Abstract
Energy metabolism is traditionally considered a reactive homeostatic system addressing stage-specific cellular energy needs. There is however growing appreciation of metabolic pathways in the active control of vital cell functions. Case in point, the stem cell lifecycle – from maintenance and acquisition of stemness to lineage commitment and specification – is increasingly recognized as a metabolism-dependent process. Indeed, metabolic reprogramming is an early contributor to the orchestrated departure from or reacquisition of stemness. Recent advances in metabolomics have helped decipher the identity and dynamics of metabolic fluxes implicated in fueling cell fate choices by regulating the epigenetic and transcriptional identity of a cell. Metabolic cues, internal and/or external to the stem cell niche, facilitate progenitor pool restitution, long-term tissue renewal or ensure adoption of cytoprotective behavior. Convergence of energy metabolism with stem cell fate regulation opens a new avenue in understanding primordial developmental biology principles with future applications in regenerative medicine practice.
Keywords: Glycolysis, oxidative metabolism, nuclear reprograming, induced pluripotent stem cells, embryonic stem cells, hematopoietic stem cells, mitochondria, metabolic remodeling
1. Introduction
The resurgent interest in intermediary metabolism combined with advances in techniques for both the high throughput examination of the global metabolome and stable isotope tracing of specific metabolic pathways, has begun to resolve the metabolic fingerprint of the stem cell life cycle. While energy metabolism has long been considered a homeostatic system, responsive to match with high fidelity stage specific energetic demands, recent studies have revealed that metabolism can also feedback to regulate the epigenetic and transcriptional identity of a cell [1]. Therefore changes in energy metabolism and metabolite content may represent a physiological mechanism by which stem cells interact both locally within their niche and respond to changes in their systemic environment. Understanding these interactions will contribute to define how modulation of energy metabolism can be harnessed to regulate stem cell self-renewal and lineage-specific differentiation, critical concepts for cell biology with ramifications in regenerative medicine. This review highlights the major metabolic pathways associated with stem cell function and how they may fuel the transition between cellular fates.
2 Metabolism driving cell fate conversions
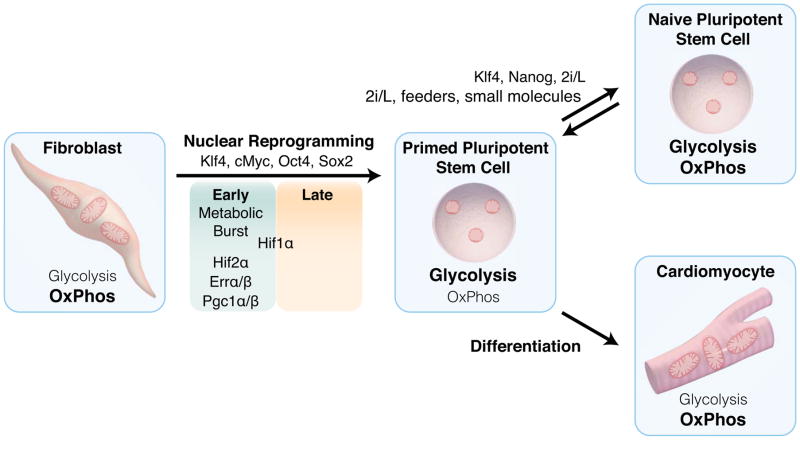
Inherent plasticity in the ability of cells to shift from one metabolic pathway to another, allows selection of the most appropriate pathway to match current energetic requirements. However an increasing body of evidence indicates that metabolic remodeling occurs early during transitions between cell states, prior to the establishment of the particular cell fate. The early transitions in energy metabolism may be required to energetically prime the cells to ensure they are competent to meet the metabolic demands of emerging cell fates, as well as to establish the epigenetic state required for a particular cell type. Several recent studies examining the temporal changes in the global gene and protein expression during nuclear reprogramming of somatic cells back to the pluripotent ground state have enabled the further characterization of this metabolism-dependent process. Indeed the first wave of gene induction during reprogramming is enriched for genes related to metabolism and proliferation in both mouse [2] and human systems, which precedes induction of embryonic genes and the core pluripotency network [3]. Temporal proteomic profiling has confirmed similar waves of protein expression changes during reprogramming, including stoichiometric changes of the mitochondrial electron transport chain within the initial wave [4]. This is characterized by a reduction in subunit expression of complex I and IV, and an increase expression of complex II, III, V of the mitochondrial electron transport chain [4, 5], which complements the early upregulation of glycolytic genes that precede induction of pluripotency genes [5–7].
