Abstract
Gene regulation in response to intracellular calcium is mediated by the calcineurin-activated transcription factor Prz1 in the fission yeast Schizosaccharomyces pombe. Genome-wide studies of the Crz1 and CrzA fungal orthologs have uncovered numerous target genes involved in conserved and species-specific cellular processes. In contrast, very few target genes of Prz1 have been published. This article identifies an extensive list of genes using transcriptome and ChIP-chip analyses under inducing conditions of Prz1, including CaCl2 and tunicamycin treatment, as well as a ∆pmr1 genetic background. We identified 165 upregulated putative target genes of Prz1 in which the majority contained a calcium-dependent response element in their promoters, similar to that of the Saccharomyces cerevisiae ortholog Crz1. These genes were functionally enriched for Crz1-conserved processes such as cell-wall biosynthesis. Overexpression of prz1+ increased resistance to the cell-wall degradation enzyme zymolyase, likely from upregulation of the O-mannosyltransferase encoding gene omh1+. Loss of omh1+ abrogates this phenotype. We uncovered a novel inhibitory role in flocculation for Prz1. Loss of prz1+ resulted in constitutive flocculation and upregulation of genes encoding the flocculins Gsf2 and Pfl3, as well as the transcription factor Cbf12. The constitutive flocculation of the ∆prz1 strain was abrogated by the loss of gsf2+ or cbf12+. This study reveals that Prz1 functions as a positive and negative transcriptional regulator of genes involved in cell-wall biosynthesis and flocculation, respectively. Moreover, comparison of target genes between Crz1/CrzA and Prz1 indicate some conservation in DNA-binding specificity, but also substantial rewiring of the calcineurin-mediated transcriptional regulatory network.
Keywords: fission yeast, calcium, transcription factor, cell wall, flocculation
CALCINEURIN is a highly conserved phosphatase central to Ca2+ signaling. In metazoans, calcineurin regulates a wide array of Ca2+-dependent processes including T-cell activation (Clipstone and Crabtree 1992), cardiac hypertrophy (Molkentin et al. 1998), neutrophil motility (Hendey et al. 1992), apoptosis (Wang et al. 1999), angiogenesis (Graef et al. 2001), and memory development (Mansuy et al. 1998). One of the primary effectors of calcineurin is the NFAT family of transcription factors, which translocates into the nucleus to regulate target genes when dephosphorylated (reviewed in Macian 2005). In fungi, the activity of the Crz1 C2H2 zinc-finger transcription factor is modulated by calcineurin in a similar way (reviewed in Thewes 2014). Crz1 orthologs have been identified in various fungal species and their function appears conserved in cell-wall-related processes and resistance to external stressors (Thewes 2014).
The Saccharomyces cerevisiae Crz1 is localized in the cytosol under optimal growth conditions, but is activated and rapidly translocated into the nucleus through dephosphorylation by calcineurin in response to exogenous Ca2+ (Stathopoulos-Gerontides et al. 1999). In addition to exogenous Ca2+, Crz1 is activated by numerous external stresses including high salt, prolonged exposure to α-factor, alkaline pH, antifungal compounds, blue light, nutrient deprivation, heavy metals, and ethanol (Matheos et al. 1997; Stathopoulos and Cyert 1997; Edlind et al. 2002; Serrano et al. 2002; Zakrzewska et al. 2005; Zhang and Rao 2007; Ruiz et al. 2008; Araki et al. 2009; Ferreira et al. 2012; Bodvard et al. 2013). Transcriptome profiling of CRZ1 was initially performed with Ca2+ or Na+ treatment that identified 163 target genes (Yoshimoto et al. 2002), and this list has been expanded subsequently with similar profiling under alkaline stress, nutrient deprivation, and transcription factor overexpression (Viladevall et al. 2004; Chua et al. 2006; Ruiz et al. 2008). The Crz1 target genes are known to function in ion homeostasis, small-molecule transport, cell-wall maintenance, lipid and sterol metabolism, and vesicle transport. Many of these target genes contain the calcineurin-dependent response element (CDRE) motif [5′-GNGGC(G/T)CA-3′] in their promoter (Yoshimoto et al. 2002). Crz1 binds this motif, which was originally discovered in the promoter of FKS2, and is sufficient to drive the transcriptional activation of a reporter gene (Stathopoulos and Cyert 1997; Yoshimoto et al. 2002).
In Schizosaccharomyces pombe, the Ppb1 calcineurin catalytic subunit dephosphorylates the transcription factor Prz1 in response to elevated Ca2+ levels (Hirayama et al. 2003). Similar to other Crz1 orthologs, dephosphorylation of Prz1 causes nuclear translocation and transcriptional regulation of its target genes through binding of a CDRE-like motif (5′-AGCCTC-3′) (Deng et al. 2006) or a Ca2+-dependent response element (5′-CAACT-3′) (Hamasaki-Katagiri and Ames 2010). Loss of prz1+ produces a normal phenotype under optimal growth conditions, but results in hypersensitivity to Ca2+ and reduced mating efficiency (Hirayama et al. 2003; Sun et al. 2013). In contrast, the calcineurin ∆ppb1 strain exhibits a more severe phenotype with additional defects in cytokinesis, cell polarity, and chloride hypersensitivity (Yoshida et al. 1994; Hirayama et al. 2003). These defects are not suppressed by prz1+overexpression, indicating that Prz1 is not the sole target of calcineurin (Hirayama et al. 2003). In addition to Ca2+, activation of Prz1 occurs upon exposure to NaCl, DTT and tunicamycin (ER stressors), micafungin (a β-glucanase inhibitor), and heat shock, when assayed by a CDRE-regulated reporter, nuclear translocation, or prz1+ messenger RNA levels (Hirayama et al. 2003; Deng et al. 2006). In addition, prz1+ overexpression activates the CDRE-regulated reporter, thus indicating positive autoregulation (Koike et al. 2012). The response to diverse external stimuli indicates that Prz1 must regulate genes involved in multiple cellular processes as observed in other fungal orthologs. However, the current target gene list of Prz1 is far from complete. Only five target genes have been identified: pmc1+ (Hirayama et al. 2003), pmr1+ (Maeda et al. 2004), ncs1+ (Hamasaki-Katagiri and Ames 2010), cmk1+ (Cisneros-Barroso et al. 2014), and prz1+ (Deng et al. 2006). This is in contrast to other fungal Crz1 orthologs that have more extensive target gene lists, ranging from dozens up to several hundred genes, functioning in cellular processes such as cell-wall biosynthesis, ion transport, lipid metabolism, and vesicle transport (Yoshimoto et al. 2002; Karababa et al. 2006; Hagiwara et al. 2008; Chen et al. 2012; Soriani et al. 2008).
Here, we substantially expand the number of putative target genes for Prz1 by transcriptome and ChIP-chip profiling and uncover novel biological roles in reproduction, cell-wall structure, and flocculation. We discovered that the DNA-binding specificity of the calcineurin-responsive transcription factors is conserved between budding and fission yeasts, but considerable differences exist among orthologous target genes. The role in cell-wall biosynthesis is conserved between Prz1 and its orthologs, and several putative target genes for this role were identified. Finally, we show that Prz1 directly represses target genes implicated in flocculation.
Materials and Methods
Yeast strains, media, and general methods
Supplemental Material Table S1, contains a list of yeast strains used in this study. Strains were grown in yeast extract with supplements (YES) or EMM and supplemented with adenine (225 mg/liter), leucine (225 mg/liter), uracil (225 mg/liter), thiamine (15 µM), geneticin (150 mg/liter), and nourseothricin (100 mg/liter) when required. Calcium chloride and tunicamycin (T7765: Sigma Aldrich, St. Louis) were added to YES medium at 0.15 M and 2.5 µg/ml, respectively. Cell-wall sensitivity was assayed with 0.5 µg/ml micafungin (A13270-1: AdooQ Bioscience, Irvine, CA) and 25 U/ml Zymolyase 100T (E1005: Zymo Research, Irvine, CA). Deletion and epitope-tagged strains were constructed by a PCR-based stitching method as described in Kwon et al. (2012). All constructed strains were verified by colony PCR and sequencing of the amplicons. For deletion strains, the entire open reading frame (ORF) was replaced with the KanMX6 or NatMX4 cassettes, while for endogenously tagged prz1 strains, GFP and HA epitopes were PCR-amplified from pYM27 and pYM14 plasmids, respectively (Janke et al. 2004) and inserted in-frame at the C-terminal end of the prz1+ ORF. Functionality of the prz1-HA and prz1-GFP strains was determined by comparing their growth to wild type on 0.15 M CaCl2-containing medium. Overexpression of prz1+ with the nmt1 or nmt41 promoter was accomplished by cloning the ORF into the pREP1/pREP2 and pREP41 vectors, respectively. Standard genetic and molecular methods were performed as described in Moreno et al. (1991).
