The project exceeded expectations, detecting an estimated 75% of SNVs with an allele frequency of >0.1%. This approach had limited power to detect rare variants (<0.1% frequency) and SVs (irrespective of their allele frequency). For diseases where rare variants or SVs are known to play an important role (e.g., epilepsy, intellectual disability, autism, and schizophrenia) (Hoischen et al. 2014), larger and deeper datasets, such as the Exome Aggregation Consortium (ExAC) database for SNV mutations within coding sequence (Song et al. 2015) and SV databases developed from thousands of population controls (Coe et al. 2014; MacDonald et al. 2014), are critically important.
The second limitation is that not all genetic variation has been equally ascertained even after conditioning on the allele frequency. Detailed targeted sequencing of regions of the human genome suggests that indels should occur at approximately one-tenth of the frequency of SNVs (Bhangale et al. 2005), suggesting that the current catalog may be missing at least 30–40% of all indels. Detection of indels associated with short tandem repeat (STR) sequences is particularly challenging and specialized methods have been developed to discover and accurately genotype these from next-generation sequencing datasets (Karakoc et al. 2012; Narzisi et al. 2014; Willems et al. 2014; Chaisson et al. 2015a). Sensitivity for indel variant discovery is generally much lower than for SNVs. A comparison of 170 genomes sequenced to high coverage by an orthogonal sequencing platform (Complete Genomics) suggests that less than 75% of indels with an allele frequency of 0.5% were detected. The sensitivity of indel detection drops precipitously as the allele frequency dips below 0.3% (Auton et al. 2015).
The situation for SVs is, in fact, much bleaker with respect to sensitivity and specificity. This stems from the fact that discovery of these variants is largely indirect, depending on mapping short-read sequencing data using read-depth or read-pair detection methods. Thus, unlike SNVs where discovery and sequence resolution occur simultaneously, deletions, duplications, and inversions are often inferred based on specific signatures, with breakpoint resolution occurring post hoc. Not surprisingly, almost half of the 68,000 SVs (46%) detected as part of the 1000 Genomes Project have no or limited breakpoint resolution (Sudmant et al. 2015b). Moreover, the majority of SV callsets are restricted to those with less than a 5% false discovery rate. This translates into a large fraction of SVs not being reported because it is currently impractical to experimentally validate all events.
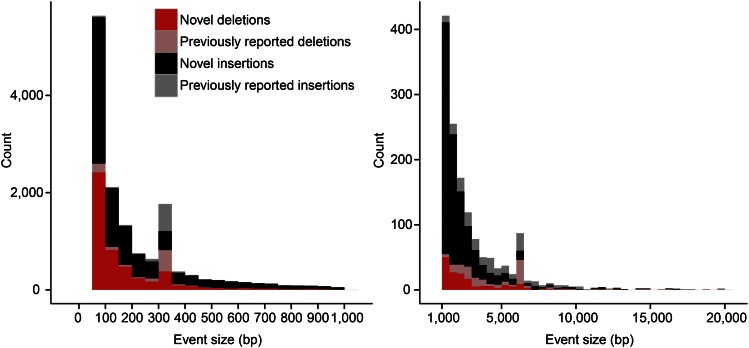
Sensitivity estimates vary considerably depending on the type of SV. It has been estimated, for example, that 68% of inversions and 35% of duplication events are unrecognized, in contrast to deletions where sensitivity estimates are as high as 80% (Sudmant et al. 2015b). This bias against particular classes of structural variation affects both common and rare genetic variation. Sensitivity also varies as a function of size, with both ends of the SV spectrum adversely affected. Comparisons with SVs resolved using long-read sequencing technologies [e.g., single-molecule, real-time (SMRT) or Pacific Biosciences sequencing technology] suggest that the majority (>80%) of insertions and deletions between 50 bp and 1 kbp in length are missed using short-read sequencing technologies (Figure 1) (Chaisson et al. 2015a), irrespective of frequency. These results argue that most widely used sequencing technologies are insufficient, because short reads fail to detect and accurately genotype a large fraction of SVs.
Figure 1.