Functionally, a metabolic burst of both oxidative and glycolytic metabolism occurs early in the reprogramming process in support of a temporary hyperenergetic state, following which oxidative metabolism declines, while glycolysis continues to increase [8]. The estrogen-related nuclear receptors, ERRα and ERRβ, and their co-activator PGC-1α/β are transiently expressed early in the reprogramming process and support the burst in oxidative metabolism. A Sca1−CD34− subpopulation with 10 and 7-fold higher expression of ERRγ and PGC-1β, respectively, display a 50-fold greater efficiency at generating iPSCs than the remaining cells [8]. Transcriptional analysis demonstrated a global upregulation of metabolic genes with oxidative phosphorylation genes representing the most significantly altered pathway in Sca1-CD34-, further supporting the importance of the hyperenergetic state for induction of pluripotency. Recent evidence also demonstrates a role of the oocyte factors Tcl1 and Tcl1b1 in supporting metabolic remodeling and enhancing reprogramming efficiency independent of changes in cell proliferation [9]. Mechanistically, Tcl1 increases Akt1 activity and may support an increased glycolysis, while Tcl1b1 suppresses the mitochondrial polynucleotide phosphorylase, thus impairing mitochondrial biogenesis and contributing to the switch from oxidative metabolism to glycolysis during reprogramming [9]. Remodeling of energy metabolism also plays a role in the transition between naïve and primed pluripotent stem cells (PSCs). Human naïve PSCs display higher oxygen utilization compared to their primed counterparts, displaying a similar bivalent use of both glycolysis and oxidative metabolism as mouse embryonic stem cells (ESCs) [10, 11]. Indeed the transition between these pluripotent states is associated with a remodeling of the metabolome, including changes in glycolysis, fatty acid and amino acid metabolism. This includes an elevated abundance of 1-methylnicotinamide, a product of nicotinamide N-methyltransferase, which is upregulated in the naïve state and consumes S-adenosylmethionine (SAM) making it unavailable for histone methylation reactions [12]. By competing for SAM availability, this suppresses methylation of repressive histone 3 lysine 27, thus linking cellular metabolism with the naïve state. Therefore, it appears that metabolic remodeling during acquisition of pluripotency is not simply a consequence of transition between cell identities, but may represent an initiating event.
3 Metabolic pathways fueling stem cell function and fate
3.1 Glycolysis
High metabolic flux through glycolysis is a common feature across a number of stem cell populations [13–21]. Indeed, stimulation of glycolysis itself or regulators of glycolysis facilitates the acquisition of the pluripotent state during nuclear reprogramming [5, 22–30] and supports the maintenance of stemness [16, 31–35]. Generation of induced pluripotent stem cells (iPSCs) stimulates a significant remodeling of the glycolytic pathway early during the reprogramming process [5], including an up regulation of genes for the initial steps of glucose uptake and phosphorylation, as well as the distal portions of the pathway [36]. Proteomic analysis has demonstrated a progressive increase in glycolysis during nuclear reprogramming [4], with greater expression of the majority of glycolytic enzymes observed in the resultant iPSCs compared to parental cells, and resemble their embryonic counterparts [5]. PSCs also display an isoform switch from hexokinase I to II [5], which supports elevated glycolytic flux due to the twin catalytic domains of this isoform as well as the preferential access to mitochondrial ATP production and reduction in product inhibition by glucose-6-phosphate due to its mitochondrial localization [20, 37].
The glycolytic phenotype is not limited to PSCs but is also characteristic of a number of tissue specific stem cell populations, such as long-term hematopoietic stem cells (LT-HSCs) [17, 35] and mesenchymal stem cells (MCSs) [18]. LT-HSCs are maintained in a quiescent slow cycling state to ensure long-term tissue renewal by protecting them from life-long accumulation of cellular damage and mutations [38]. Compared either to whole bone marrow or their committed progenitors, HSCs display a metabolic profile consistent with elevated glycolytic flux including high levels of fructose-1,6-bisphosphate and pyruvate, the products of the rate limiting steps of glycolysis, in line with high pyruvate kinase activity [17, 35]. MSCs also have a greater dependence on glycolysis due to elevated expression of glycolytic enzymes compared to their differentiated osteoblasts [18]. HSCs are dependent on expression of pyruvate dehydrogenase kinase (PDK) 2 and 4, which phosphorylate and inactivate pyruvate dehydrogenase thus preferentially shunting pyruvate to lactate production instead of entering the tricarboxylic acid cycle for further oxidation. Indeed deletion of PDK2 and 4 results in reduced dependence on glycolysis and leads to HSC functional exhaustion, including loss of quiescence and transplantation capacity [35]. Restoration of glycolysis in deficient HSCs through either PDK overexpression or pharmacologic inhibition of mitochondrial pyruvate entry was sufficient to restore glycolysis in defective HSCs and reinitiate cell cycle quiescence and reconstitution capacity [35], thus supporting the critical importance of glycolysis for maintenance of HSC function and fate.