Microarray expression profiling
Wild-type and ∆prz1 cultures were concurrently grown in 100 ml liquid YES at 30° for 16–20 hr to a matching cell density of ∼8 × 106 cells/ml before harvesting. Calcium chloride and tunicamycin treatment were 0.15 M for 0.5 hr and 2.5 µg/ml for 1.5 hr, respectively, prior to harvesting. The ∆pmr1 and ∆prz1 cultures were also grown concurrently in 100 ml liquid YES at 30° for 16–20 hr to a matching cell density of ∼8 × 106 cells/ml. For prz1+ overexpression, cultures of the prz1OE strain and empty vector control were concurrently grown in 100 ml of EMM (supplemented with adenine and uracil) without thiamine at 30° for 18–22 hr to ∼8 × 106 cells/ml and then harvested. The same procedure was used in the heterologous overexpression of S. cerevisiae CRZ1 with the nmt1 or nmt41 promoter in the ∆prz1 strain. Sample preparation for hybridization to 8 × 15,000 Agilent S. pombe expression microarrays and scanning were carried out as described in detail in Kwon et al. (2012). All microarray experiments were performed with a single dye-swap and normalized using the R Limma package with Lowess scaling (Smyth and Speed 2003), and the eBayes method was used to combine the replicates by fitting to a linear model (Smyth 2004). Hierarchical clustering was performed using the uncentered Pearson correlation with Cluster 3.0 (Eisen et al. 1999), and the tree image was generated using Java Treeview (Saldanha 2004).
ChIP-chip profiling
The endogenous C-terminal-tagged prz1-HA strain was grown in 200 ml liquid YES medium at 30° for 16–20 hr and treated with calcium chloride or tunicamycin as described above. A detailed description in sample preparation for hybridization to 4 × 44,000 Agilent S. pombe Genome ChIP-on-chip microarrays is found in Kwon et al. (2012). All ChIP-chip experiments were performed with dye swaps for two biological replicates. The data were normalized using the median correction method, and replicates were combined with eBayes from the R limma package (Smyth and Speed 2003; Smyth 2004). Significant peaks were identified using chIPOTle (Buck et al. 2005). To detect potential promoter occupancy, the peak was considered only when within 1000 bp (the average length of the sonicated DNA fragments) of the promoter region, defined as 0–1500 bp upstream of the start codon.
Motif and functional enrichment analyses
Motif searching for the DNA-binding specificity of Prz1 was carried out in MEME using promoter sequences consisting of 1000 bp upstream of the start codon (Bailey and Elkan 1994). The maximum size of the motif was set at 10 bp and at zero or one motif per promoter sequence. Functional enrichment was performed with the Princeton GO term finder (Boyle et al. 2004).
Crz1 target genes
A list of Crz1 target genes in S. cerevisiae was assembled from a literature review of genome-wide, as well as smaller scale, studies (Matheos et al. 1997; Yoshimoto et al. 2002; Chua et al. 2006; Fardeau et al. 2007; Hu et al. 2007; Cai et al. 2008; Ruiz et al. 2008). In most cases, the Crz1 target genes identified as significant were used. For the Chua et al. (2006) study, no specific cutoff was defined, and a log10-fold change of 3 was selected. The Crz1/CrzA target genes used for Candida albicans, Candida glabrata, Aspergillus nidulans, and Aspergillus fumigatus were from individual studies of each organism that performed transcriptome analysis using microarray experiments (Karababa et al. 2006; Hagiwara et al. 2008; Soriani et al. 2008; Chen et al. 2012).
The list of S. pombe genes with orthologs in S. cerevisiae was obtained from the V2.18 ortholog list (Wood et al. 2012). The orthologs in the other species were determined using Inparanoid data sets (Sonnhammer and Östlund 2015) and from the Fungal Orthogroups Repository (Wapinski et al. 2007), except for A. fumigatus, which relied solely on Inparanoid data.
Cell-wall degradation assays
Strains were grown with their respective controls as described for the expression microarray experiments. Wild-type and ∆prz1 strains were grown in liquid YES medium, while genetic backgrounds containing nmt1-driven prz1+ or empty vector were grown in liquid EMM minus thiamine for 18–24 hr. The cells were washed twice with 0.9% saline solution and resuspended in TE buffer at ∼1.2 × 107 cells/ml. Three milliliters of cell suspension was transferred to test tubes in the presence and absence of 25 U/ml Zymolyase 100T (Zymo Research) and shaken at 37°. OD600 readings were taken every 15 min to assess the degree of cell-wall degradation. The significance of the different treatments was assessed at the 2-hr time point with an ANOVA followed by a two-tailed t-test.
Flocculation assays
Constitutive flocculation was assessed by inoculating cells in liquid YES at an initial cell density of ∼107 cells/ml and growing at 30° in a shaking incubator. After 24 hr, 10 ml of culture was transferred to a 90-mm plastic petri dish and rotated slowly on an orbital low-speed shaker (Labnet International, Woodridge, NJ) for 10 min at room temperature. Images of flocs were acquired with a SPImager (S&P Robotics Inc., Toronto).
Fluorescence microscopy
The intracellular localization of natively regulated Prz1-GFP was determined in the prz1-GFP strain treated with CaCl2 or tunicamycin and in the ∆pmr1 prz1-GFP strain. Cells were grown and treated as described for the expression microarray experiments. Images of live prz1-GFP cells were captured with a Zeiss Imager Z1 microscope and AxioCam MRM digital camera (Zeiss, Thornwood, NY). The proportion of cells containing Prz1-GFP predominantly in the cytoplasm, nucleus, or both compartments was determined manually in ∼300 cells from three biological replicates.
Data availability
The microarray expression data have been submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus Database (GSE77761). The ChIP-chip data also have been submitted to the NCBI Gene Expression Omnibus Database (GSE77761).
Results
Chemical and genetic activation of Prz1
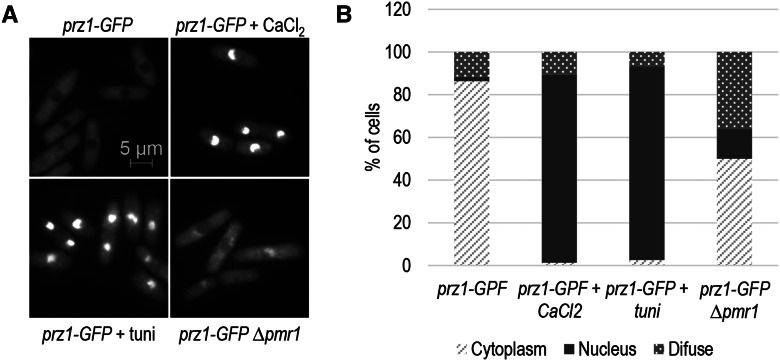
Identification of target genes using genome-wide approaches requires that the transcription factor be in an active state. Dephosphorylation of Prz1 by calcineurin results in nuclear translocation and regulation of its target genes. Therefore, we first determined whether several types of chemical treatment and a change in genetic background resulted in activation of Prz1 by promoting its translocation into the nucleus. Several studies have examined the intracellular localization of GFP-tagged Prz1, but expression of the fusion protein was controlled by the nmt1 promoter. The elevated expression in this strain could potentially increase the nuclear localization of Prz1 through mass action. As a result, we constructed a strain expressing a Prz1-GFP fusion protein under control of the native promoter to examine its nuclear localization in response to CaCl2 and tunicamycin treatment, as well as deletion of pmr1+. The prz1-GFP strain exhibited sensitivity to CaCl2 comparable to wild type, indicating that the fusion protein was functional. When cells were grown in rich medium, Prz1-GFP was primarily localized in the cytoplasm and not in the nucleus in the majority (>85%) of cells (Figure 1, A and B). Only ∼2% of these cells exhibited predominantly nuclear localization. In contrast, Prz1-GFP exhibited mostly nuclear localization (∼90%) when exposed to 0.15 M CaCl2 and 2.5 µg/ml tunicamycin for 0.5 and 1.5 hr, respectively (Figure 1, A and B). These results indicate that Prz1 is activated when treated with CaCl2 and tunicamycin, which is in agreement with previous studies (Hirayama et al. 2003; Deng et al. 2006). We also investigated the intracellular localization of Prz1-GFP when pmr1+, which encodes a Golgi Ca2+/Mn2+ ATPase, is deleted. Loss of pmr1+ is synthetic lethal with the ∆prz1 strain (Ryan et al. 2012), suggesting that Prz1 function is required in the ∆pmr1 background. Moreover, calcium homeostasis may be disrupted in the ∆pmr1 strain (Maeda et al. 2004), which could result in activation of Prz1. The frequency of nuclear localization of Prz1-GFP in ∆pmr1 cells (∼14%) was greater than unperturbed wild-type cells (Figure 1, A and B). Interestingly, a substantial proportion of ∆pmr1 cells (∼38%) displayed levels of Prz1-GFP present in both the nucleus and cytoplasm. These results suggest a robust activation of Prz1 in response to CaCl2 or tunicamycin treatments and intermediate activation in ∆pmr1 cells compared to untreated wild type.