Sensitivity of SV detection as a function of length. Sequence-resolved insertions and deletions from single-molecule, real-time (SMRT) sequencing of a haploid human genome (CHM1) (Chaisson et al. 2015a) are compared to those discovered by other approaches (Conrad et al. 2010; Kidd et al. 2010; Mills et al. 2011; Sudmant et al. 2015a,b). Previously identified variants (pink or gray) are contrasted to those exclusively discovered by SMRT sequencing. Of the variants between 50 bp and 1 kbp, 88% are novel in contrast to 75% of variants between 1 kbp and 20 kbp in length. An exception occurs for Alu and LINE insertions where 73% of the events have been previously discovered because of specialized methods to detect mobile element insertions.
At the other end of the spectrum, the largest common SVs in the human genome are segmental duplications (duplicated sequences >1 kbp and >90% sequence identity). Approximately half of the copy number variants between two humans larger than 1 kbp map to this 5% of the human genome (Sudmant et al. 2015a). Structural variation in these regions is frequently complex and associated with multicopy number states (sometimes referred to as multiallelic or mCNVs). mCNVs are currently approximated based on mapping short-read sequences to a reference genome and estimating the diploid copy to the nearest integer (e.g., 1, 2, 3, 4, 5 copies, etc.). As the size of the mCNV and whole-genome sequence coverage decreases, so too does the accuracy of copy number estimates (Sudmant et al. 2010). Since different chromosomal haplotype combinations (e.g., 2 copies on one chromosome and 3 copies on another vs. 4 copies on one chromosome and 1 on another) may arrive at the same diplotype copy number (e.g., 5 copies), imputation for this type of variation becomes increasingly problematic as copy number increases (Handsaker et al. 2015). mCNV breakpoints frequently are flanked by high-identity repetitive sequence, further limiting imputation and association of this form of genetic variation with human phenotypes. Haplotype-resolved sequencing of these regions has consistently shown that such inferential genotyping underestimates the genetic complexity of the underlying genetic variation of these regions (Boettger et al. 2012; Steinberg et al. 2012; Antonacci et al. 2014; O’Bleness et al. 2014). Several lines of evidence suggest that this missing variation will be critical to interpreting the “missing heritability” of human disease. First, the genes and regions associated with this variation are hotspots of recurrent mutation directly or indirectly associated with human disease and the emergence of novel genes associated with the evolution of human phenotypes (Chaisson et al. 2015a; Florio et al. 2015). The HLA locus is perhaps the most well-cited example of this (Raymond et al. 2005) but many more examples of regions of comparable complexity have emerged over the last few years (Boettger et al. 2012; Steinberg et al. 2012; Antonacci et al. 2014; O’Bleness et al. 2014). Second, SVs have been estimated to be enriched 50-fold for expression quantitative trait loci (eQTL) when compared to SNVs (relative to the number of events tested) (Sudmant et al. 2015b). Similarly, indel variants were the top associated eQTL 26–40% of the time (Auton et al. 2015). These data confirm intuition that this variation, because of its size, is likely to have a greater impact on gene expression than SNVs. Similarly, genome-wide association loci are estimated to be enriched almost threefold for common SVs (Sudmant et al. 2015b). This estimate must be considered a lower bound because large swathes of SVs are more difficult to impute based on linkage disequilibrium with nearby SNVs obtained from whole-genome sequencing data. Only 44% of duplications, for example, with an allele frequency >0.1% could be imputed by the best flanking single-nucleotide polymorphism (r2 > 0.6) (Sudmant et al. 2015b) after considering all nonsingleton SNVs in 1 Mbp flanking each SV. The proportion of untagged duplications remained similar at higher allele frequencies perhaps as a result of recurrent mutational events and the paucity of reliable SNVs in close proximity. Many of these genetic variants thus represent terra incognita with respect to ongoing genetic association studies and, therefore, the causative variants have yet to be discovered.