3.1.1 Hypoxia and Hypoxia-inducible factors in regulating glycolysis in stem cells
A number of transcriptional and signaling pathways converge to regulate glycolysis in stem cell populations, with hypoxia and hypoxia-inducible factor 1α (HIF1α) and 2α (HIF2α) pathway being the well characterized (Ruohola-Baker this issue). Under normoxic conditions, oxygen-regulated prolyl hydroxylases hydroxylate proline residues in HIFs, thereby priming these proteins to undergo ubiquitination and proteasomal degradation [39, 40]. Thus hypoxia stabilizes HIF1α and 2α though suppression of prolyl hydroxylation, allowing them to form nuclear heterodimers with HIF1β and bind to hypoxia response elements in the promoter regions of genes, including many glycolytic genes such as GLUT1, LDHA and PDKs [38–40]. Indeed, many tissue specific stem cells reside within in vivo niches with low oxygen tensions, which maintain these cells in a glycolytic quiescent state to ensure tissue regenerative capacity by limiting oxidative metabolism generation of reactive oxygen species (ROS) and subsequent accumulation of ROS-induce cellular damage [32, 38, 41, 42]. In the hematopoietic system, loss of HIF1α results in decreased glycolytic flux leading to impaired HSC quiescence and eventual HSC exhaustion [43], implicate the importance in HIF1α for HSC maintenance. This phenotype can be rescued by overexpression of PDK2/4 or using 1-aminoethylphosphonic acid to promote glycolysis and reestablish stem cell quiescence [35]. HIF1α can also be transcriptionally activated in HSCs by MEIS1 leading to elevated HIF activity and downstream functions [17]. Some of HIF1α action on glycolysis may also be mediated via transcriptional activation of CRIPTO, which binds its receptor GRP78 to stimulate glycolytic proteins [44, 45]. MSCs and neuronal stem cells, which normally reside within an in vivo hypoxic niche, also lose hypoxia-dependent quiescence when cultured in normoxia resulting in accelerated proliferation associated with oxidative metabolism and lost of stem cell function due to increased cellular senescence [46–48].
Embryonic stem cells (ESCs) within the blastocyst inner cell mass also reside in a hypoxic environment of the lumen of the oviduct and uterus with oxygen tension ranging from 1.5 to 9% [49]. Although PSCs can be propagated in high oxygen in vitro, a hypoxic environment promotes glycolysis [50] and contributes to their stemness maintenance and self-renewal capacity [31, 32, 51, 52]. Indeed hypoxia alone is sufficient to de-differentiate early lineage committed progeny from ESCs and iPSCs back to a self-renewing pluripotent state and is associated with enrichment of HIF1α/HIF2α target genes and restoration of a glycolytic phenotype [53]. Consistent with this observation, stabilization of HIF1α and HIF2α during early reprogramming of somatic cells to iPSCs supports an early glycolytic shift through activation of their target genes PDK1-3 and pyruvate kinase isoform M2 [6, 7]. However, HIF2α stabilization after day 12 of reprogramming impairs reprogramming efficiency partially due to upregulation of TNF-related apoptosis-inducing ligand [6]. HIF1α-induced glycolytic genes are also critical for the transition between ESCs and epiblast stem cells (EpiSCs), the later of which display a characteristic HIF1α gene expression profile associated with a greater reliance on glycolysis for energy production [11]. Stabilization of HIF1α chemically or through expression of a non-degradable isoform in mouse ESCs shifted metabolism to a more glycolytic phenotype and increased the percentage of EpiSC-like colonies [11]. Therefore hypoxia and signaling through the HIF pathway appears to be a critical rheostat in directing the stem cell fate and function through regulation of glycolysis.