Figure 1.
Intracellular localization of endogenously controlled Prz1-GFP. (A) Wild-type cells expressing endogenously controlled Prz1-GFP were exponentially grown in YES medium (top left) and treated with 0.15 M CaCl2 (top right) or 2.5 µg/ml tunicamycin (bottom left) for 0.5 and 1.5 hr, respectively. The intracellular localization of endogenously controlled Prz1-GFP in ∆pmr1 cells grown in rich medium (bottom right). (B) Bar graph showing the percentage of cells in each category of Prz1-GFP localization from A. The data are from three replicates of ∼100 cells each.
Identification of Prz1 target genes by genome-wide analyses
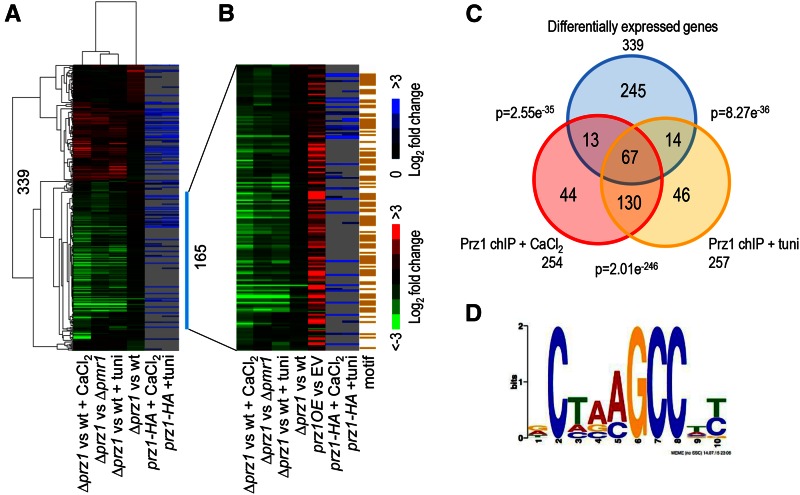
There are currently only a handful of known direct target genes of Prz1. To expand the list of Prz1 target genes, we performed transcriptome profiling and ChIP-chip analysis under the inducing conditions determined by our Prz1-GFP intracellular localization studies. Transcriptomes were compared between the ∆prz1 mutant with wild type exposed to 0.15 M CaCl2 or 2.5 µg/ml tunicamycin, as well as between the untreated ∆prz1 and the ∆pmr1 strains (Table S2). As a control, transcriptome profiling was performed on the ∆prz1 and wild-type strains grown in rich medium (Table S2). We identified 339 genes that were differentially regulated by more than twofold with a P < 0.001 in at least one of the four expression microarray experiments (Figure 2A). Lower expression levels in the ∆prz1 strain, relative to the wild-type or ∆pmr1 strain, represented positively regulated target genes of Prz1, while higher expression indicated negatively regulated targets. CaCl2 treatment resulted in the most differentially expressed genes (150 lower and 67 higher in ∆prz1), followed by the ∆pmr1 strain (109 lower and 67 higher in ∆prz1) and the tunicamycin treatment (94 lower and 51 higher in ∆prz1) (Figure 2A). In contrast, only 17 and 51 genes had lower and higher expression, respectively, in the ∆prz1 strain relative to wild type. This is consistent with previous observations that transcriptome profiling of most transcription-factor deletion strains does not uncover many direct target genes in S. pombe under optimal growth conditions (Chua 2013; Vachon et al. 2013). Hierarchical clustering revealed that the tunicamycin treatment and the ∆pmr1 strain expression profiles were the most similar (Figure 2A), which is in agreement with the observations that both tunicamycin and pmr1+ are involved in ER stress (Dürr et al. 1998; Deng et al. 2006) and therefore may activate Prz1 in a similar way.
Figure 2.
Identification of Prz1 target genes by transcriptome and ChIP-chip profiling. (A) The heat map shows two-dimensional hierarchical clustering of 339 genes that were differentially expressed by at least twofold in at least one of the microarray experiments. The first four columns of the heat map compare transcriptomes of the following conditions: the ∆prz1 strain and wild type, the ∆prz1 strain and wild type supplemented with 0.15 M CaCl2 for 0.5 hr, the ∆prz1 strain and wild type supplemented with 2.5 µg/ml tunicamycin for 1.5 hr, and the ∆prz1 strain compared to the ∆pmr1 strain. All of the above experiments were performed in rich medium. In the heat map, genes upregulated and downregulated in the ∆prz1 strain relative to the control are indicated in red and green, respectively. The two rightmost columns in the heat map show ChIP-chip analysis of a prz1-HA strain treated with 0.15 M CaCl2 or 2.5 µg/ml tunicamycin for 0.5 and 1.5 hr, respectively. (B) The heat map shows the expression profiles of 165 putative target genes that are positively regulated by Prz1. The first four columns of the heat map match the expression data from A while the fifth column shows the expression profiles of the same target genes upregulated in a prz1+ overexpression strain compared to the empty vector (EV) control. The next two columns in the heat map show ChIP-chip analysis of a prz1-HA strain treated with 0.15 M CaCl2 or 2.5 µg/ml tunicamycin for 0.5 and 1.5 hr, respectively. The rightmost column of the heat map shows the 91 genes containing the CDRE motif within their promoter in orange. The color bars indicate relative expression and ChIP enrichment ratios between experimental and control strains. All microarray expression and ChIP-chip experiments were performed in replicate with dye reversal. (C) The Venn diagram shows the overlap between the 339 differentially expressed genes in the transcriptome experiments and the genes identified from the ChIP-chip analysis with Prz1 promoter occupancy in the presence of CaCl2 or tunicamycin. The significance of the overlap is indicated as P-values that were determined using a hypergeometric distribution. (D) A DNA motif generated by MEME from promoter analysis of the 165 putative target genes of Prz1. This motif is similar to the CDRE motif (5′-AGCCTC-3′) previously discovered in Deng et al. (2006).
ChIP-chip profiling of a natively regulated Prz1-HA strain was performed to further confirm that the differentially expressed genes retrieved from the transcriptome studies are putative target genes of Prz1. These ChIP-chip experiments were carried out under inducing conditions of Prz1 in the presence of CaCl2 or tunicamycin with the same dosages as the transcriptome studies. We found that Prz1 bound to the promoters of 254 and 257 genes during CaCl2 and tunicamycin treatments, respectively, and the overlap of genes (197 genes or ∼77%) between both treatments was substantial (Figure 2, B and C). Moreover, we detected Prz1 occupancy in its own promoter in response to both CaCl2 and tunicamycin treatments (Table S3 and Table S4), which is in agreement with other studies (Deng et al. 2006). For the CaCl2 treatment, 80 of the 254 promoter-bound genes (31.4%) were differentially expressed in the ∆prz1 strain (P = 2.55e-35 using a hypergeometric distribution) (Figure 2, B and C). In response to tunicamycin treatment, 81 of the 257 promoter-bound genes (31.5%) were differentially expressed in the ∆prz1 strain (P = 8.27e-36) (Figure 2, B and C). Overall, 94 of the 339 genes (27.7%) differentially expressed in the transcriptome experiments were bound in one of the ChIP-chip experiments (Figure 2, B and C). This amount of overlap between transcriptome and ChIP-chip analyses of Prz1 is comparable to a similar study on the fission yeast CSL transcription factors Cbf11 and Cbf12 (Převorovský et al. 2015).
