Abstract
The origin of the flower was a key innovation in the history of complex organisms, dramatically altering Earth’s biota. Advances in phylogenetics, developmental genetics, and genomics during the past 25 years have substantially advanced our understanding of the evolution of flowers, yet crucial aspects of floral evolution remain, such as the series of genetic and morphological changes that gave rise to the first flowers; the factors enabling the origin of the pentamerous eudicot flower, which characterizes ∼70% of all extant angiosperm species; and the role of gene and genome duplications in facilitating floral innovations. A key early concept was the ABC model of floral organ specification, developed by Elliott Meyerowitz and Enrico Coen and based on two model systems, Arabidopsis thaliana and Antirrhinum majus. Yet it is now clear that these model systems are highly derived species, whose molecular genetic-developmental organization must be very different from that of ancestral, as well as early, angiosperms. In this article, we will discuss how new research approaches are illuminating the early events in floral evolution and the prospects for further progress. In particular, advancing the next generation of research in floral evolution will require the development of one or more functional model systems from among the basal angiosperms and basal eudicots. More broadly, we urge the development of “model clades” for genomic and evolutionary-developmental analyses, instead of the primary use of single “model organisms.” We predict that new evolutionary models will soon emerge as genetic/genomic models, providing unprecedented new insights into floral evolution.
Keywords: ABC model, basal angiosperms, evo-devo, fading borders model: floral diversity, flower evolution, Pentapetalae
THE origin of the flower during the late Jurassic to early Cretaceous eras (most recent estimates are between 150 and 190 MYA; Magallón et al. 2015) was a key evolutionary innovation that profoundly altered the Earth’s biota. Flowering plants (angiosperms), with reproductive security and speed conferred by the flower, replaced other seed plants in most ecosystems. Diversification of flowers and the resulting fruit spurred coevolutionary change in pollinators and dispersers, with subsequent wide-ranging effects on herbivores, mycorrhizae, and other interacting organisms. The development of human civilization during the past 10 millenia was likewise closely linked to the flower, as seeds and fruit—especially grains—are the basis of agriculture in both agrarian and modern society. Thus, elucidation of the genetic basis of the origin and evolution of the flower has fundamental implications for both our understanding of organismal evolution and our ability to increase food production through bioengineering of key angiosperm crops.
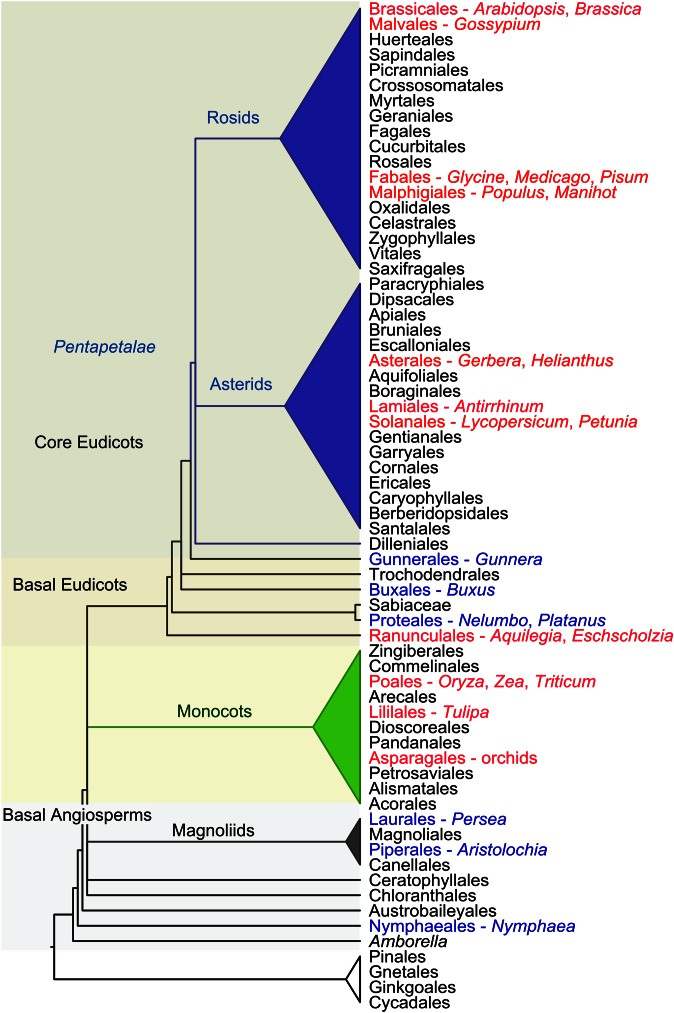
A broad spectrum of multidisciplinary research involving phylogenetics, developmental genetics, and genomics spurs this work and facilitates revised views of floral evolution. One key element has been the development of a robust phylogenetic framework for the angiosperm branch of the Tree of Life (e.g., Soltis et al. 2011) to place the developmental genetics of the flower in the appropriate evolutionary context (evolutionary-developmental biology, evo-devo). Accordingly, the original genetic models used to unravel flower developmental genetics, Arabidopsis thaliana and Antirrhinum majus (Coen and Meyerowitz 1991), are highly derived rosid and asterid species, respectively, embedded within the core eudicot clade of angiosperms (Figure 1). Key aspects of the genetics of Arabidopsis and Antirrhinum flower development also operate in genetic models of the highly derived grass family (Poaceae), including Oryza and Zea (Mena et al. 1996; Ambrose et al. 2000), which are nested within the monocot clade. Despite this apparent conservation across much of angiosperm diversity, a synthesis of comparative molecular studies suggests that the floral genetic programs of Arabidopsis and Antirrhinum are evolutionarily derived, and a new paradigm (described below) is necessary to describe the early evolution of flowers (Soltis et al. 2002, 2006a,b, 2007, 2009a; Kanno et al. 2003; Albert et al. 2005; Kim et al. 2005; Chanderbali et al. 2006, 2009, 2010; Shan et al. 2006; Kramer et al. 2007; Theissen and Melzer 2007; Broholm et al. 2008; Specht and Bartlett 2009; Rasmussen et al. 2009; Yoo et al. 2010a,b; Brockington et al. 2013; Ronse de Craene and Brockington 2013; Hileman 2014a,b; Specht and Howarth 2015; Glover et al. 2015; Galimba and Di Stilio 2015).
Figure 1.
Summary tree of seed plant phylogeny showing the main lineages of flowering plants and the sister group, the extant gymnosperms. Species with established resources for flower developmental genetics, indicated in red, are distributed predominantly among the asterid and rosid clades of the Pentapetalae. Additional “evolutionary models,” shown in blue, are needed to address questions regarding the genetic basis of major transitions in floral evolution.
In this paper, we will review central tenets of floral developmental evolution, linking specific floral innovations to known genetic programs and propose new directions for understanding the genetic bases for evolutionary diversification of flowers. We focus on three scales of evolutionary innovation: the origin of the first flowers; the origin of flowers of Pentapetalae, a major subclade representing ∼70% of living angiosperm species; and the origin of specific floral innovations. See Table 1 for a glossary of terms related to this article.