3.1.2 Glycolysis fuels stemness
While the reliance on glycolysis in tissue specific stem cells fits well with the lower energetic demands of the quiescence state and helps to limit oxidative metabolism-dependent generation of ROS and mitigate accumulation of ROS-induce cellular damage to ensure life-long tissue renewal, the importance of utilizing glycolysis in highly proliferative PSCs is less obvious. Yet, a glycolytic phenotype appears to be a consistent feature of highly proliferative cells [54, 55]. While glycolysis is inherently less efficient in terms of energy generation, producing only a fraction of the 36 ATP that can be generated via complete oxidation of glucose, it enables a faster rate of ATP generation and in some cases can outpace ATP production from oxidative metabolism [56, 57]. Generation of ATP does not appear to be limiting in proliferating cells as glycolytic cells maintain high ATP/ADP ratios even when then are stimulated to divide at an accelerated rate [58, 59]. Consistent with this observation is that cancer cells can utilize an alternative pathway to metabolize phosphoenolpyruvate to pyruvate that supports anabolism yet uncouples glycolysis from PKM-dependent ATP production [55, 60]. Therefore in the presence of abundant resources, for instance in cell culture where stem cells are bathed in high levels of glucose, utilization of glycolysis may be advantageous as it maintains pools of carbon intermediates required for biosynthesis of cellular contents to enable the generation of new daughter cells [54, 55]. Glycolytic and pentose phosphate pathway intermediates serve as precursors for a number of essential biosynthetic pathways including purine and pyrimidine nucleotides, amino acids and triacyclglycerols (Fig. 1). At least half of the carbon atoms required for de novo purine and pyrimidine synthesis are derived from the pentose phosphate pathway intermediate ribose-5-phosphate, which is subsequently activated to 5-phosphoribosyl-α-pyrophosphate. Meanwhile, carbon atoms for non-essential amino acid synthesis are directly derived from glycolytic intermediates, with cysteine, glycine and serine being synthesized from 3-phosphoglycerate and alanine synthesized from pyruvate. Dihydroxyacetone phosphate also serves as a precursor for glycerol-3-phosphate, which is critical for triacylglycerol and phospholipid biosynthesis. The detailed anabolic requirements of proliferating cells and the economics of macromolecule biosynthesis have been extensively reviewed elsewhere [55].
Fig. 1.
Metabolic remodeling of glycolysis and oxidative metabolism fuels acquisition, maintenance and departure from pluripotency. Nuclear reprogramming of somatic cells through induction of reprogramming factors induces dramatic metabolic remodeling, characterized by an initial metabolic burst of both glycolysis and oxidative metabolism, following which oxidative metabolism declines, while glycolysis continues to increase. Conversion between the primed and naïve pluripotent states in human cells is also associated with changes in the metabolic infrastructure in support of a transition from predominantly glycolytic to bivalent metabolism utilizing both glycolysis and oxidative metabolism. In contrast, upon a differentiation stimulus, pluripotent stem cells activate mitochondria biogenesis to generate a robust network of mitochondria in support of oxidative metabolism to match lineage-specific energetic demands.
3.2 Hexosamine Biosynthesis Pathway
Alternatively a small proportion of glucose can be shunted into the hexosamine biosynthesis pathway to generate UDP-N-acetylglucosamine (UDP-GlcNAc), a substrate for post-translational modification of proteins with O-linked-N-acetylglucosamine (O-GlcNAc) [61]. This pathway acts as a site of convergence of carbohydrate, amino acid, fatty acid and nucleotide metabolism, making it a critical nutrient sensing pathway to link metabolic status with cell function and fate. Members of the core pluripotency network and nuclear reprogramming factors, including Oct4, Sox2 and cMyc, actively undergo O-GlcNAcylation [62, 63]. These modifications have direct impact on stem cell function as PSC self-renewal and nuclear reprogramming is impaired by either reducing UDP-GlcNAc generation by limiting glucose availability or blocking O-GLcNAc transferase (OGT), while stem cell differentiation is blocked by increases in global O-GlcNAc levels [62]. OGT may also mediate its impact on ESCs by binding and GlcNAcylation of ten-eleven translocation (TET) proteins TET1 and TET2 [64, 65], which catalyze the conversion of 5-methylcytosine (5mC) to 5-hydroxylmethylcytosine (5hmC) and leads to DNA demethylation, particularly in CpG rich promoter regions [64]. The OGT-TET complex may also help to recruit repressive complexes, such as Sin3A and NuRD, to suppress expression of developmental genes and maintain the pluripotent state [65]. Hematopoietic cells may also utilize glucose dependent UDP-GlcNAc for N-glycosylation to regulate growth factor surface expression to coordinate glucose and glutamine metabolism and stimulate cell growth and proliferation [66].