Among the 339 differentially regulated genes, 165 genes were consistently lower in the ∆prz1 strain relative to wild type when treated with CaCl2 or tunicamycin, indicating that Prz1 may positively regulate these target genes (Figure 2B). We next subjected these 165 putative target genes of Prz1 to gene ontology analysis using the Princeton GO Term Finder. The gene products were enriched for biological process categories such as reproduction (P = 5.1e-4), cell-wall organization, or biogenesis (P = 4.9e-3), as well as components of membranes (P = 2.2e-6) and the Golgi apparatus (P = 1.1e-5) (Table 1). Moreover, the vast majority of these target genes were also upregulated in the prz1+ overexpression strain, and Prz1 was found to be associated with the promoters of 37 of these genes (P = 2.3e-12) (Figure 2B). All the known target genes of Prz1 (prz1+, pmc1+, pmr1+, ncs1+, and cmk1+) were also found within these 165 genes (Table 1). Strikingly, promoter analysis by MEME revealed a common motif in 91 of the 165 putative Prz1 target genes that closely resembled the CDRE sequence 5′-AGCCTC-3′ (Deng et al. 2006) (Figure 2D and Table S5). Altogether, these results indicate that the majority of these 165 genes are likely direct targets that are positively regulated by Prz1.
Table 1. Gene ontology enrichment among the 165 target genes positively regulated by Prz1 using Princeton GO Term Finder.
| Gene ontology term | P-value | Gene list |
|---|---|---|
| Reproduction (GO:0000003) | 5.1e−4 | cdc1+, dic1+, gpa1+, gwt1+, isp3+, krp1+, mam2+, map1+, matPc+, mcp2+, mcp5+, mde6+, mei2+, meu13+, meu17+, meu22+, meu27+, mfm2+, mst2+, mug108+, mug133+, mug136+, mug63+, mug8+, ncs1+, pmp31+, ppk35+, rec10+, scd2+, set3+, SPAPB1A10.08, ste11+, ste4+, bgs1+, cfr1+, pvg5+ |
| Cell-wall organization or biogenesis (GO:0071554) | 4.9e−3 | bgs1+, cfr1+, pvg5+, cfh2+, gas2+, gmh2+, omh1+, pun1+, pvg1+, rga5+, SPAC13C5.05c, SPAC9G1.10c, SPBC1198.07c, SPBC19C7.05, SPBC21B10.07 |
| Membrane (GO:0016020) | 2.18e−6 | gda1+, sen54+, SPAC977.02, mug133+, ima1+, pet2+, SPBC21B10.07, git3+, SPBC1271.03c, ppk35+, ncs1+, pun1+, SPAC23C11.06c, SPCPB1C11.02, gas2+, krp1+, SPAC14C4.07, dnf1+, SPBC15C4.06c, pmp31+, imt2+, mfm2+, cki2+, frp1+, SPAC18B11.03c, SPCC1529.01, ost2+, mam2+, omh1+, fur4+, SPAC212.01c, bgs1+, isp5+, ggc1+, cfh2+, rsn1+, psd2+, SPAC630.04c, erg1+, SPBC1348.03, meu22+, tco1+, bst1+, SPAC869.03c, meu17+, SPAC5D6.04, SPAC750.04c, ppr3+, SPAC1687.08, SPBC19C7.05, mac1+, gwt1+, yip5+, SPBC1198.07c, itr2+, gga21+, pet1+, SPCC553.12c, lcb4+, pvg1+, SPAC869.05c, SPCC794.03, SPAC4C5.03, str1+, tvp15+, gmh2+, pmr1+, SPCC4B3.02c, mfs1+, SPAC23D3.12, gpa1+, imt1+, pvg5+ |
| Golgi apparatus (GO:005794) | 1.08e−5 | fmd2+, gda1+, tco1+, bst1+, cfr1+, mug133+, SPAC869.05c, pet2+, SPAC869.03c, meu17+, SPAC3A11.10c, omh1+, tvp15+, fur4+, SPBC19C7.05, gmh2+, pun1+, yip5+, SPCC4B3.02c, gga21+, SPAC23D3.12, krp1+, pet1+, SPAC14C4.07, dnf1+, lcb4+, imt1+, mug136+, pvg5+, pvg1+, imt2+ |
The target genes that have the CDRE motif in their promoter (Figure 2D) are indicated in boldface type and those with an ortholog that is regulated by Crz1 in S. cerevisiae are underlined. Only the genes with enriched gene ontology terms are shown.
Interestingly, 92 genes were upregulated at least twofold in the ∆prz1 strain during CaCl2 or tunicamycin treatment (Figure S1), indicating that Prz1 may also negatively regulate a different set of target genes. In addition, 46 of these genes were downregulated in the prz1+ overexpression strain, and Prz1 was associated with the promoters of 40 of the 92 putative target genes (P = 5.2e−25) (Figure S1). However, unlike the positively regulated genes, no common binding motif was detected by MEME. Gene ontology analysis of these 92 genes detected functional enrichment in ion transmembrane transport (P = 4.7e−4) and small molecule catabolic processes (P = 3.0e−4) (Table 2 and Table S6). Direct transcriptional repression of target genes has not been detected for Prz1 or confirmed in fungal Crz1 orthologs (Thewes 2014).
Table 2. Gene ontology enrichment among the 92 target genes negatively regulated by Prz1 using Princeton GO Term Finder.
| Gene ontology term | P-value | Gene list |
|---|---|---|
| Small-molecule catabolic process (GO:0044282) | 3.0e−4 | SPAC2F3.05c, car1+, SPAC139.05, SPAC4H3.08, gut2+, gpd3+, atd1+, tms1+, SPCC1223.09, eno102+ |
| Ion transmembrane transport (GO:0034220) | 4.7e−4 | gti1+, tgp1+, pho1+, mfs3+, mug86+, SPCPB1C11.03, SPBPB2B2.01, SPBC36.02c, SPBC1683.01, pho84+, mae1+, SPBPB10D8.01, SPBC3H7.02, pgt1+, SPCC794.04c |
The genes that have an ortholog regulated by Crz1 in S. cerevisiae are underlined. Only the genes with enriched gene ontology terms are shown.
Orthologous target genes between S. pombe Prz1 and other fungal Crz1/CrzA
The known target genes of Prz1 are well conserved in S. cerevisiae. All of the known Prz1 target genes (prz1+, pmc1+, pmr1+, ncs1+, and cmk1+) have budding yeast orthologs regulated by Crz1 with the exception of ncs1+ (Hamasaki-Katagiri and Ames 2010). However, the proportion of the remaining Prz1 putative target genes that have Crz1 target gene orthologs was considerably lower. There were 24 Crz1 target gene orthologs among the 165 putative target genes upregulated by Prz1 (Table 1 and Table S5) (Matheos et al. 1997; Yoshimoto et al. 2002; Chua et al. 2006; Fardeau et al. 2007; Hu et al. 2007; Cai et al. 2008; Ruiz et al. 2008). The conservation of target genes is highest for the conserved biological process of cell-wall organization or biogenesis in which 40% of the Prz1 putative target genes annotated in this gene ontology category have Crz1 target gene orthologs (Table 1). Interestingly, there were eight putative target genes (atd1+, sua1+, bfr1+, SPBC1683.01, pho84+, fhn1+, plb1+, and SPAC513.07) seemingly repressed by Prz1 that had orthologs that were positively regulated by Crz1 (Yoshimoto et al. 2002; Chua et al. 2006; Ruiz et al. 2008). This phenomenon is further supported by the observation that the promoters of seven of these eight genes were bound by Prz1 in one or both of the ChIP-chip experiments (Table S6).