Table 1. Glossary.
| Glossary of Terms |
|---|
| Amborella: a genus that contains a single species, Amborella trichopoda, and widely regarded as the sister group of all other extant angiosperms. |
| ANA grade: the three successively basal-most branches (grade) of angiosperms; the acronym ANA derives from the names of its three constituent lineages, Amborella, Nymphaeales, and Austrobaileyales. |
| Asterids: one of the two major subclades of core eudicots, the other being the rosids. The asterids are characterized by fused petals (sympetaly). Examples include potato, tomato, and sunflowers. |
| Austrobaileyales: one of the three ANA-grade lineages. Includes the spice “star anise.” |
| Basal angiosperms: an informal name for the flowering plants outside of the large, and derived, eudicot and monocot clades. They include the ANA grade and magnoliids. |
| Basal eudicots: an informal name for a paraphyletic group comprising the eudicot lineages outside of the core eudicot clade. |
| Carpel: the female reproductive organ of a flower. |
| Core eudicots: a monophyletic group comprising all eudicots apart from the basal eudicots, ∼70% of all angiosperm species. |
| Eudicots: the eudicots are the largest clade of flowering plants, characterized by pollen grains that exhibit three colpi or grooves paralleling the polar axis (tricolpate pollen). |
| Gymnosperm cones: the reproductive structure in gymnosperms composed of a central stalk densely covered with leaf-like organs (sporophylls); female cones bear ovules on the surface of their sporophylls; the sporophylls of male cones bear pollen sacs. |
| Nymphaeales: an order with three families of aquatic plants, Hydatellaceae, Cabombaceae, and Nymphaeaceae (water lilies). It is one of the three early-diverging basal angiosperm lineages that constitute the ANA grade. |
| Magnoliids: the largest clade of basal angiosperms. Familiar species include avocado, bay laurel, black pepper, cinnamon, magnolias, nutmeg, and tulip tree. |
| Monocots: the second largest clade of flowering plants, and one of the major groups into which the flowering plants have traditionally been divided. They are characterized by seeds with a single cotyledon (embryonic leaf) and many other synapomorphies. |
| Pentapetalae: all core eudicots except Gunnerales. |
| Perianth: a collective term for all parts of the flower external to the stamens and carpels. |
| Petals: the whorl of floral organs, usually colored, that surrounds the stamens. |
| Rosids: one of the two major subclades of core eudicots, the other being the asterids. In contrast to the fused petals of asterids, the petals of rosids are free. Examples include many familiar plants, such as roses, peaches, and the legumes (e.g., peanuts). |
| Sepals: the outer, often leaf-like, floral organs that surround the petals, stamens, and carpels. |
| Stamen: the male reproductive organ of a flower. |
| Synorganization: the close and precise interrelationship of floral organs of the same or different kinds during development, usually involving fusion of the parts involved. |
| Whole-genome duplication: the duplication of a complete genome, for example, of a diploid genome (with two copies of each chromosome) to form a tetraploid (with four copies of each chromosome); this term is sometimes used to refer to the process of duplication (i.e., polyploidization) and sometimes in reference to the state of having multiple, duplicate genomes (i.e., polyploidy). |
Conservation and Divergence in Floral Morphology and Developmental Genetics
Origin and evolution of floral developmental genetics
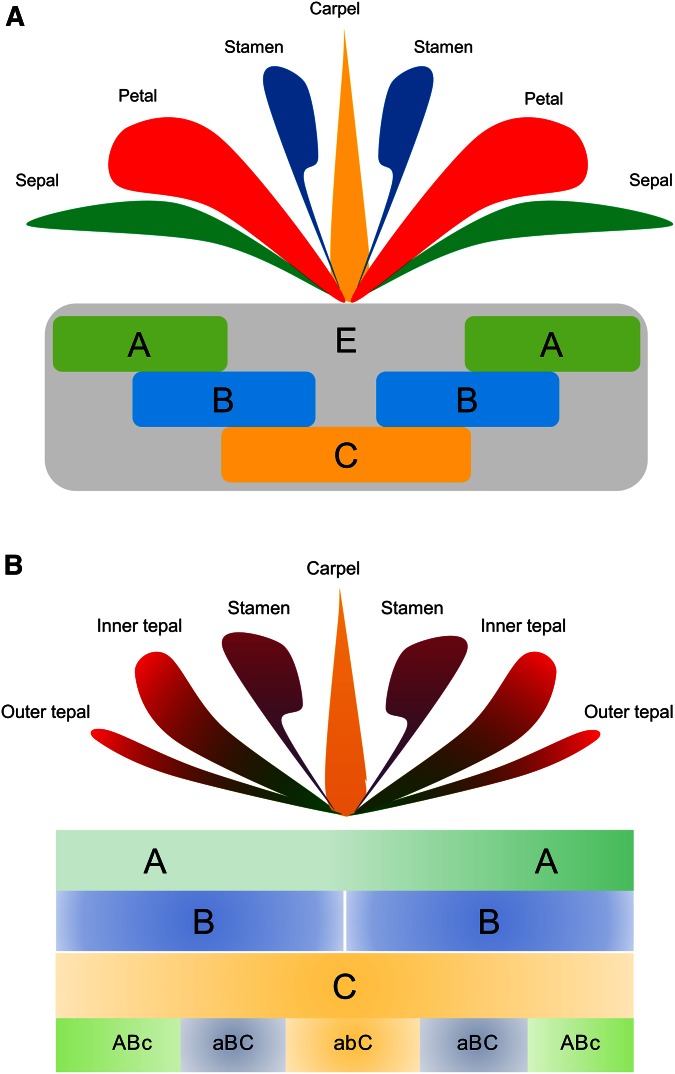
Twenty-five years ago, a combinatorial genetic model for the specification of floral organ identity, the so-called ABC model, was proposed by Enrico Coen and Elliot Meyerowitz (Coen and Meyerowitz 1991), based on studies of two of the major plant model systems of the time, Arabidopsis thaliana and Antirrhinum majus. Per this model, floral organ identities are specified through the action of three key gene functions (Figure 2) such that A function alone specifies sepals, A and B functions together determine petals, combined B and C functions specify stamens, and C function alone determines carpels (e.g., Bowman et al. 1989; Irish and Sussex 1990; Schwarz-Sommer et al. 1990; Coen and Meyerowitz 1991; Ma 1994). Subsequently, D and E functions were described and added to the model, with D controlling aspects of ovule development and E interacting with A, B, and C functions to specify organ identity (e.g., Colombo et al. 1995; Pelaz et al. 2000; Honma and Goto 2001; Ma 2005). Given the requirement of E-class genes for floral organ specification, the ABC model is now often referred to as the ABCE model. In Arabidopsis, the A-function genes are APETALA1 (API) and APETALA2 (AP2), B function is provided by APETALA3 (AP3) and PISTILLATA (PI), C function by AGAMOUS (AG), and E function by multiple SEPALLATA genes (SEP1–4). All but one (AP2) of the ABCE genes are members of the MADS-box gene family (Jofuku et al. 1994; Ma and dePamphilis 2000; Becker and Theissen 2003).
Figure 2.
Classic ABCE model of floral organ identity vs. fading borders model. (A) The classic ABCE model specifies four morphologically discrete floral organs: sepals are produced where only A function acts, petals are produced where A and B functions overlap, stamens occur where B and C functions overlap, and carpels are produced where C function acts alone. (B) In contrast, in the fading borders model, the borders between A, B, and C functions are blurred to produce a gradual transition of organ identity programs across the floral meristem. Hence, floral organs are influenced by “ABc,” “aBC,” and “abC” activities, where lowercase font indicates lower functional influence. These three combinations of gene activities promote the development of morphologically intergrading petaloid organs (tepals), stamens, and carpels, respectively. Modified from Chanderbali et al. (2010).
The ABCE model was developed through observations of homeotic conversion of floral organs in genetic mutants. For example, sepals replace petals and carpels replace stamens in B-function mutants, while C-function mutants exhibit homeotic conversion of stamens into petals (Bowman et al. 1989; Coen and Meyerowitz 1991). Recognition that such dramatic changes in floral structure can be rapidly obtained by disrupting individual ABCE functions soon led to the suggestion that evolutionary changes in floral form might involve shifts of ABCE functions across spatial domains of the flower, or the “shifting boundary” hypothesis (van Tunen et al. 1993; Bowman 1997; Albert et al. 1998; Kramer et al. 2003). For example, instead of a dimorphic perianth that is differentiated into sepals and petals, an entirely petaloid perianth could develop through the activation of B function in an organ that would be positionally homologous to a sepal. This genetic mechanism appears to operate in some basal eudicots, such as Aquilegia (columbines, Ranunculaceae; Kramer et al. 2003), and perhaps many monocots, including Tulipa (tulips, Liliaceae; van Tunen et al. 1993; Kanno et al. 2003), in which sepals and petals differ only (or primarily) in position rather than in morphology.