Given its importance, what is the solution to improving our understanding of the more complex forms of human genetic variation? There are three obvious steps. First, sequence genomes from populations much more deeply (e.g., >30-fold sequence coverage) to increase sensitivity of detection of SVs and indels at the individual genome level. This should be done in the context of families in order to understand transmission properties and mutation rates, which are expected to vary by orders of magnitude for SVs when compared to SNVs. While there are currently many initiatives that have been launched to sequence genomes, most of these are associated with specific clinical phenotypes and have restricted use and data access. It is important that the sequence data and the variants be made publicly available without restriction to have the broadest impact. The genetic resources collected as part of the 1000 Genomes Project are an obvious first choice because cell lines, high-quality DNA, and consents are already in place (Auton et al. 2015). They are also ideal because a large fraction of the 3500 samples collected from the 1000 Genomes Project exist in the form of parent–child trios, although few related individuals were sequenced as part of the project.
Second, characterize genomes using orthogonal technologies (Berlin et al. 2015; Chaisson et al. 2015a; Mak et al. 2016), specifically long-read sequencing technologies such as SMRT and Oxford Nanopore Technologies sequencing that increase power to detect both complex and intermediate-size (50–2000 bp) SVs. Longer reads will also improve physical phasing of SVs and SNVs enhancing future association studies (especially for more complex SVs such as mCNVs). It should be noted, however, that long-read sequencing technology delivers sequence reads that are still too short (<70 kbp) to completely resolve the most complex SVs within segmental duplications, so continued investment into mapping and sequencing technologies that resolve molecules up to 1 Mbp in length should be a priority (Chaisson et al. 2015b).
Third, combine computational and experimental methods to resolve the physical haplotype structure of human genomes as opposed to relying on inferential methods (English et al. 2015; Pendleton et al. 2015). This is especially relevant with respect to STRs and mCNVs where the frequency of recurrent mutation is expected to be high and direct observation and resolution of the sequence structure will be key to associating variants with phenotype. Such an endeavor could begin with a small number (n = 20–50) of human reference genomes completely resolved at the single haplotype level, including the structure and organization of the copy number polymorphic segmental duplications. Data from these new references could be used in the short term to computationally improve imputation for mCNVs and other complex SVs. When long-read sequencing technology becomes affordable and ubiquitous, haplotype-resolved sequenced genomes will likely become the standard for studies of human genetic disease and phenotype. This, of course, requires that we start thinking about a six-billion as opposed to a three-billion base-pair genome. While many of these next steps may have seemed an impossibility 5 years ago, rapid advances in genomic technology make a complete understanding of human genetic variation a real possibility that can now be pursued.
Acknowledgments
We are grateful to A. Auton, G. Abecasis, and H. M. Kang for access to underlying summary data from the 1000 Genomes Project. We thank T. Brown for manuscript assistance. This work was supported, in part, by a grant from the National Institutes of Health (R01HG002385 to E.E.E.). E.E.E. is an investigator of the Howard Hughes Medical Institute. Competing financial interests: E.E.E. is on the scientific advisory board of DNAnexus, Inc. and is a consultant for Kunming University of Science and Technology as part of the 1000 China Talent Program.