The number of orthologous target genes decreases with more distantly related fungi (Figure S2). Comparison with a study in C. albicans found that, of the 65 Crz1 targets, only 5 overlap with the 165 positively regulated targets and 2 overlap with the negatively regulated target genes in S. pombe (Karababa et al. 2006). A similar overlap was also observed between the 34 target genes of C. glabrata Crz1 (Chen et al. 2012). CrzA target genes in A. nidulans and A. fumigatus showed even less overlap with the S. pombe Prz1 target genes (Hagiwara et al. 2008; Soriani et al. 2008). Cell-wall organization is positively regulated by Prz1, as well as its orthologs in fungi, which would indicate conservation among the target genes implicated in that process. However, this conservation was not as extensive as expected. Only two Prz1 target genes with cell-wall functions (SPBC21B10.07 and bgs1+) share orthologs in S. cerevisiae and C. albicans. Other cell-wall genes like gas2+ and SPAC9G1.10c were conserved in the more distantly related species C. glabrata and A. fumigatus, respectively, while not sharing an ortholog in S. cerevisiae.
The difference in the number of orthologs suggests an increasingly distant relationship between Prz1 and its Crz1/CrzA orthologs over evolutionary time. However, it may reflect incomplete data because, unlike S. cerevisiae, the other targets are each determined from a single study (Karababa et al. 2006; Hagiwara et al. 2008; Soriani et al. 2008; Chen et al. 2012). The curation of the ortholog lists may also play a role in the overlap observed. The ortholog list between S. cerevisiae and S. pombe has been extensively refined (Wood et al. 2012), whereas with the other species orthology was determined using Inparanoid data sets (Sonnhammer and Östlund 2015) and the Fungal Orthogroups Repository (Wapinski et al. 2007). Although the overlap with these other species offers an interesting look at conserved elements, it is clearly an incomplete picture.
Prz1 activates target genes functioning in cell-wall synthesis and structure
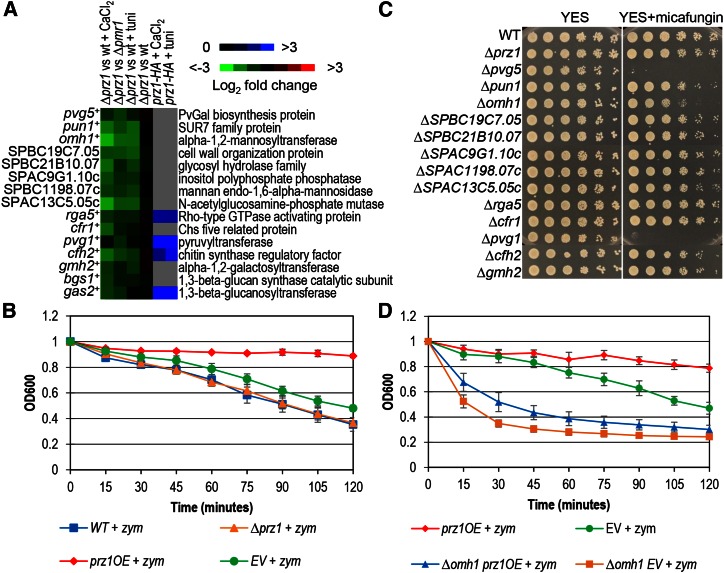
The activation of Prz1 upregulates 15 putative target genes implicated in the biosynthesis and structure of the cell wall (Figure 3A; Table 1). Among these 15 genes, 9 contained a CDRE motif in the promoter while Prz1 occupancy was detected in 4 (Figure 3A; Table 1; Table S5). To further investigate this, we first determined whether ∆prz1 and prz1OE strains possessed an altered cell-wall structure. The ∆prz1 and prz1OE strains were tested for resistance to cell-wall degradation by zymolyase (a β-glucanase) relative to their controls. The short duration of the zymolyase assay was ideal because of the reduced fitness exhibited by the prz1OE strain (Koike et al. 2012; Vachon et al. 2013). We found that overexpression of prz1+ with the nmt1 promoter confers increased resistance to zymolyase (P = 1.0e-4), suggesting that the upregulation of these target genes could enhance the strength of the cell wall (Figure 3B). In contrast, no change in resistance to zymolyase was observed in the ∆prz1 strain compared to wild type (Figure 3B). We next attempted to identify the putative target genes that could be responsible for the increased resistance to zymolyase in the prz1OE strain. Loss of these target genes could potentially result in sensitivity to cell-wall-perturbing agents. Thirteen deletion strains of the Prz1 putative target genes implicated in cell-wall processes were assayed for growth sensitivity to the antifungal micafungin, which inhibits β-1,3-glucan production. The remaining two genes, gas2+ and bgs1+, the latter being an essential gene, were not available in the Bioneer deletion collection. Among the 13 deletion strains, only ∆pvg1, ∆pvg5, and ∆omh1 were sensitive to micafungin (Figure 3C). Both pvg1+ and pvg5+ function in the synthesis of pyruvated galactose residues in N-linked glycans (Andreishcheva et al. 2004; Yoritsune et al. 2013), and omh1+ encodes a putative α1,2-mannosyltransferase for synthesis of O-linked glycans (Ikeda et al. 2009). The ∆prz1 strain, in addition to not being sensitive to zymolyase, was not sensitive to micafungin (Figure 3C). The lack of sensitivity is perhaps due to redundancy in the regulation of these cell-wall target genes by a transcription factor other than Prz1. We then overexpressed prz1+ with the nmt1 promoter in the ∆pvg1, ∆pvg5, and ∆omh1 strains to determine if the zymolyase resistance exhibited in the prz1OE strain was affected by loss of these genes. Consistent with the micafungin assay, the ∆omh1 strain was more susceptible to degradation by zymolyase than the control after 2 hr of treatment (P = 1.2e-2), while the ∆pvg1 and ∆pvg5 strains did not show a significant increase in sensitivity (Figure 3D and Figure S3). Indeed, loss of omh1+ almost completely abrogated the zymolyase resistance caused by prz1+ overexpression (P = 5.0e-4). A similar genetic interaction was also observed in the ∆pvg1 background except the degree of abrogation was less (P = 6.8e-3) (Figure S3). Together, these results indicate that the cell-wall function of Prz1 involves activation of its target genes omh1+ and pvg1+.
Figure 3.
Characterization of putative target genes of Prz1 implicated in cell-wall-related processes. (A) The heat map shows relative expression and Prz1 promoter occupancy for 15 putative target genes annotated to function in cell-wall organization or biogenesis. The color bars indicate relative expression and ChIP enrichment ratios between experimental and control strains. All microarray expression and ChIP-chip experiments were performed in replicate with dye reversal. (B) Cell-wall degradation assays. Wild-type and ∆prz1 strains were grown in liquid YES medium, while nmt1-driven prz1+ or empty vector (EV) were grown in liquid EMM minus thiamine for 18–24 hr. The samples were adjusted to matching cell densities and transferred to test tubes in the presence of 25 U/ml Zymolyase 100T. The samples were shaken at 37°, and OD600 readings were taken every 15 min to assess the degree of cell-wall degradation. Overexpression of prz1+ caused resistance to the cell-wall-degrading enzyme zymolyase (P = 1.0e−4) while Δprz1 cells did not show significant sensitivity to zymolyase compared to wild type. (C) Spot dilution for micafungin sensitivity of deletion strains of the putative Prz1 target genes involved in cell-wall-related processes. Exponentially growing wild-type and deletion strains were pinned on solid YES medium containing 0.5 µg/ml micafungin and incubated at 30° for 3 days. (D) Cell-wall degradation assays. The Δomh1 strain was more sensitive to zymolyase treatment than wild type (P = 1.6e−2). The zymolyase-resistant phenotype from overexpression of prz1+ was abrogated by loss of omh1+ (P = 5.0e−4). The zymolyase experiments were repeated in triplicate, and error bars represent the standard error. The P-values were determined with ANOVA followed by a two-tailed t-test after 2 hr of zymolyase treatment.