The basal angiosperm lineages (the ANA grade of Amborella, Nymphaeales, and Austrobaileyales; Chloranthales; magnoliids) accommodate only a few percent of angiosperms (Drinnan et al. 1994) but exhibit tremendous floral diversity, particularly in the number and arrangement of floral organs (Figure 3), and are pivotal to questions about floral development in the earliest angiosperms. Developmental genetic studies conducted for basal angiosperms indicate that they have a broader pattern of expression of B- (and to a lesser extent, C- and E-) function homologs than do eudicots (Kim et al. 2005; Soltis et al. 2009a) and prompted the formulation of a new model, the “fading borders” model (e.g., Buzgo et al. 2004, 2005; Soltis et al. 2006b, 2007; Chanderbali et al. 2010). The fading borders model interprets the gradual transition in floral morphology observed in basal angiosperms, magnoliids, and basal eudicots as reflecting a gradient in expression levels of floral organ identity genes across the developing floral meristem, where weak expression at the margin of the range of activity of a given gene overlaps with the expression of another regulator in adjacent organs. Gradually fading influence toward the periphery of broadly expressed organ identity functions, of B function in particular, imparts some features of one set of organs onto adjacent floral organs. Subsequent restriction of expression (and function) to specific regions of the floral meristem resulted in discrete whorls of morphologically distinct floral organs that together characterize the core eudicots. Genetic models are currently lacking among basal angiosperms, but petals in an Arabidopsis mutant that exhibits reduced B function are morphologically intermediate between petals and sepals (Bowman et al. 1989), offering genetic support for the fading borders concept.
Figure 3.
Floral variation in ANA grade, magnoliid, and basal eudicot angiosperms. Although comprising only a few percent of extant angiosperm species, these lineages exhibit enormous floral variation compared to the more canalized flowers of core eudicots and monocots. (A) Nymphaea caerulea (Nymphaeaceae; basal angiosperm). (B) Austrobaileya scandens (Austrobaileyaceae; basal angiosperm). (C) Persea americana (Lauraceae; magnoliid). (D) Piper neesiasnum (Piperaceae; magnoliid). (E) Aristolochia veraguensis (Aristolociaceae; magnoliid). (F) Asimina incana (Annonaceae; magnoliid). (G) Magnolia champaca (Magnoliaceae; magnoliid). (H) Argemone albiflora (Papaveraceae; basal eudicot). (I) Anemone canadensis (Ranunculaceae; basal eudicot). (J) Ranunculus ficaria (Ranunculaceae; basal eudicot). A is courtesy of Deborah Chanderbali; B is courtesy of Douglas Soltis; C is courtesy of Andre Chanderbali; and D–J are courtesy of Walter Judd.
Recognition that the B and C components of the ABCE system are conserved in the specification of reproductive organ development in gymnosperms (Winter et al. 1999; Theissen and Becker 2004) led to hypotheses of how genetic changes in the gymnosperm system could have produced the first flowers (Frohlich 2003; Baum and Hileman 2006; Theissen and Melzer 2007). Given that gymnosperm cones are unisexual, these evolutionary scenarios involve the modification of separate genetic programs for male (BC) and/or female (C) gymnosperm reproductive organs that would transform a unisexual cone into a bisexual structure with male organs (stamens) below female organs (carpels). Hypothesized genetic mechanisms include shifting gradients of organ identity functions in male (“out of male”) or female (“out of female”) gymnosperm cones (Baum and Hileman 2006; Theissen and Melzer 2007), or ectopic imposition of established female organ identity programs onto male cones in the “mostly male” scenario (Frohlich 2003). Further genetic transformations are also needed to produce a recognizable flower. These changes include condensation of the initial cone-like axis such that stamens appear to surround carpels, the origin of an external envelope of sterile perianth appendages surrounding the sexual organs (producing a structure similar to basal angiosperm flowers), and perianth dimorphism into sepals and petals to produce the typical flower specified by the ABCE model (Baum and Hileman 2006).
Clearly, the genetic regulators of floral organ specification are shared across angiosperms and were likely inherited from gymnosperm ancestors. Modifications to the activities of these key floral regulators may underlie the origin of the flower; can alter floral morphology, in some cases dramatically; and drive divergence in floral form. Given current evidence, the ancestral flower likely had a fading borders type of floral developmental program with broadly overlapping expression domains that produced morphologically intergrading floral organs, similar to those seen in a number of extant basal angiosperms. Over evolutionary time, restriction of ABCE function to specific regions of the floral meristem resulted in discrete whorls of the four morphologically distinct floral organs (sepals, petals, stamens, carpels) that characterize the Pentapetalae (Figure 2). Positive autoregulatory control and obligate heterodimerization are possible molecular mechanisms through which the ABCE model was refined via subsequent evolution (Theissen and Becker 2004; Theissen and Melzer 2007). Thus, although the ABCE model, given its priority and influence, has been considered the default floral developmental program, with variants viewed as derivatives of this program, comparative studies conducted in a phylogenetic context demonstrate instead that the model is evolutionarily derived.
Origin of the Pentapetalae flower
Sometime after the origin of the flower, a novel floral ground plan was established in the most species-rich major clade of angiosperms—the Pentapetalae (as defined in Cantino et al. 2007) (see Figure 1). The changes to floral organization that occurred with the origin of Pentapetalae include concentric whorls of floral organs, with parts typically in fives or multiples thereof (pentamery), and morphologically distinct perianth organs (sepals and petals). The genetic basis for the origin of this canonical floral ground plan represents one of the major unresolved mysteries of flowering plant evolution. Independent studies have identified three evolutionary events that correspond closely with the origin of Pentapetalae, but their precise roles are unclear. First, two whole-genome duplication (WGD) events are believed to have occurred in close succession prior to the origin of the Pentapetalae, but their exact positions are uncertain (Jiao et al. 2012). Second, it is well documented that duplicate copies of most flower development genes, which may have originated through these WGDs, are maintained in the genomes of Pentapetalae species (e.g., Kramer and Irish 2000; Soltis and Soltis 2004; Kim et al. 2004; Zahn et al. 2005; Howarth and Donoghue 2006, 2009; Soltis et al. 2009a; Boyden et al. 2012; Pabón-Mora et al. 2014). Together, these genes pattern the development of morphological traits such as organ identity, symmetry, fusion, polarity, elongation, and growth, and thus have functions that could have contributed to the origin of a whorled pentamerous flower. Third, there appears to have been a shift from the fading borders model of floral developmental gene expression of basal angiosperms and basal eudicots to the canalized ABCE model in Pentapetalae (Chanderbali et al. 2009, 2010; Voelckel et al. 2010; Yoo et al. 2010b), but the precise phylogenetic location of this transition is uncertain.
Although flowers of Pentapetalae are considerably canalized (i.e., fixed in the arrangement, merosity, and morphology of its organs) compared to early-diverging lineages of angiosperms (Endress 1996, 2006; Soltis et al. 2002), they often exhibit extensive modifications to their floral organs. Although it is not clear whether such modifications originated directly via natural selection or as side effects of developmental changes (the spandrels of San Marco phenomenon; Gould and Lewontin 1979), the canalization of the Pentapetalae flower may have facilitated some such alterations through “synorganization”—a close association, fused or otherwise, among floral organs. Synorganization is hypothesized to have led to the evolution of novel morphologies and functions and is dependent on a whorled phyllotaxis with a fixed number of floral parts, as was established in Pentapetalae (Endress 1990, 1996, 2006). A notable example of synorganization is the fusion of petals (sympetaly) into the tubular corolla—recognized as a morphological innovation for centuries (de Jussieu 1789; Reichenbach 1827)—that characterizes the large asterid clade of Pentapetalae and has itself been further modified multiple times during asterid evolution (Stuurman et al. 2004; Wu et al. 2007).
The flowers of monocots are also very stable in number and arrangement of floral organs and are typically trimerous. Although monocots do not exhibit the extent of synorganization present in Pentapetalae, the orchids provide a parallel example of this phenomenon (Endress 2015). Studies of floral modifications offer a wealth of next-generation research possibilities that promise new insights into angiosperm floral innovations. Much progress has been made, for example, in understanding the genetic basis of sympetaly (Zhong and Preston 2015), petal differentiation (Huang and Irish 2015), and floral symmetry (Hileman 2014a,b). The available genetic data suggest that these innovations are regulated by genes that operate in parallel with, or downstream from, the ABCE organ identity program.