3.3 Mitochondrial function and metabolism
3.3.1 Mitochondria structure and function in stem cells
Associated with the greater dependence on glycolysis for stemness maintenance, stem cells display immature mitochondrial infrastructure and may repurpose their mitochondria away from canonical ATP generators. In this context, stem cells largely express low copy numbers of mitochondrial DNA (St. John this issue) and display a sparse mitochondria infrastructure that has been proposed to represent a marker of stemness, consisting of immature spherical structures with poorly developed cristae that are predominantly localized in the perinuclear space [13, 15, 18, 67–73]. As a consequence of this immature mitochondrial infrastructure, stem cells have lower levels of mitochondria respiration and reduced oxidative reserve capacity compared to their differentiated counterparts [5, 14–19]. Tissue specific stem cells may minimize their reliance on mitochondria function due to lower energetic demands of the quiescent state and to reduce the potentially toxic effect of ROS. LT-HSCs can be isolated from whole bone marrow by gating for low mitochondrial membrane potential, and display greater colony formation capacity and bone marrow reconstitution capacity compared to the low potential subpopulation [17]. Consistent with a lower requirement for oxidative metabolism in HSCs, depletion of the mitochondrial protein tyrosine phosphatase (PTPMT1) leads to reduced oxidative capacity and an expansion of the HSCs pool and an associated block in stem cell differentiation that leads to hematopoietic failure [74]. While simulation of mitochondrial biogenesis through deletion of the tuberous sclerosis complex results in impaired HSCs quiescence and HSC pool maintenance, as well as defective hematopoiesis, which can be restored with rapamycin treatment [75]. Indeed, accumulation of mitochondrial DNA mutations due to expression of a proofreading defective mitochondrial DNA polymerase (POLG), leads to impaired mitochondrial function associated with a premature aging hematopoietic phenotype [76]. While the mtDNA mutations result in distinct differentiation blocks, it had little functional effect on maintenance of the HSC pool, further supporting the divergent role of mitochondria in HSCs versus progenitors [76]. However, some level of mitochondrial function is required for maintenance of HSC function, as loss of FOXO3 function, resulting in impaired mitochondrial respiration and elevated glycolysis, ultimately leads to loss of HSC quiescence [77]. Mitochondrial features may be a consistent indication of the differentiation capacity of tissue specific stem cells as cardiac progenitor cells display efficient cardiomyocyte differentiation when abundant mitochondria are available compared to poor differentiation capacity in cells with few mitochondria [78]. Manipulation of the mitochondrial infrastructure by stimulating mitochondria biogenesis was sufficient to overcome this differentiation block [78].
While mitochondrial structure and function appear to be critical regulators of stem cell function and fate, they may play divergent roles in pluripotent versus tissue specific stem cells. Despite the immature nature of stem cell mitochondrial structure/function and evidence that autophagic clearance of mitochondria is critical for induction of pluripotency [79], impaired mitochondrial homeostasis is associated with loss of stemness properties. For example, knockdown of POLG in ESCs leads to loss of pluripotency and induction of differentiation [80], while knockdown of growth factor erv1-like in ESCs induces expression of GTPase dynamin-related 1 (Drp1) leading to excessive mitochondrial fission and loss of pluripotency, impaired differentiation capacity and a reduction in cell viability [81]. In addition, stem-like cells asymmetrically segregate their mitochondria between daughter cells, with daughter cells receiving a greater proportion of young mitochondria maintaining their stem cell traits, while impaired segregation caused by disruption of mitochondria fission results in the loss of stem cell characteristics [82]. In addition, high mitochondrial membrane potential has been linked with stem cell properties [15, 71, 83, 84] and may be actively maintained through hydrolysis of glycolytically-derived ATP by ATP synthase as electron transport activity is reduced in PSCs [85]. The role of high mitochondrial membrane potential plays in maintenance of the pluripotent state remains unresolved, but it has been proposed to help maintain a more fragmented mitochondrial network [86, 87], maintain optimal redox potential for lipid and amino acid synthesis [88] or to prime stem cells to quickly respond to the energetic demands of cellular differentiation [21]. Indeed, mitochondrial potential may represent a potential tool to select reprogramming intermediates [5] and robust stem cell populations for cell therapy [17, 89]. ESCs sorted into low and high mitochondrial membrane potential subpopulations have similar expression of pluripotent markers yet display distinct differentiation capacity with the high subpopulation more efficiently forming teratomas (a defining trait of PSCs) than their low subpopulation counterparts [90]. In addition, cells undergoing nuclear reprogramming also obtain a high mitochondrial potential associated with the pluripotent state [5]. Therefore PSCs appear to actively regulate mitochondrial distribution and activity in support of mitochondrial functions beyond canonical oxidative metabolism that are critical for stemness maintenance.