Prz1 repression of flocculation
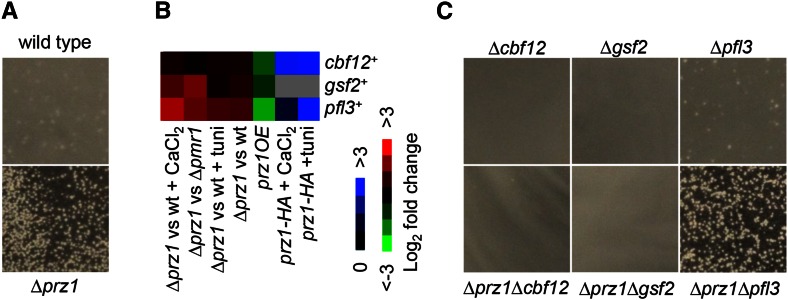
The ∆prz1 strain exhibited a slightly crumbly texture on solid media that is often present in flocculent strains. To determine if ∆prz1 cells are flocculent, we cultured the strain in liquid YES and EMM with an initial density of ∼107 cells/ml for 24 hr. The ∆prz1 cells, but not wild type, formed large flocs in both EMM and YES media (Figure 4A). In contrast, overexpression of prz1+ did not suppress the flocculation of wild-type cells in flocculation-inducing medium (data not shown). We next examined whether putative target genes in our genome-wide data could be responsible for the constitutive flocculent phenotype of ∆prz1 cells. Interestingly, several genes implicated in triggering flocculation were upregulated in the ∆prz1 strain. These upregulated genes (1.6- to 5.3-fold) encoded the flocculins Gsf2 and Pfl3 and the transcription factor Cbf12, which is known to activate gsf2+ (Převorovský et al. 2015; Kwon et al. 2012) (Figure 4B). In addition, these three genes were downregulated in the prz1OE strain, and the promoters of pfl3+ and cbf12+ were bound by Prz1 in both CaCl2 and tunicamycin treatments (Figure 4B). These results suggest that Prz1 inhibits flocculation by repression of gsf2+, pfl3+, or cbf12+. To further confirm this hypothesis, we examined whether the constitutive flocculent phenotype of the ∆prz1 strain in YES medium could be abrogated in a ∆gsf2, ∆pfl3, or ∆cbf12 genetic background. Indeed, we discovered that loss of gsf2+ or cbf12+, but not pfl3+, could abrogate the constitutive flocculation observed in ∆prz1 cells (Figure 4C).
Figure 4.
Constitutive flocculation of the ∆prz1 strain. (A) Wild type and the ∆prz1 strain were grown at an initial cell density of ∼107 cells/ml in liquid YES medium for 24 hr at 30° and assayed for flocculation. (B) Negative regulation of flocculation genes by Prz1. The heat map shows relative expression and Prz1 promoter occupancy for the flocculation genes cbf12+, gsf2+, and pfl3+. The color bars indicate relative expression and ChIP enrichment ratios between experimental and control strains. All microarray expression and ChIP-chip experiments were performed in replicate with dye reversal. (C) The ∆prz1 flocculation phenotype was abolished by loss of cbf12+ or gsf2+. Cells were assayed for flocculation as described for A.
Discussion
In this study, we substantially expanded the target gene list and implicated new functional roles for the calcineurin-responsive transcription factor Prz1 in S. pombe. Moreover, we presented genetic evidence that links putative target genes to these new functions of Prz1. Our results also demonstrate that identification of target genes by genome-wide approaches requires that the transcription factor be active. Transcriptome profiling of the ∆prz1 strain grown in rich medium did not identify many direct target genes of Prz1. We used several inducing conditions that promote Prz1 activity, including chemicals (Ca2+ and tunicamycin), overexpression of prz1+, and the ∆pmr1 genetic background. Both Ca2+ and tunicamycin and have been shown to activate a CDRE-regulated reporter, and Ca2+ has also been shown to cause Prz1 to translocate to the nucleus (Hirayama et al. 2003; Deng et al. 2006). Systematic overexpression of yeast transcription factors, combined with transcriptome profiling, has been used effectively to identify direct target genes (Chua et al. 2006; Vachon et al. 2013; Chua 2013). The prz1OE strain displays reduced fitness, which is common for transcription factors overexpressed in fission yeast (Vachon et al. 2013). This reduced fitness is likely caused by increased promoter occupancy and aberrant expression of target genes. When overexpressed, Prz1 is still largely localized in the cytoplasm (Hirayama et al. 2003). Despite this, Prz1 overproduction allows some of the transcription factors to override the normal cytoplasmic retention signals and enter the nucleus, resulting in differential expression in a large number of putative target genes (893 upregulated and 532 downregulated by more than twofold).
In addition, Prz1 activation was observed in the ∆pmr1 genetic background. The pmr1+ gene encodes a Golgi P-type Ca2+/Mn2+-ATPase that functions to reduce abnormally high levels of cytosolic Ca2+ (Cortés et al. 2004). Therefore, loss of pmr1+ is expected to cause elevated cytosolic Ca2+ relative to wild type and result in the downstream activation of Prz1. Our results show that nuclear localization of Prz1-GFP was increased in ∆pmr1 cells compared to unperturbed wild-type cells (Figure 1). Although the Prz1-GFP nuclear localization in ∆pmr1 cells was less than wild-type cells treated with Ca2+ or tunicamycin, the putative target genes of Prz1 could still be recovered by comparing the transcriptome profiles of ∆pmr1 and ∆prz1 strains (Figure 2). Interestingly, synthetic lethality is observed in the ∆pmr1 ∆prz1 double mutant (Ryan et al. 2012), indicating that Prz1 activity is required in the ∆pmr1 strain. This observation is intriguing as it suggests that synthetic-lethal interactions could be used to identify genetic backgrounds that contain transcription factors in their active state (genetic activation). Subsequently, the comparison of transcriptomes between the transcription factor deletion strain and a deletion strain with which it shares a synthetic-lethal interaction could conceivably identify direct target genes. We are currently exploring whether this approach is indeed effective in identifying target genes of transcription factors in S. pombe.
The annotated functions of the positively regulated 165 putative target genes of Prz1 align well with certain functions of the fungal Crz1 orthologs: ion homeostasis and transport, lipid metabolism, and cell-wall biosynthesis. In S. pombe, functional enrichment was detected only for genes implicated in reproduction and cell-wall organization or biogenesis. There were 15 putative target genes of Prz1 that have a role in cell-wall processes, 6 of which have orthologs in S. cerevisiae. These genes are implicated in β-glucan synthesis (bgs1+/FKS1 and rga5+/SAC7), mannosidase activity (SPBC1198.07c/DFG5), cell-wall integrity (pun1+/PUN1), as well as chitin deposition (SPBC19C7.05/RCR1) and transglycosylation (SPBC21B10.07/UTR2). In contrast, two S. pombe-specific target genes (pvg1+ and pvg5+) were involved in the synthesis of pyruvylated galactose residues that are not found in the cell wall of S. cerevisiae (Andreishcheva et al. 2004; Yoritsune et al. 2013). The upregulation of omh1+, which encodes a putative mannosyltransferase, and pvg1+ by prz1+ overexpression contributes to the zymolyase-resistant phenotype of the prz1OE strain (Figure 3; Figure S3).
In addition, 33 target genes implicated in reproduction were identified as positively regulated by Prz1. These genes involved in reproduction are consistent with the decreased mating efficiency observed in ∆prz1 mutants (Sun et al. 2013). In S. cerevisiae, Crz1 is required for survival in prolonged exposure to mating pheromones (Stathopoulos and Cyert 1997), but pheromone genes have not been identified as targets of Crz1. The function of Prz1 in the mating process is likely different since very few of the target genes have orthologs in S. cerevisiae (Table 1). Interestingly, we detected Prz1 promoter occupancy of ste11+, which encodes the primary transcriptional activator of the mating response (Sugimoto et al. 1991).
Our expanded list of 165 putative target genes upregulated by Prz1 identified a CDRE motif in 91 promoters similar to the 5′-AGCCTC-3′ motif found by Deng et al. (2006). This motif is also similar to the S. cerevisiae CDRE motif [5′-TG(C/A)GCCNC-3′] (Stathopoulos and Cyert 1997). The apparent conservation in binding specificity of Crz1 and Prz1 indicates the possibility of heterologous complementation. To test this, we overexpressed CRZ1 with the nmt1 or nmt41 promoter in the ∆prz1 strain. In contrast to prz1+ overexpression, heterologous overexpression of CRZ1 did not cause reduced fitness or suppress the calcium sensitivity of the ∆prz1 strain (data not shown). Transcriptome profiling of nmt1-driven CRZ1 in the ∆prz1 strain did not result in differential regulation of the Prz1 target genes. The inability of Crz1 to regulate Prz1 target genes may be due to the absence of binding to the CDRE motifs or required trans-acting factors. In contrast, the C. albicans Crz1 was able suppress the Ca2+ sensitivity of the S. cerevisiae ∆crz1 strain, as well as drive expression of the CDRE reporter (Karababa et al. 2006).