Origins of floral development genes
Phylogenomic analyses of the evolutionary history of functionally validated genetic regulators of flower development (232 Arabidopsis genes) suggest that ∼70% belong to orthogroups (sets of homologous genes representing narrowly defined gene lineages) that originated in nonflowering plants, ∼10% originated in the most recent common ancestor of angiosperms, and the remaining 20% evolved during angiosperm diversification (Amborella Genome Project 2013). Importantly, an ancient WGD event that occurred in the common ancestor of all angiosperms (Jiao et al. 2011) would have been the source of many new genes that contributed to the origin of the flower and other important angiosperm innovations (Buzgo et al. 2005; Zahn et al. 2005; De Bodt et al. 2005; Amborella Genome Project 2013). For example, many of the floral genes exist as paralogous gene lineages, likely due to this WGD event, in extant angiosperms. Gene Ontology (GO) annotations related to reproduction (flower development, reproductive developmental process, pollination, and similar terms) and several MADS-box gene lineages are overrepresented in this set of new genes (Amborella Genome Project 2013). They include the B, C, and E components of the ABCE program (i.e., AP3/PI, AG/STK, SEP1/SEP3). On the other hand, many novel gene lineages arose through multiple rounds of WGD during angiosperm diversification (Soltis et al. 2009b, 2010), and some have acquired new functions in specific floral organs within evolutionarily derived angiosperm lineages (Irish 2006; Soltis et al. 2006b; Zahn et al. 2006).
Thus, it seems reasonable to conclude that: (1) orthologs of most floral genes existed long before their specific roles in flowering were established; (2) novel gene lineages first appeared with the origin of the angiosperms and probably contributed to the origin of the flower; and (3) after a functional flower evolved, genetic innovations continued as new genes originated and/or were recruited into floral genetic programs.
Moving Forward
Next-generation tools for next-generation evo-devo studies
Advances in next-generation sequencing (NGS) have revolutionized much of plant evolutionary biology (e.g., Egan et al. 2012; Godden et al. 2012; Soltis et al. 2013), as well as biological research in general. It is now possible to obtain enormous amounts of genomic and transcriptomic sequence data for virtually any plant system that poses intriguing evolutionary questions and to do so at low cost. For example, a wealth of new data has been generated through projects such as the 1KP project (Matasci et al. 2014), the Floral Genome Project (Albert et al. 2005), and many clade-focused phylogenetic projects from the past decade. These technological advances provide unprecedented research opportunities to characterize and compare floral genetic programs to elucidate the genetic basis of novel floral ground plans. Toward this end, investigations of developmental gene regulatory networks (GRNs) that underlie floral diversity will be equally as valuable. Similar to GRN evolution in animal development (Levine and Davidson 2005; Peter and Davidson 2011), in plants, there is increasing evidence that: (1) most changes in floral morphology result from altered timing, location, and/or level(s) of GRN activity; and (2) similar GRNs are repeatedly coopted as similar adaptive traits are gained and lost (Specht and Howarth 2015). The GRNs and molecular mechanisms underlying the formation of Arabidopsis flowers have been studied in some detail, and technological advances such as translating ribosome affinity purification (TRAP) (Jiao and Meyerowitz 2010) promise even greater resolution in the future (Ó’Maoiléidigh et al. 2014). Improved knowledge of GRNs involved in floral organ identity, symmetry, cell type, floral color, and synorganization has become more attainable with technological advances in recent years, and further clarification of GRNs should be a goal in the study of flower developmental genetics.
Growing numbers of tools are now available to study gene function in nonmodel plants. Virus-induced gene silencing (VIGS) uses the plant’s innate defense response to invading viruses to silence specific genes inserted in modified viral genomes (Dinesh-Kumar et al. 2003). This technique overcomes the time-consuming steps of genetic transformation (specific genes can be silenced in a few weeks) and is limited primarily by the susceptibility of plants to infection by VIGS vectors (Senthil-Kumar and Mysore 2014). VIGS has been applied successfully in a diverse range of eudicot and monocot species (Becker and Lange 2010), including functional investigations of genes involved in floral organ identity and symmetry in basal eudicots and core eudicots (e.g., Gould and Kramer 2007; Sharma et al. 2011; Hidalgo et al. 2012; Gonçalves et al. 2013; Preston et al. 2014).
Another recently developed avenue of research is genome editing via the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas (CRISPR/associated) system (Sorek et al. 2013). Whereas VIGS reduces post-transcriptional gene activity, the CRISPR/Cas system facilitates genome editing by taking advantage of the Cas9 nuclease and a single guide RNA (sgRNA) to target a specific sequence in the genome. In addition to genome editing, which facilitates efficient and precise reverse genetics, genome engineering, and targeted transgene integration experiments, the CRISPR/Cas technology is suitable for studies of gene regulation through both transcriptional activation and repression. Moreover, the type III-B CRISPR/Cas system facilitates post-transcriptional silencing of gene expression in a manner that may be more specific than VIGS-mediated RNA interference (Bortesi and Fischer 2015). The CRISPR/Cas system can be delivered through Agrobacterium-mediated transformation of tissue culture preparations and may be applied in any angiosperm species amenable to these techniques. Thus far, CRISPR/Cas has been successfully applied in several eudicot and monocot species, including A. thaliana, Citrus sinensis, Nicotiana benthamiana, Oryza sativa, Solanum lycopersicum, Sorghum bicolor, Triticum aestivum, and Zea mays (Bortesi and Fischer 2015).
From model species to model clades
As next-generation techniques for assaying gene expression and function become more accessible, it may be increasingly feasible to apply these methods to multiple species within a clade of interest. Using a “model-clade” approach (i.e., not just a single species but a group of species within a defined clade targeted for gene expression, functional, and/or classical genetics studies), research questions can focus on related taxa that often encompass multiple trait gains, losses, or modifications (see Howarth and Dunn 2016). For instance, a model clade could include a single shift in a trait of interest as well as multiple independent iterations of similar shifts. Moreover, a model clade can include transitional forms of the trait of interest, facilitating investigations of the stepwise evolution of evolved gene function and phenotypic change. A model clade would therefore possess a number of features that would make it useful for floral evo-devo studies: (1) a well-supported species phylogeny; (2) high diversity of floral traits; and (3) multiple species, spread throughout the clade, that are easy to obtain and amenable to gene expression and/or functional techniques. These species need not be sister taxa, but are chosen to represent phenotypic variation across the clade. Studies of model clades therefore could rapidly provide clues about the genetic basis of evolutionary change that would not be achievable via the analysis of a single species from that clade. This model clade approach is well exemplified by evo-devo studies of orchid flower development, derived from comparisons of wild-type and peloric terata (Mondragón-Palomino 2013). Other examples include the evolution of zygomorphy in Zingiberales (Bartlett and Specht 2010, 2011), Dipsacales (Howarth et al. 2011) and Malpighiaceae (Zhang et al. 2010; Zhang et al. 2013a), petal evolution in Ranunculaceae (Zhang et al. 2013b) and Aizoaceae (Caryophyllales; Brockington et al. 2012, 2013), and the origins of petaloid bracts in Cornus (Cornaceae; Feng et al. 2012).
NGS technology can thus be applied to generate the data required to characterize floral GRNs; these GRNs, in turn, can be compared to identify candidate regulatory changes underlying floral evolutionary shifts. These “candidate genes” may then be functionally investigated in planta via CRISPER/Cas and related systems. Efforts to develop such integrative research programs are needed. Here, we advocate for avenues of research where these tools can be relatively easily developed and applied toward a more complete understanding of the genetic programs that underlie evolutionary changes in flowers.