3.3.2 Tricarboxylic acid cycle and catapleurosis
Typical of a proliferative phenotype, PSCs avidly need anabolic precursors to ensure the de novo synthesis of cell constituents, which competes with complete substrate oxidation within the mitochondrial tricarboxylic acid cycle (TCA) [55]. To support stemness, stem cells can commission cataplerosis, extracting partially oxidized mitochondria substrates for anabolic purposes [91]. In fact, ESCs incompletely oxidize pyruvate in the TCA to generate exportable intermediates [92]. In this way, citrate is exported from mitochondria, processed by ATP-citrate lyase to form cytosolic acetyl-CoA, which serves as substrate for protein/histone acetylation along with fatty acid/cholesterol biosynthesis [93]. Initial ESCs differentiation is linked to reduced acetyl-CoA production and loss of histone H3K9 and H3K27 acetylation, suggesting that TCA-derived cytosolic acetyl-CoA facilitates histone acetylation and an open chromatin state [92]. Inhibition of ATP citrate lyase compromises acetyl-CoA content and histone acetylation enabling myogenic differentiation [94], while acetate excess blunts early differentiation and histone deacetylation [92]. Not exclusive to pluripotency, ATP-citrate lyase also contributes to adipogenic differentiation proficiency [93].
Glutamine is also a major energy substrate for stem cells and can also contribute its carbons to the TCA cycle in support of stem cell function and fate regulation. Case in point, in hematopoietic lineage specification, use of glutaminolysis and nucleotide biosynthesis versus glucose catabolism selects for erythroid versus myeloid fates [95]. Glutaminolysis contributes to early erythroid commitment but is not required for lineage maintenance, suggesting that specific metabolic states enable and initiate differentiation along specific lineages, not simply matching the energetic demands of the lineage destination [95, 96]. Both glucose and glutamine be utilized by ESCs to generate alpha-ketoglutarate – a distinct metabolite-linking metabolism with stemness regulation [97]. Alpha-ketoglutarate serves as a substrate for a number of enzymes, including HIFα prolyl hydroxylase and alpha-ketoglutarate dependent dioxygenases that include Jumonji C-domain-containing histones demethylases and the ten-eleven translocation family of DNA demethylases. A high alpha-ketoglutarate/succinate ratio thus favors demethylation of repressive chromatin marks including H3K9/K23. How these TCA cycle intermediates contribute to stemness acquisition during nuclear reprogramming, where changes in DNA methylation and histone marks are robust, remains unexplored.
3.3.3 Thr/single carbon metabolism
The metabolic state of PSCs is associated with a high requirement for the catabolism of specific amino acids. For example removal of threonine from cell culture media [98] or pharmacologic inhibition of threonine dehydrogenase [99] leads to loss of stemness, cell cycle arrest and cell death in mouse PSCs, while threonine dehydrogenase induction promotes pluripotent induction through nuclear reprogramming [100]. Indeed, threonine dehydrogenase and downstream enzymes in threonine catabolism, including glycine C-acetyltransferase (GCAT) and glycine decarboxylase (GLDC) are highly expressed in mouse PSCs and are quickly suppressed during stem cell differentiation [98, 101]. This pathway is critical for coupling the breakdown of threonine with supplying single carbon equivalents to the folate pool. The folate pool can then donate these carbon equivalents to a number of anabolic pathways including the biosynthesis of purine nucleotides, which is consistent with the observation that DNA synthesis is suppressed when threonine is removed during mouse ESC culture [98]. In addition, threonine catabolism also helps maintains a high SAM to S-adenosylhomocysteine ratio to promote histone 3 lysine 4 methylation, and ultimately support proliferation and self-renewal of mouse PSCs [101]. In an analogous fashion, human PSCs rely directly on methionine catabolism by methionine adenosyltransferase to support the SAM productio as threonine dehydrogenase is only expressed as a non-functional pseudogene in humans [102].