We have also uncovered a novel repressive role of Prz1 in flocculation. Loss of prz1+ causes constitutive flocculation in high-density cultures and the induction of both the transcription factor gene cbf12+ and the dominant flocculin gene gsf2+ (Figure 4, A and B). The constitutive flocculation is dependent on the presence of cbf12+ and gsf2+ (Figure 4C). Prz1 promoter occupancy was also detected for cbf12+ and the pfl3+ flocculin gene (Figure 4B). Although CDRE motifs were not detected in the promoters of these genes, five copies of the Ca2+-dependent response element (5′-CAACT-3′) were present in the cbf12+ promoter. Prz1 has been shown to bind to this motif in the ncs1+ promoter (Hamasaki-Katagiri and Ames 2010). The repressive flocculation function of Prz1 and the Ca2+-dependent response element have not been discovered in other fungal Crz1 orthologs.
Previous studies on Prz1 orthologs have predominantly focused on positively regulated functions and target genes. One exception was 3 genes identified as negatively regulated by C. glabrata Crz1 (Chen et al. 2012). In A. fumigatus, 31 genes were also identified as being downregulated in response to calcium treatment, and some may represent direct targets of CrzA (Soriani et al. 2008). The low number of identified negatively regulated target genes of Prz1 orthologs could be the result of limitations in previous studies, which do not contain genome-wide binding data.
The repression of target genes by Prz1 could be the result of S. pombe-specific rewiring of the transcriptional regulatory network due to an alteration in the amino acid sequence of the transcription factor. In contrast to S. cerevisiae Crz1, S. pombe Prz1 does not contain a polyglutamine tract domain that is often associated with transcriptional activation (Hirayama et al. 2003). It could be the absence of this domain that provides Prz1 with its transcriptional repressive function. Interestingly, Schaefer et al. (2012) observed that considerably fewer S. pombe genes (3) compared to S. cerevisiae genes (79) contain this domain. In addition, we examined whether the repressed putative targets of Prz1 could be the result of transcriptional interference where a transcribed gene can repress the transcription of the adjacent gene (Martens et al. 2004). Among the 92 negatively regulated putative target genes of Prz1, only 13 (14.1%) are located adjacent to a putative activated target gene. This indicates that the vast majority of putative negatively regulated target genes of Prz1 are not a result of transcriptional interference. One notable example, SPAC513.07, which encodes a flavonol reductase, is negatively regulated by Prz1 in S. pombe, but its orthologs are activated by Crz1 in S. cerevisiae, C. albicans, and C. glabrata. This may indicate a rewiring of the transcriptional regulation of this gene between fungal species. The specific mechanism of Prz1-mediated negative regulation remains unknown, and more experimentation will be needed to determine if there is nucleosome remodeling occurring at these sites or some undiscovered cofactor involved.
Our genome-wide analysis of Prz1 has uncovered gene regulation in conserved and species-specific functions among fungal Crz1 orthologs. Preliminary comparison with other Crz1 target genes among different fungi shows substantial rewiring within the transcriptional regulatory network controlling calcineurin-mediated processes. This rewiring is evident even in well-conserved processes such as cell-wall biogenesis, where distinct fungi accomplish a similar function by positive regulation of different target genes. Moreover, this study revealed that the calcineurin-mediated transcriptional regulatory network of S. pombe has undergone substantial rewiring to include negative regulation of target genes implicated in species-specific processes such as flocculation.
Acknowledgments
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada and Canada Foundation for Innovation (to G.C.).
Footnotes
Communicating editor: A. P. Mitchell
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.184218/-/DC1.
Literature Cited
- Andreishcheva E. N., Kunkel J. P., Gemmill T. R., Trimble R. B., 2004. Five genes involved in biosynthesis of the pyruvylated Galβ1,3-epitope in Schizosaccharomyces pombe N-linked glycans. J. Biol. Chem. 279: 35644–35655. [DOI] [PubMed] [Google Scholar]
- Araki Y., Wu H., Kitagaki H., Akao T., Takagi H., et al. , 2009. Ethanol stress stimulates the Ca2+-mediated calcineurin/Crz1 pathway in Saccharomyces cerevisiae. J. Biosci. Bioeng. 107: 1–6. [DOI] [PubMed] [Google Scholar]
- Bailey T. L., Elkan C., 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Sec. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36. [PubMed] [Google Scholar]
- Bodvard K., Jörhov A., Blomberg A., Molin M., Käll M., 2013. The yeast transcription factor Crz1 is activated by light in a Ca2+/calcineurin-dependent and PKA-independent manner. PLoS One 8: e53404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E. I., Weng S., Gollub J., Jin H., Botstein D., et al. , 2004. GO:TermFinder-open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M. J., Nobel A. B., Lieb J. D., 2005. ChIPOTle: a user-friendly tool for the analysis of chIP-chip data. Genome Biol. 6: R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Dalal C. K., Elowitz M. B., 2008. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature 455: 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-L., Konieczka J. H., Springer D. J., Bowen S. E., Zhang J., et al. , 2012. Convergent evolution of calcineurin pathway roles in thermotolerance and virulence in Candida glabrata. G3 (Bethesda) 2: 675–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua G., 2013. Systematic genetic analysis of transcription factors to map the fission yeast transcription-regulatory network. Bioc Soc T 41: 1696–1700. [DOI] [PubMed] [Google Scholar]
- Chua G., Morris Q. D., Sopko R., Robinson M. D., Ryan O., et al. , 2006. Identifying transcription factor functions and targets by phenotypic activation. Proc. Natl. Acad. Sci. USA 103: 12045–12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisneros-Barroso E., Yance-Chávez T., Kito A., Sugiura R., Gómez-Hierro A., et al. , 2014. Negative feedback regulation of calcineurin-dependent Prz1 transcription factor by the CaMKK-CaMK1 axis in fission yeast. Nucleic Acids Res. 42: 9573–9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clipstone N. A., Crabtree G. R., 1992. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature 357: 695–697. [DOI] [PubMed] [Google Scholar]
- Cortés J. C. G., Katoh-Fukui R., Moto K., Ribas J. C., Ishiguro J., 2004. Schizosaccharomyces pombe Pmr1p is essential for cell wall integrity and is required for polarized cell growth and cytokinesis. Eukaryot. Cell 3: 1124–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Sugiura R., Takeuchi M., Suzuki M., Ebina H., et al. , 2006. Real-time monitoring of calcineurin activity in living cells: evidence for two distinct Ca2+-dependent pathways in fission yeast. Mol. Biol. Cell 17: 4790–4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr G., Strayle J., Plemper R., Elbs S., Klee S. K., et al. , 1998. The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum-associated protein degradation. Mol. Biol. Cell 9: 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlind T., Smith L., Henry K., Katiyar S., Nickels J., 2002. Antifungal activity in Saccharomyces cerevisiae is modulated by calcium signalling. Mol. Microbiol. 46: 257–268. [DOI] [PubMed] [Google Scholar]
- Eisen M. B., Spellman P. T., Brown P. O., Botstein D., 1999. Cluster analysis and display of genome-wide exopresion patterns. Proc. Natl. Acad. Sci. USA 95: 12930–12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardeau V., Lelandais G., Oldfield A., Salin H., Lemoine S., et al. , 2007. The central role of PDR1 in the foundation of yeast drug resistance. J. Biol. Chem. 282: 5063–5074. [DOI] [PubMed] [Google Scholar]
- Ferreira R. T., Courelas Silva A. R., Pimentel C., Batista-Nascimento L., Rodrigues-Pousada C., et al. , 2012. Arsenic stress elicits cytosolic Ca2+ bursts and Crz1 activation in Saccharomyces cerevisiae. Microbiol 158: 2293–2302. [DOI] [PubMed] [Google Scholar]
- Graef I. A., Chen F., Chen L., Kuo A., Crabtree G. R., 2001. Signals transduced by Ca2+/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell 105: 863–875. [DOI] [PubMed] [Google Scholar]