Evolution of floral developmental genetic programs
Evolutionary genetic scenarios for the origin of the flower involve shifting gradients of gene expression/function in gymnosperm cones (out of male or out of female), reminiscent of the fading borders program of basal angiosperm flowers, or ectopic imposition of established female identity programs on male organs (mostly male) similar to the shifting boundary concept (see Theissen and Melzer 2007). Elucidation of the genetic programs of flowers that appear to represent intermediate steps in the transition from bisexual cone to flower, as seen in basal angiosperms and basal eudicots (Figure 2) is pivotal to understanding the origin and evolution of flowers, and floral diversification generally. For example, what is the evolutionary origin of the ancestral floral perianth? How are the spatial distribution and relative strengths of organ identity functions in basal angiosperm flowers determined? How are these determinants of organ identity functions manifested in monocots and basal eudicots vs. core eudicots such as Arabidopsis? To address such questions, new evolutionary model systems among the basal angiosperms and basal eudicots are necessary.
Among basal angiosperms, there are a few phylogenetically pivotal species with good potential to be tractable genetic models. Several members of Nymphaea (water lilies; Nymphaeaceae) have small genomes (e.g., Nymphaea thermarum, Nymphaea caerulea, and Nymphaea capensis; Pellicer et al. 2013) and are easily cultivated in aquaria or tubs. Collections of mutant water lilies could be maintained in greenhouse research facilities to facilitate forward genetic screens. N. thermarum has already been proposed as a potential model (Povilus et al. 2014), on account of its rapid life cycle (5–6 months), small size, an apparently selfing breeding system, and relatively small genome- its haploid genome size (1C value) is 0.51 pg (∼500 Mb), which is approximately three times the size of that of Arabidopsis. Likewise, a small genome (1C = 0.58), ease of culture, and ongoing genome sequencing as part of the Amborella Genome Project make N. caerulea another attractive target among the water lilies. Cabomba caroliniana has also been proposed as a possible genetic model for basal angiosperms (Vialette-Guiraud et al. 2011). It is also easily cultivated, but has a large genome with a 1C value of 3.55 pg (Pellicer et al. 2013), which is considerably larger than either of the above-mentioned species of Nymphaea. Importantly, Nymphaea species have been interbred by horticulturalists for >100 years to obtain hybrids with desirable floral characteristics (Les et al. 2004), indicating that this group could be used as a model clade in classical genetic studies.
Among magnoliids, Aristolochia fimbriata (pipevine; Aristolochiaceae) has been proposed as a potential experimental system and has numerous features that facilitate genetic studies (Bliss et al. 2013). Whereas most magnoliids are woody, this species is herbaceous, easily cultured with a rapid life cycle (3 months), transformable, and can be regenerated via tissue culture. Notably, flowers of Aristolochia exhibit synorganization in the perianth and are zygomorphic (bilaterally symmetrical), representing a derived floral state among the basal angiosperms (Soltis et al. 2005; Judd et al. 2007). The genome size is also small (1C = 0.45; Bliss et al. 2013), but no genome sequencing is underway. Persea americana (the avocado; Lauraceae) is also a potential candidate among magnoliids. This commercially valuable crop species is amenable to genetic transformation, tissue culture, in vitro mutagenesis, and related technologies such as cryopreservation, as well as in vitro and ex vitro micrografting to circumvent the long juvenile period (reviewed in Chanderbali et al. 2008). A draft nuclear genome sequence (1C = 0.92; Arumuganathan and Earle 1991) is also close at hand (L. Herrera-Estrella, V. A. Albert, A. Herrera-Estrella, M. Rendon, and E. Ibarra-Laclette, personal communication). As tissue culture protocols are already in place for both of these magnoliid species, it should be possible to conduct CRISPR/Cas-mediated genetic manipulation of the ABCE genes and other candidates that emerge from RNA-sequencing (RNA-Seq) and GRN analyses.
Among basal eudicots, genome sequences are currently available for members of Ranunculales (Aquilegia coerulea; Kramer 2009) and Proteales (Nelumbo nucifera; Ming et al. 2013), but similar efforts are needed for representatives of other basal eudicot lineages. Exemplars from economically important groups (e.g., poppies, Papaveraceae; plane tree, Platanaceae; boxwood, Buxaceae) could be targeted for complete genome sequencing. A genome sequence for a basal core eudicot would also be an important evolutionary reference, with Gunnera (Gunneraceae), the sister lineage of Pentapetalae, as a logical candidate. Indeed, efforts are underway to obtain a genome sequence for Gunnera manicata (L. Fay-Wei, personal communication). All of these resources would be invaluable to investigations of the evolutionary transition from basal eudicots to Pentapetalae. Promising target species for CRISPR/Cas-mediated genome editing and VIGS are members of Ranunculales, in which the application of reverse genetics is relatively well advanced, as in Aquilegia (Gould and Kramer 2007; Rasmussen et al. 2009; Kramer and Hodges 2010; Galimba and Di Stilio 2015) and Eschscholzia (Tekleyohans et al. 2013).
Summary: New Evo-Devo Approaches for Understanding Flower Developmental Genetics
Studies of genetic model plants, primarily Arabidopsis, have identified a limited number of transcription factors with crucial roles in floral development and led to the formulation of the ABCE model. This elegantly simple model appears to have its evolutionary roots in a gymnosperm “BC” system that has been modified and elaborated during flowering plant evolution. At the inception of the ancestral floral development program, specification of floral organ identity was likely deployed through a fading borders program, which still appears to specify the morphologically intergrading floral organs of basal angiosperms such as Amborella and water lilies. Sharpened borders of organ identity functions likely underlie the origin of the canalized Pentapetalae flower, as seen in Arabidopsis. Moreover, shifting borders of petal identity functions appear to promote the development of identical petaloid organs in the perianth of some flowers (e.g., tulips). Imposed on these fundamental changes in floral form, still further modifications are evident in individual lineages of flowering plants, including multiple origins of bilateral symmetry and synorganization with its attendant novelties.
Comparative studies of orthologs and/or homologs of known floral regulators across angiosperms often suggest conserved roles in specific floral traits; however, they also highlight ample opportunities for neo- or subfunctionalization of duplicated genes as a consequence of multiple WGDs during the diversification of angiosperms. These findings underscore the likelihood that the regulation of flower development in distantly related angiosperms might involve genes that are not orthologous to known candidate genes, and/or the regulatory networks may be substantially different. To advance from a comparative approach based on candidate genes to a more mechanistic account of floral diversity, the establishment of collections of mutant phenotypes in phylogenetically relevant nonmodel plant species would be especially valuable. Although these resources are currently not available, forward genetic approaches and/or high-throughput transcriptome sequencing combined with reverse genetic screening may increasingly be feasible in select basal angiosperm and basal eudicot species. The development of these systems will herald a new generation of multidisciplinary evo-devo research during which many new plant systems can be the focus of study—species that afford the opportunity to address questions of floral evolution and organization that cannot be addressed with the current set of model systems. The development of these approaches would rapidly elucidate evolutionary changes in the regulatory networks underlying floral development.
Acknowledgments
We thank two anonymous reviewers and Jim Doyle for insightful comments on an earlier version of this manuscript and Walter Judd and Deborah Chanderbali for photographs used in Figure 3. We also thank Michael Turelli for the invitation to contribute this article and Adam Wilkins for excellent editorial suggestions. This work was supported by National Science Foundation grants IOS-1121301, IOS-0922742, DEB-1455601, and DEB-1457440.