Footnotes
Communicating editor: M. Johnston
Literature Cited
- 1000 Genomes Project Consortium, 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonacci F., Dennis M. Y., Huddleston J., Sudmant P. H., Steinberg K. M., et al. , 2014. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat. Genet. 46: 1293–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A., Brooks L. D., Durbin R. M., Garrison E. P., Kang H. M., et al. , 2015. A global reference for human genetic variation. Nature 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K., Koren S., Chin C. S., Drake J. P., Landolin J. M., et al. , 2015. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33: 623–630. [DOI] [PubMed] [Google Scholar]
- Bhangale T. R., Rieder M. J., Livingston R. J., Nickerson D. A., 2005. Comprehensive identification and characterization of diallelic insertion-deletion polymorphisms in 330 human candidate genes. Hum. Mol. Genet. 14: 59–69. [DOI] [PubMed] [Google Scholar]
- Boettger L. M., Handsaker R. E., Zody M. C., McCarroll S. A., 2012. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat. Genet. 44: 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M. J., Huddleston J., Dennis M. Y., Sudmant P. H., Malig M., et al. , 2015a Resolving the complexity of the human genome using single-molecule sequencing. Nature 517: 608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M. J., Wilson R. K., Eichler E. E., 2015b Genetic variation and the de novo assembly of human genomes. Nat. Rev. Genet. 16: 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe B. P., Witherspoon K., Rosenfeld J. A., van Bon B. W., Vulto-van Silfhout A. T., et al. , 2014. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 46: 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D. F., Pinto D., Redon R., Feuk L., Gokcumen O., et al. , 2010. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English A. C., Salerno W. J., Hampton O. A., Gonzaga-Jauregui C., Ambreth S., et al. , 2015. Assessing structural variation in a personal genome-towards a human reference diploid genome. BMC Genomics 16: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M., Albert M., Taverna E., Namba T., Brandl H., et al. , 2015. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347: 1465–1470. [DOI] [PubMed] [Google Scholar]
- Genome of the Netherlands Consortium , 2014. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 46: 818–825. [DOI] [PubMed] [Google Scholar]
- Handsaker R. E., Van Doren V., Berman J. R., Genovese G., Kashin S., et al. , 2015. Large multiallelic copy number variations in humans. Nat. Genet. 47: 296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoischen A., Krumm N., Eichler E. E., 2014. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat. Neurosci. 17: 764–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakoc E., Alkan C., O’Roak B. J., Dennis M. Y., Vives L., et al. , 2012. Detection of structural variants and indels within exome data. Nat. Methods 9: 176–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd J. M., Graves T., Newman T. L., Fulton R., Hayden H. S., et al. , 2010. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell 143: 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald J. R., Ziman R., Yuen R. K., Feuk L., Scherer S. W., 2014. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 42: D986–D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak A. C., Lai Y. Y., Lam E. T., Kwok T. P., Leung A. K., et al. , 2016. Genome-wide structural variation detection by genome mapping on nanochannel arrays. Genetics 202: 351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R. E., Walter K., Stewart C., Handsaker R. E., Chen K., et al. , 2011. Mapping copy number variation by population-scale genome sequencing. Nature 470: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narzisi G., O’Rawe J. A., Iossifov I., Fang H., Lee Y. H., et al. , 2014. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat. Methods 11: 1033–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bleness M., Searles V. B., Dickens C. M., Astling D., Albracht D., et al. , 2014. Finished sequence and assembly of the DUF1220-rich 1q21 region using a haploid human genome. BMC Genomics 15: 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton M., Sebra R., Pang A. W., Ummat A., Franzen O., et al. , 2015. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods 12: 780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond C. K., Subramanian S., Paddock M., Qiu R., Deodato C., et al. , 2005. Targeted, haplotype-resolved resequencing of long segments of the human genome. Genomics 86: 759–766. [DOI] [PubMed] [Google Scholar]
- Song W., Gardner S. A., Hovhannisyan H., Natalizio A., Weymouth K. S., et al. , 2015. Exploring the landscape of pathogenic genetic variation in the ExAC population database: insights of relevance to variant classification. Genet. Med. DOIhttp//dx..org/10.1038/gim.2015.180. [DOI] [PubMed] [Google Scholar]
- Steinberg K. M., Antonacci F., Sudmant P. H., Kidd J. M., Campbell C. D., et al. , 2012. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat. Genet. 44: 872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant P. H., Kitzman J. O., Antonacci F., Alkan C., Malig M., et al. , 2010. Diversity of human copy number variation and multicopy genes. Science 330: 641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant P. H., Mallick S., Nelson B. J., Hormozdiari F., Krumm N., et al. , 2015a Global diversity, population stratification, and selection of human copy-number variation. Science 349: aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant P. H., Rausch T., Gardner E. J., Handsaker R. E., Abyzov A., et al. , 2015b An integrated map of structural variation in 2,504 human genomes. Nature 526: 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UK10K Consortium; K. Walter, J. L. Min, J. Huang, L. Crooks et al, 2015 The UK10K project identifies rare variants in health and disease. Nature 526: 82–90. [DOI] [PMC free article] [PubMed]
- Willems T., Gymrek M., Highnam G., Mittelman D., Erlich Y., 2014. The landscape of human STR variation. Genome Res. 24: 1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]