- Hagiwara D., Kondo A., Fujioka T., Abe K., 2008. Functional analysis of C2H2 zinc finger transcription factor CrzA involved in calcium signaling in Aspergillus nidulans. Curr. Genet. 54: 325–338. [DOI] [PubMed] [Google Scholar]
- Hamasaki-Katagiri N., Ames J. B., 2010. Neuronal calcium sensor-1 (Ncs1p) is up-regulated by calcineurin to promote Ca2+ tolerance in fission yeast. J. Biol. Chem. 285: 4405–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendey B., Klee C. B., Maxfield F. R., 1992. Inhibition of neutrophil chemokinesis on vitronectin by inhibitors of calcineurin. Science 258: 296–299. [DOI] [PubMed] [Google Scholar]
- Hirayama S., Sugiura R., Lu Y., Maeda T., Kawagishi K., et al. , 2003. Zinc finger protein Prz1 regulates Ca2+ but not Cl− homeostasis in fission yeast: identification of distinct branches of calcineurin signaling pathway in fission yeast. J. Biol. Chem. 278: 18078–18084. [DOI] [PubMed] [Google Scholar]
- Hu Z., Killion P. J., Iyer V. R., 2007. Genetic reconstruction of a functional transcriptional regulatory network. Nat. Genet. 39: 683–687. [DOI] [PubMed] [Google Scholar]
- Ikeda Y., Ohashi T., Tanaka N., Takegawa K., 2009. Identification and characterization of a gene required for α1,2-mannose extension in the O-linked glycan synthesis pathway in Schizosaccharomyces pombe. FEMS Yeast Res. 9: 115–125. [DOI] [PubMed] [Google Scholar]
- Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., et al. , 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21: 947–962. [DOI] [PubMed] [Google Scholar]
- Karababa M., Valentino E., Pardini G., Coste A. T., Bille J., et al. , 2006. CRZ1, a target of the calcineurin pathway in Candida albicans. Mol. Microbiol. 59: 1429–1451. [DOI] [PubMed] [Google Scholar]
- Koike A., Kato T., Sugiura R., Ma Y., Tabata Y., et al. , 2012. Genetic screening for regulators of Prz1, a transcriptional factor ccting downstream of calcineurin in fission yeast. J. Biol. Chem. 287: 19294–19303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon E. J. G., Laderoute A., Chatfield-Reed K., Vachon L., Karagiannis J., et al. , 2012. Deciphering the transcriptional-regulatory network of flocculation in Schizosaccharomyces pombe. PLoS Genet. 8: e1003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F., 2005. NFAT proteins: key regulators of T-cell development and function. Nat. Rev. Immunol. 5: 472–484. [DOI] [PubMed] [Google Scholar]
- Maeda T., Sugiura R., Kita A., Saito M., Deng L., et al. , 2004. Pmr1, a P-type ATPase, and Pdt1, and Nramp homologue, cooperatively regulate cell morphogenesis in fission yeast: the importance of Mn2+ homeostasis. Genes Cells 9: 71–82. [DOI] [PubMed] [Google Scholar]
- Mansuy I. M., Winder D. G., Moallem T. M., Osman M., Mayford M., et al. , 1998. Inducible and reversible gene expression with the rtTA system for the study of memory. Neuron 21: 257–265. [DOI] [PubMed] [Google Scholar]
- Martens J. A., Laprade L., Winston F., 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574. [DOI] [PubMed] [Google Scholar]
- Matheos D. P., Kingsbury T. J., Ahsan U. S., Cunningham K. W., 1997. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 11: 3445–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin J. D., Lu J.-R., Antos C. L., Markham B., Richardson J., et al. , 1998. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93: 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P., 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194: 795–823. [DOI] [PubMed] [Google Scholar]
- Převorovský M., Oravcová M., Tvarůžková J., Zach R., Folk P., et al. , 2015. Fission yeast CSL transcription factors: mapping their target genes and biological roles. PLoS One 10: e0137820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A., Serrano R., Ariño J., 2008. Direct regulation of genes involved in glucose utilization by the calcium/calcineurin pathway. J. Biol. Chem. 283: 13923–13933. [DOI] [PubMed] [Google Scholar]
- Ryan C. J., Roguev A., Patrick K., Xu J., Jahari H., et al. , 2012. Hierarchical modularity and the evolution of genetic interactomes across species. Mol. Cell 46: 691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A. J., 2004. Java Treeview: extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- Schaefer M. H., Wanker E. E., Andrade-Navarro M. A., 2012. Evolution of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res. 40: 4273–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R., Ruiz A., Bernal D., Chambers J. R., Ariño J., 2002. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: evidence for calcium-mediated signalling. Mol. Microbiol. 46: 1319–1333. [DOI] [PubMed] [Google Scholar]
- Smyth G. K., 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. 3: 1–25. [DOI] [PubMed] [Google Scholar]
- Smyth G. K., Speed T., 2003. Normalization of cDNA microarray data. Methods 31: 265–273. [DOI] [PubMed] [Google Scholar]
- Sonnhammer E. L., Östlund G., 2015. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 43: D234–D239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriani F. M., Malavazi I., Da Silva Ferreira M. E., Savoldi M., Von Zeska Kress M. R., et al. , 2008. Functional characterization of the Aspergillus fumigatus CRZ1 homologue, CrzA. Mol. Microbiol. 67: 1274–1291. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A. M., Cyert M. S., 1997. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 11: 3432–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos-Gerontides A., Guo J. J., Cyert M. S., 1999. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev. 13: 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto A., Iino Y., Maeda T., Watanabe Y., and M. Yamamoto, 1991. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 5: 1990–1999. [DOI] [PubMed] [Google Scholar]
- Sun L.-L., Li M., Suo F., Liu X.-M., Shen E.-Z., et al. , 2013. Global analysis of fission yeast mating genes reveals new autophagy factors. PLoS Genet. 9: e1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thewes S., 2014. Calcineurin-Crz1 signaling in lower eukaryotes. Eukaryot. Cell 13: 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachon L., Wood J., Kwon E. J. G., Laderoute A., Chatfield-Reed K., et al. , 2013. Functional characterization of fission yeast transcription factors by overexpression analysis. Genetics 194: 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viladevall L., Serrano R., Ruiz A., Domenech G., Giraldo J., et al. , 2004. Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J. Biol. Chem. 279: 43614–43624. [DOI] [PubMed] [Google Scholar]
- Wang H., Pathan N., Ethell I. M., Krajewski S., Yamaguchi Y., et al. , 1999. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science 284: 339–343. [DOI] [PubMed] [Google Scholar]
- Wapinski H., Pfeffer A., Friedman N., Regev A., 2007. Automatic genome-wide reconstruction of phylogenetic gene trees. Bioinformatics 23: i549–i558. [DOI] [PubMed] [Google Scholar]
- Wood V., Harris M. A., McDowall M. D., Rutherford K., Vaughan B. W., et al. , 2012. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 40: D695–D699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoritsune K., Matsuzawa T., Ohashi T., Takegawa K., 2013. The fission yeast Pvg1p has galactose-specific pyruvyltransferase activity. FEBS Lett. 587: 917–921. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Toda T., Yanagida M., 1994. A calcineurin-like gene ppb1+ in fission yeast: mutant defects in cytokinesis, cell polarity, mating and spindle pole body positioning. J. Cell Sci. 107: 1725–1735. [DOI] [PubMed] [Google Scholar]
- Yoshimoto H., Saltsman K., Gasch A. P., Li H. X., Ogawa N., et al. , 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277: 31079–31088. [DOI] [PubMed] [Google Scholar]
- Zakrzewska A., Boorsma A., Brul S., Hellingwerf J. K., Klis F. M., 2005. Transcriptional response of Saccharomyces cerevisiae to the plasma membrane-perturbing compound chitosan. Eukaryot. Cell 4: 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.-Q., Rao R., 2007. Global disruption of cell cycle progression and nutrient response by the antifungal agent amiodarone. J. Biol. Chem. 282: 37844–37853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The microarray expression data have been submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus Database (GSE77761). The ChIP-chip data also have been submitted to the NCBI Gene Expression Omnibus Database (GSE77761).