Footnotes
Communicating editor: A. S. Wilkins
Literature Cited
- Albert, V. A., M. H. G. Gustafsson, and L. D. Laurenzio, 1998 Ontogenetic systematics, molecular developmental genetics, and the angiosperm petal, pp. 349–374 in Molecular Systematics of Plants II, edited by D. E. Soltis, P. S. Soltis, and J. J. Doyle. Springer-Verlag, Vienna. [Google Scholar]
- Albert, V. A., D. E. Soltis, J. E. Carlson, W. G. Farmerie, P. K. Wall et al., 2005 Floral gene resources from basal angiosperms for comparative genomics research. BMC Plant Biol. 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amborella Genome Project , 2013. The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Ambrose B. A., Lerner D. R., Ciceri P., Padilla C. M., Yanofsky M. F., et al. , 2000. Molecular and genetic analyses of the Silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5: 569–579. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K., Earle E. D., 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208–218. [Google Scholar]
- Bartlett M. E., Specht C. D., 2010. Evidence for the involvement of Globosa-like gene duplications and expression divergence in the evolution of floral morphology in the Zingiberales. New Phytol. 187: 521–541. [DOI] [PubMed] [Google Scholar]
- Bartlett M. E., Specht C. D., 2011. Changes in expression pattern of the teosinte branched1-like genes in the Zingiberales provide a mechanism for evolutionary shifts in symmetry across the order. Am. J. Bot. 98: 227–243. [DOI] [PubMed] [Google Scholar]
- Baum, D. A., and L. C. Hileman, 2006 A developmental genetic model for the origin of the flower, pp. 1–27 in Annual Plant Reviews 20: Flowering and Its Manipulation, edited by C. Ainsworth. Blackwell Scientific, Oxford. [Google Scholar]
- Becker A., Theissen G., 2003. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29: 464–489. [DOI] [PubMed] [Google Scholar]
- Becker A., Lange M., 2010. VIGS: genomics goes functional. Trends Plant Sci. 15: 1–4. [DOI] [PubMed] [Google Scholar]
- Bliss B. J., Wanke S., Barakat A., Ayyampalayam S., Wickett N., et al. , 2013. Characterization of the basal angiosperm Aristolochia fimbriata: a potential experimental system for genetic studies. BMC Plant Biol. 13: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortesi L., Fischer R., 2015. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33: 41–52. [DOI] [PubMed] [Google Scholar]
- Bowman J. L., 1997. Evolutionary conservation of angiosperm flower development at the molecular and genetic levels. J. Biosci. 22: 515–527. [Google Scholar]
- Bowman J. L., Smyth D. R., Meyerowitz E. M., 1989. Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden G. S., Donoghue M. J., Howarth D. G., 2012. Duplications and expression of RADIALIS-like genes in Dipsacales. Int. J. Plant Sci. 173: 971–983. [Google Scholar]
- Brockington S. F., Rudall P. J., Frohlich M. W., Oppenheimer D. G., Soltis P. S., et al. , 2012. “Living stones” reveal alternative petal identity programs within the core eudicots. Plant J. Cell. Mol. Biol. 69: 193–203. [DOI] [PubMed] [Google Scholar]
- Brockington S., Santos P. D., Glover B., Craene L. R. D., 2013. Androecial evolution in Caryophyllales in light of a paraphyletic Molluginaceae. Am. J. Bot. 100: 1757–1778. [DOI] [PubMed] [Google Scholar]
- Broholm S. K., Tähtiharju S., Laitinen R. A. E., Albert V. A., Teeri T. H., et al. , 2008. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. USA 105: 9117–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzgo M., Soltis P. S., Soltis D. E., 2004. Floral developmental morphology of Amborella trichopoda (Amborellaceae). Int. J. Plant Sci. 165: 925–947. [Google Scholar]
- Buzgo M., Soltis P., Kim S., Soltis D. E., 2005. The making of the flower. Biologist 52: 149–154. [Google Scholar]
- Cantino P. D., Doyle J. A., Graham S. W., Judd W. S., Olmstead R. G., et al. , 2007. Towards a phylogenetic nomenclature of Tracheophyta. Taxon 56: 1E–44E. [Google Scholar]
- Chanderbali A., Kim S., Buzgo M., Zheng Z., Oppenheimer D., et al. , 2006. Genetic footprints of stamen ancestors guide perianth evolution in Persea (Lauraceae). Int. J. Plant Sci. 167: 1075–1089. [Google Scholar]
- Chanderbali A. S., Albert V. A., Ashworth V. E. T. M., Clegg M. T., Litz R. E., et al. , 2008. Persea americana (avocado): bringing ancient flowers to fruit in the genomics era. BioEssays 30: 386–396. [DOI] [PubMed] [Google Scholar]
- Chanderbali A. S., Albert V. A., Leebens-Mack J., Altman N. S., Soltis D. E., et al. , 2009. Transcriptional signatures of ancient floral developmental genetics in avocado (Persea americana; Lauraceae). Proc. Natl. Acad. Sci. USA 106: 8929–8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanderbali A. S., Yoo M.-J., Zahn L. M., Brockington S. F., Wall P. K., et al. , 2010. Conservation and canalization of gene expression during angiosperm diversification accompany the origin and evolution of the flower. Proc. Natl. Acad. Sci. USA 107: 22570–22575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E. S., Meyerowitz E. M., 1991. The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37. [DOI] [PubMed] [Google Scholar]
- Colombo L., Franken J., Koetje E., van Went J., Dons H. J., et al. , 1995. The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7: 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S., Maere S., Van de Peer Y., 2005. Genome duplication and the origin of angiosperms. Trends Ecol. Evol. 20: 591–597. [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar S. P., Anandalakshmi R., Marathe R., Schiff M., Liu Y., 2003. Virus-induced gene silencing. Methods Mol. Biol. 236: 287–294. [DOI] [PubMed] [Google Scholar]
- Drinnan A. N., Crane P. R., Hoot S. B., 1994. Patterns of floral evolution in the early diversification of non-magnoliid dicotyledons (eudicots), pp. 93–122 in Plant Systematics and Evolution Supplement 8: Early Evolution of Flowers, edited by Endress P. K., Friis E. M. Springer-Verlag, Vienna. [Google Scholar]
- Egan A. N., Schlueter J., Spooner D. M., 2012. Applications of next-generation sequencing in plant biology. Am. J. Bot. 99: 175–185. [DOI] [PubMed] [Google Scholar]
- Endress P. K., 1990. Patterns of floral construction in ontogeny and phylogeny. Biol. J. Linn. Soc. Lond. 39: 153–175. [Google Scholar]
- Endress P. K., 1996. Diversity and Evolutionary Biology of Tropical Flowers. Cambridge University Press, Cambridge. [Google Scholar]
- Endress P. K., 2006. Angiosperm floral evolution: morphological developmental framework, pp. 1–61 in Advances in Botanical Research 44: Developmental Genetics of the Flower, edited by Soltis D. E., Leebens-Mack J. H., Soltis P. S. Elsevier, San Diego. [Google Scholar]
- Endress P. K., 2015. Development and evolution of extreme synorganization in angiosperm flowers and diversity: a comparison of Apocynaceae and Orchidaceae. Ann. Bot. (Lond.) DOI: 10.1093/aob/mcv119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C.-M., Liu X., Yu Y., Xie D., Franks R. G., et al. , 2012. Evolution of bract development and B-class MADS box gene expression in petaloid bracts of Cornus s. l. (Cornaceae). New Phytol. 196: 631–643. [DOI] [PubMed] [Google Scholar]
- Frohlich M. W., 2003. An evolutionary scenario for the origin of flowers. Nat. Rev. Genet. 4: 559–566. [DOI] [PubMed] [Google Scholar]
- Galimba K. D., Di Stilio V. S., 2015. Sub-functionalization to ovule development following duplication of a floral organ identity gene. Dev. Biol. 405: 158–172. [DOI] [PubMed] [Google Scholar]
- Glover B. J., Airoldi C. A., Brockington S. F., Fernández-Mazuecos M., Martínez-Pérez C., et al. , 2015. How have advances in comparative floral development influenced our understanding of floral evolution? Int. J. Plant Sci. 176: 307–323. [Google Scholar]
- Godden G. T., Jordon-Thaden I. E., Chamala S., Crowl A. A., García N., et al. , 2012. Making next-generation sequencing work for you: approaches and practical considerations for marker development and phylogenetics. Plant Ecol. Divers. 5: 427–450. [Google Scholar]