3.3.4 Fatty acid metabolism
Beyond glucose and glutamine, the metabolism of fatty acids can contribute to both catabolic energy generation and anabolic precursor generation. Although fatty acid oxidation is critical for embryonic development and for specific cell lineages, its role in stem cell biology has been relatively unexplored. Initial work has demonstrated that peroxisome proliferator-activated receptor (PPAR)δ is highly expressed in HSCs and is associated with high rates of fatty acid oxidation, which is critical for maintaining the balance between HSC self-renewal and lineage specification [103]. Indeed genetic depletion of PPARδ or its upstream regulator promyelocytic leukaemia protein (PML) or pharmacologic inhibition of fatty acid oxidation stimulated the exit of HSCs from quiescence and lead to their symmetric commitment into lineage specified daughter cells. This leads to a short-term expansion of HSCs followed by exhaustion of this compartment, which ultimately impairs the reconstitution capacity of these cells. In contrast, activation of this pathway using PPARδ agonists promotes asymmetric cell division to support HSC maintenance and self-renewal and was sufficient to rescue the quiescence and maintenance defects in PML−/− cells [103]. Therefore regulation of fatty acid oxidation may play a critical role partitioning cells between self-renewal and lineage specification.
Lipid biosynthesis also appears critical to maintain stem and progenitor cell proliferation in the adult brain [104]. Fatty acid synthase (FAS), the rate-limiting enzyme for fatty acid synthesis, is highly expressed in neural stem/progenitor cells (NSPCs) within active regions of neurogenesis, and endows these cells with a distinct metabolic phenotype in support of de novo lipogenesis and the generation of new lipid membranes required for cell division. Deletion or inhibition of FAS impairs NSPC proliferation resulting in fewer NSPCs in neurogenic zones and less newly formed neurons [104]. Rates of lipid synthesis are dependent both on FAS activity, as well as the generation of its substrate, malonyl-CoA, by acetyl-CoA carboxylase (ACC). Indeed, Mid1-interacting protein (Mig12), an activator of ACC, and Spot14/thyroid hormone responsive protein, which dimerizes with Mig12 to prevent ACC activation, are also critical for regulating neurogenic NSPCs, thus further supporting the importance of de novo lipogenesis. Overexpression of Spot14 impairs NSPCs proliferation, associated with reduced malonyl-CoA levels and impaired lipogenesis from glucose and acetate, however this phenotype can be rescued by overexpressing Mig12 [104]. Therefore Spot14 may ensure neural stem cell quiescence by reducing lipogenesis and placing a metabolism-dependent brake on neurogenesis [105]. Interestingly, malonyl-CoA also regulates long-chain fatty acid β-oxidation by serving as an endogenous inhibitor of carnitine palmitoyl transferase 1 (CPT1), the rate-limiting enzyme for long-chain fatty acid transport into the mitochondria. Therefore by regulating the equilibrium between fatty acid oxidation and lipogenesis, malonyl-CoA may serve as a rheostat of stem cell function and fate [105]. In this context, naïve PSCs oxidize fatty acids as a source of energy, while their primed counterparts do not oxidize fatty acids and shift to fatty acid synthesis [12].
4 Summary
Recent advances in metabolomics have unmasked a fundamental role for intermediary metabolism in securing the fitness of stemness acquisition and maintenance. This underappreciated dimension of metabolism leverages an inherent metabolic plasticity to pre-emptively ‘condition’ the epigenetic and transcriptional landscape for cell fate conversions and matches the anabolic and catabolic demands of the destination cell fate. In turn, modulation of energy metabolism emerges as a legitimate target to optimize stem cell self-renewal and lineage-specific differentiation. The impact of metabolism on stem cell function and fate has potential implications across aging and disease, thus this initial dissection of underlying metabolic mechanisms paves the way to a more comprehensive appreciation of stem cell biology with potential applications for regenerative medicine.
Fig. 2.
Common intermediary metabolic pathways that proliferating stem cells utilize to match the anabolic demands of cell division. Rapid proliferation places unique demands on the cell, with the requirement for anabolic precursors, such as nucleotides, amino acids and lipids, outpacing that of ATP. Therefore, stem cells extract glycolytic intermediates and repurpose their mitochondria from efficient energy generators to cataplerotic engines, in order to supply hydrocarbons for the generation of cellular biomass. ACP acyl carrier protein, CPT1 carnitine palmitoyl transferas 1, dATP deoxyadenosine triphosphate, dCTP deoxycytidine triphosphate, dGTP deoxyguanosine triphosphate, dTTP deoxythymidine triphosphate, IMP inosine monophosphate, PRPP 5-phosphoribosyl-α-pyrophosphate, UMP uridine monophosphate, THF tetrahydrofolate.