- Gonçalves B., Nougué O., Jabbour F., Ridel C., Morin H., et al. , 2013. An APETALA3 homolog controls both petal identity and floral meristem patterning in Nigella damascena L. (Ranunculaceae). Plant J. Cell. Mol. Biol. 76: 223–235. [DOI] [PubMed] [Google Scholar]
- Gould S. J., Lewontin R. C., 1979. The Spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proc. R. Soc. Lond. B Biol. Sci. 205: 581–598. [DOI] [PubMed] [Google Scholar]
- Gould B., Kramer E. M., 2007. Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia (columbine, Ranunculaceae). Plant Methods 3: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo O., Bartholmes C., Gleissberg S., 2012. Virus-induced gene silencing (VIGS) in Cysticapnos vesicaria, a zygomorphic-flowered Papaveraceae (Ranunculales, basal eudicots). Ann. Bot. (Lond.) 109: 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman L. C., 2014a Bilateral flower symmetry—How, when and why? Curr. Opin. Plant Biol. 17: 146–152. [DOI] [PubMed] [Google Scholar]
- Hileman L. C., 2014b Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20130348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma T., Goto K., 2001. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529. [DOI] [PubMed] [Google Scholar]
- Howarth, D., and M. P. Dunn, 2016 A phylogenetic approach to studying developmental evolution, Encyclopedia of Evolutionary Biology, edited by K. E. Sears, and R. Kliman. Elsevier, Amsterdam, in press. [Google Scholar]
- Howarth D. G., Donoghue M. J., 2006. Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. USA 103: 9101–9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth D. G., Donoghue M. J., 2009. Duplications and expression of DIVARICATA-like genes in Dipsacales. Mol. Biol. Evol. 26: 1245–1258. [DOI] [PubMed] [Google Scholar]
- Howarth D. G., Martins T., Chimney E., Donoghue M. J., 2011. Diversification of CYCLOIDEA expression in the evolution of bilateral flower symmetry in Caprifoliaceae and Lonicera (Dipsacales). Ann. Bot. (Lond.) 107: 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Irish V. F., 2015. Gene networks controlling petal organogenesis. J. Exp. Bot. 67: 61–68. [DOI] [PubMed] [Google Scholar]
- Irish V. F., 2006. Duplication, diversification, and comparative genetics of angiosperm MADS‐Box genes, pp. 129–161 in Advances in Botanical Research 44: Developmental Genetics of the Flower, edited by Soltis D. E., Leebens-Mack J. H., Soltis P. S. Elsevier, San Diego. [Google Scholar]
- Irish V. F., Sussex I. M., 1990. Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2: 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Meyerowitz E. M., 2010. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 6: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wickett N. J., Ayyampalayam S., Chanderbali A. S., Landherr L., et al. , 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Leebens-Mack J., Ayyampalayam S., Bowers J. E., McKain M. R., et al. , 2012. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 13: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku K. D., den Boer B. G., Montagu M. V., Okamuro J. K., 1994. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd W. S., Campbell E., Kellogg E., Stevens P., 2007. Plant Systematics: A Phylogenetic Approach. Sinauer Associates, Sunderland, MA. [Google Scholar]
- de Jussieu A. L., 1789. Antonii Laurentii de Jussieu Genera plantarum :secundum ordines naturales disposita, juxta methodum in Horto regio parisiensi exaratam. Herissant et Theophilum Barrois, Paris. [Google Scholar]
- Kanno A., Saeki H., Kameya T., Saedler H., Theissen G., 2003. Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol. Biol. 52: 831–841. [DOI] [PubMed] [Google Scholar]
- Kim S., Yoo M.-J., Albert V. A., Farris J. S., Soltis P. S., et al. , 2004. Phylogeny and diversification of B-function MADS-box genes in angiosperms: evolutionary and functional implications of a 260-million-year-old duplication. Am. J. Bot. 91: 2102–2118. [DOI] [PubMed] [Google Scholar]
- Kim S., Koh J., Yoo M.-J., Kong H., Hu Y., et al. , 2005. Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. Plant J. Cell. Mol. Biol. 43: 724–744. [DOI] [PubMed] [Google Scholar]
- Kramer E. M., 2009. Aquilegia: a new model for plant development, ecology, and evolution. Annu. Rev. Plant Biol. 60: 261–277. [DOI] [PubMed] [Google Scholar]
- Kramer E. M., Irish V. F., 2000. Evolution of the petal and stamen developmental programs: evidence from comparative studies of the lower eudicots and basal angiosperms. Int. J. Plant Sci. 161: S29–S40. [Google Scholar]
- Kramer E. M., Hodges S. A., 2010. Aquilegia as a model system for the evolution and ecology of petals. Philos. Trans. R. Soc. B Biol. Sci. 365: 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E. M., Di Stilio V. S., Schluter P. M., 2003. Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. Int. J. Plant Sci. 164: 1–11. [Google Scholar]
- Kramer E. M., Holappa L., Gould B., Jaramillo M. A., Setnikov D., et al. , 2007. Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. Plant Cell 19: 750–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Les D. H., Moody M. L., Doran A. S., Phillips W. E., 2004. A genetically confirmed intersubgeneric hybrid in Nymphaea L. (Nymphaeaceae Salisb.). HortScience 39: 219–222. [Google Scholar]
- Levine M., Davidson E. H., 2005. Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 102: 4936–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., 1994. The unfolding drama of flower development: recent results from genetic and molecular analyses. Genes Dev. 8: 745–756. [DOI] [PubMed] [Google Scholar]
- Ma H., 2005. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56: 393–434. [DOI] [PubMed] [Google Scholar]
- Ma H., dePamphilis C., 2000. The ABCs of floral evolution. Cell 101: 5–8. [DOI] [PubMed] [Google Scholar]
- Magallón S., Gómez-Acevedo S., Sánchez-Reyes L. L., Hernández-Hernández T., 2015. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 207: 437–453. [DOI] [PubMed] [Google Scholar]
- Matasci N., Hung L.-H., Yan Z., Carpenter E. J., Wickett N. J., et al. , 2014. Data access for the 1,000 Plants (1KP) project. Gigascience 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M., Ambrose B. A., Meeley R. B., Briggs S. P., Yanofsky M. F., et al. , 1996. Diversification of C-function activity in maize flower development. Science 274: 1537–1540. [DOI] [PubMed] [Google Scholar]
- Ming R., VanBuren R., Liu Y., Yang M., Han Y., et al. , 2013. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 14: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón-Palomino M., 2013. Perspectives on MADS-box expression during orchid flower evolution and development. Front. Plant Sci. 4: 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ó’Maoiléidigh , D. S., E. Graciet, and F. Wellmer, 2014. Gene networks controlling Arabidopsis thaliana flower development. New Phytol. 201: 16–30. [DOI] [PubMed] [Google Scholar]
- Pabón-Mora N., Wong G. K.-S., Ambrose B. A., 2014. Evolution of fruit development genes in flowering plants. Front. Plant Sci. 5: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta G. S., Baumann E., Wisman E., Yanofsky M. F., 2000. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203. [DOI] [PubMed] [Google Scholar]
- Pellicer J., Kelly L. J., Magdalena C., Leitch I. J., 2013. Insights into the dynamics of genome size and chromosome evolution in the early diverging angiosperm lineage Nymphaeales (water lilies). Genome 56: 437–449. [DOI] [PubMed] [Google Scholar]
- Peter I. S., Davidson E. H., 2011. Evolution of gene regulatory networks controlling body plan development. Cell 144: 970–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povilus R. A., Losada J. M., Friedman W. E., 2014. Floral biology and ovule and seed ontogeny of Nymphaea thermarum, a water lily at the brink of extinction with potential as a model system for basal angiosperms. Ann. Bot. (Lond.) 115: 211–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J. C., Barnett L. L., Kost M. A., Oborny N. J., Hileman L. C., 2014. Optimization of virus-induced gene silencing to facilitate evo-devo studies in the emerging model species Mimulus guttatus (Phrymaceae). Ann. Mo. Bot. Gard. 99: 301–312. [Google Scholar]
- Rasmussen D. A., Kramer E. M., Zimmer E. A., 2009. One size fits all? Molecular evidence for a commonly inherited petal identity program in Ranunculales. Am. J. Bot. 96: 96–109. [DOI] [PubMed] [Google Scholar]
- Reichenbach, H. G. L., 1827 Dr. Joh. Christ. Mössler’s Handbuch der Gewächskunde. J. F. Hammerich, Altona, Germany. [Google Scholar]
- Ronse de Craene L. P., Brockington S. F., 2013. Origin and evolution of petals in angiosperms. Plant Ecol. Evol. 146: 5–25. [Google Scholar]
- Schwarz-Sommer Z., Huijser P., Nacken W., Saedler H., Sommer H., 1990. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250: 931–936. [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M., Mysore K. S., 2014. Tobacco rattle virus–based virus-induced gene silencing in Nicotiana benthamiana. Nat. Protoc. 9: 1549–1562. [DOI] [PubMed] [Google Scholar]
- Shan H., Su K., Lu W., Kong H., Chen Z., et al. , 2006. Conservation and divergence of candidate class B genes in Akebia trifoliata (Lardizabalaceae). Dev. Genes Evol. 216: 785–795. [DOI] [PubMed] [Google Scholar]
- Sharma B., Guo C., Kong H., Kramer E. M., 2011. Petal-specific subfunctionalization of an APETALA3 paralog in the Ranunculales and its implications for petal evolution. New Phytol. 191: 870–883. [DOI] [PubMed] [Google Scholar]
- Soltis D. E., Soltis P. S., Albert V. A., Oppenheimer D. G., dePamphilis C. W., et al. , 2002. Missing links: the genetic architecture of flower and floral diversification. Trends Plant Sci. 7: 22–31. [DOI] [PubMed] [Google Scholar]
- Soltis D. E., Soltis P. S., 2004. Amborella not a “basal angiosperm”? Not so fast. Am. J. Bot. 91: 997–1001. [DOI] [PubMed] [Google Scholar]
- Soltis D. E., Soltis P. S., Endress P. K., Chase M. W., 2005. Phylogeny and Evolution of Angiosperms. Sinauer Associates, Sunderland, MA. [Google Scholar]
- Soltis D. E., Leebens-Mack J. H., Soltis P. S. (Editors), 2006a Advances in Botanical Research 44: Developmental Genetics of the Flower. Elsevier, San Diego. [Google Scholar]
- Soltis P. S., Soltis D. E., Kim S., Chanderbali A., Buzgo M., 2006b Expression of floral regulators in basal angiosperms and the origin and evolution of ABC‐function, pp. 483–506 in Advances in Botanical Research 44: Developmental Genetics of the Flower, edited by Soltis D. E., Leebens-Mack J. H., Soltis P. S. Elsevier, San Diego. [Google Scholar]
- Soltis D. E., Ma H., Frohlich M. W., Soltis P. S., Albert V. A., et al. , 2007. The floral genome: an evolutionary history of gene duplication and shifting patterns of gene expression. Trends Plant Sci. 12: 358–367. [DOI] [PubMed] [Google Scholar]
- Soltis P. S., Brockington S. F., Yoo M.-J., Piedrahita A., Latvis M., et al. , 2009a Floral variation and floral genetics in basal angiosperms. Am. J. Bot. 96: 110–128. [DOI] [PubMed] [Google Scholar]
- Soltis D. E., Albert V. A., Leebens-Mack J., Bell C. D., Paterson A. H., et al. , 2009b Polyploidy and angiosperm diversification. Am. J. Bot. 96: 336–348. [DOI] [PubMed] [Google Scholar]
- Soltis P. S., Burleigh J. G., Chanderbali A. S., Yoo M.-J., Soltis D. E., 2010. Gene and genome duplications in plants, pp. 269–298 in Evolution after Gene Duplication, edited by Dittmar K., Liberles D. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- Soltis D. E., Smith S. A., Cellinese N., Wurdack K. J., Tank D. C., et al. , 2011. Angiosperm phylogeny: 17 genes, 640 taxa. Am. J. Bot. 98: 704–730. [DOI] [PubMed] [Google Scholar]
- Soltis D. E., Gitzendanner M. A., Stull G., Chester M., Chanderbali A., et al. , 2013. The potential of genomics in plant systematics. Taxon 62: 886–898. [Google Scholar]
- Sorek R., Lawrence C. M., Wiedenheft B., 2013. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 82: 237–266. [DOI] [PubMed] [Google Scholar]
- Specht C. D., Bartlett M. E., 2009. Flower evolution: the origin and subsequent diversification of the angiosperm flower. Annu. Rev. Ecol. Evol. Syst. 40: 217–243. [Google Scholar]
- Specht C. D., Howarth D. G., 2015. Adaptation in flower form: a comparative evodevo approach. New Phytol. 206: 74–90. [DOI] [PubMed] [Google Scholar]
- Stuurman J., Hoballah M. E., Broger L., Moore J., Basten C., et al. , 2004. Dissection of floral pollination syndromes in Petunia. Genetics 168: 1585–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekleyohans D. G., Lange S., Becker A., 2013. Virus-induced gene silencing of the alkaloid-producing basal eudicot model plant Eschscholzia californica (California Poppy). Methods Mol. Biol. 975: 83–98. [DOI] [PubMed] [Google Scholar]
- Theissen G., Becker A., 2004. Gymnosperm orthologues of class B floral homeotic genes and their impact on understanding flower origin. Crit. Rev. Plant Sci. 23: 129–148. [Google Scholar]
- Theissen G., Melzer R., 2007. Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann. Bot. (Lond.) 100: 603–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tunen, A. J., W. Eikelboom, and G. C. Angenent, 1993 Floral organogenesis in Tulipa. Flower. News. Lett. 16: 33–38.
- Vialette-Guiraud A. C. M., Alaux M., Legeai F., Finet C., Chambrier P., et al. , 2011. Cabomba as a model for studies of early angiosperm evolution. Ann. Bot. (Lond.) 108: 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelckel C., Borevitz J. O., Kramer E. M., Hodges S. A., 2010. Within and between whorls: comparative transcriptional profiling of Aquilegia and Arabidopsis. PLoS One 5: e9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K.-U., Becker A., Münster T., Kim J. T., Saedler H., et al. , 1999. MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proc. Natl. Acad. Sci. USA 96: 7342–7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. A., Lowry D. B., Cooley A. M., Wright K. M., Lee Y. W., et al. , 2007. Mimulus is an emerging model system for the integration of ecological and genomic studies. Heredity 100: 220–230. [DOI] [PubMed] [Google Scholar]
- Yoo M.-J., Soltis P. S., Soltis D. E., 2010a Expression of floral MADS‐Box genes in two divergent water lilies: Nymphaeales and Nelumbo. Int. J. Plant Sci. 171: 121–146. [Google Scholar]
- Yoo M.-J., Chanderbali A. S., Altman N. S., Soltis P. S., Soltis D. E., 2010b Evolutionary trends in the floral transcriptome: insights from one of the basalmost angiosperms, the water lily Nuphar advena (Nymphaeaceae). Plant J. 64: 687–698. [DOI] [PubMed] [Google Scholar]
- Zahn L. M., Kong H., Leebens-Mack J. H., Kim S., Soltis P. S., et al. , 2005. The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169: 2209–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn L. M., Feng B., Ma H., 2006. Beyond the ABC‐model: regulation of floral homeotic genes, pp. 163–207 in Advances in Botanical Research 44: Developmental Genetics of the Flower, edited by Soltis D. E., Leebens-Mack J. H., Soltis P. S. Elsevier, San Diego. [Google Scholar]
- Zhang W., Kramer E. M., Davis C. C., 2010. Floral symmetry genes and the origin and maintenance of zygomorphy in a plant-pollinator mutualism. Proc. Natl. Acad. Sci. USA 107: 6388–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Steinmann V. W., Nikolov L., Kramer E. M., Davis C. C., 2013a Divergent genetic mechanisms underlie reversals to radial floral symmetry from diverse zygomorphic flowered ancestors. Front. Plant Sci. 4: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Guo C., Zhang W., Wang P., Li L., et al. , 2013b Disruption of the petal identity gene APETALA3–3 is highly correlated with loss of petals within the buttercup family (Ranunculaceae). Proc. Natl. Acad. Sci. USA 110: 5074–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Preston J. C., 2015. Bridging the gaps: evolution and development of perianth fusion. New Phytol. 208: 330–335. [DOI] [PubMed] [Google Scholar]