Acknowledgments
This work was supported by grants from the National Institutes of Health (K99-HL121079), Leducq Foundation, Marriott Foundation and Mayo Clinic Center for Regenerative Medicine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, et al. A Molecular Roadmap of Reprogramming Somatic Cells into iPS Cells. Cell. 2012;151:1617–32. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cacchiarelli D, Trapnell C, Ziller MJ, Soumillon M, Cesana M, Karnik R, et al. Integrative Analyses of Human Reprogramming Reveal Dynamic Nature of Induced Pluripotency. Cell. 2015;162:412–24. doi: 10.1016/j.cell.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, et al. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2012;2:1579–92. doi: 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–71. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathieu J, Zhou W, Xing Y, Sperber H, Ferreccio A, Agoston Z, et al. Hypoxia-inducible factors have distinct and stage-specific roles during reprogramming of human cells to pluripotency. Cell Stem Cell. 2014;14:592–605. doi: 10.1016/j.stem.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prigione A, Rohwer N, Hoffmann S, Mlody B, Drews K, Bukowiecki R, et al. HIF1alpha modulates cell fate reprogramming through early glycolytic shift and upregulation of PDK1-3 and PKM2. Stem Cells. 2014;32:364–76. doi: 10.1002/stem.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kida YS, Kawamura T, Wei Z, Sogo T, Jacinto S, Shigeno A, et al. ERRs Mediate a Metabolic Switch Required for Somatic Cell Reprogramming to Pluripotency. Cell Stem Cell. 2015;16:547–55. doi: 10.1016/j.stem.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khaw SL, Min-Wen C, Koh CG, Lim B, Shyh-Chang N. Oocyte Factors Suppress Mitochondrial Polynucleotide Phosphorylase to Remodel the Metabolome and Enhance Reprogramming. Cell Rep. 2015;12:1080–8. doi: 10.1016/j.celrep.2015.07.032. [DOI] [PubMed] [Google Scholar]
- 10.Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–69. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, et al. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–16. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nature cell biology. 2015;17:1523–35. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–71. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park do J, et al. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2006;348:1472–8. doi: 10.1016/j.bbrc.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Chung S, Dzeja PP, Faustino RS, Perez-Terzic C, Behfar A, Terzic A. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat Clin Pract Cardiovasc Med. 2007;4(Suppl 1):S60–7. doi: 10.1038/ncpcardio0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondoh H, Lleonart ME, Nakashima Y, Yokode M, Tanaka M, Bernard D, et al. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 2007;9:293–9. doi: 10.1089/ars.2006.1467. [DOI] [PubMed] [Google Scholar]
- 17.Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7:380–90. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 2008;26:960–8. doi: 10.1634/stemcells.2007-0509. [DOI] [PubMed] [Google Scholar]
- 19.Turner WS, Seagle C, Galanko JA, Favorov O, Prestwich GD, Macdonald JM, et al. Nuclear magnetic resonance metabolomic footprinting of human hepatic stem cells and hepatoblasts cultured in hyaluronan-matrix hydrogels. Stem Cells. 2008;26:1547–55. doi: 10.1634/stemcells.2007-0863. [DOI] [PubMed] [Google Scholar]
- 20.Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CA, Ramalho-Santos J, et al. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Folmes CD, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11:596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, et al. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–5. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–41. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–20. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 25.Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature. 2009;460:1085–6. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–53. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–4. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–5. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–8. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–77. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci USA. 2005;102:4783–8. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7:150–61. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Varum S, Momcilovic O, Castro C, Ben-Yehudah A, Ramalho-Santos J, Navara CS. Enhancement of human embryonic stem cell pluripotency through inhibition of the mitochondrial respiratory chain. Stem Cell Res. 2009;3:142–56. doi: 10.1016/j.scr.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–9. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takubo K, Nagamatsu G, Kobayashi CI, Nakamura-Ishizu A, Kobayashi H, Ikeda E, et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell. 2013;12:49–61. doi: 10.1016/j.stem.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prigione A, Lichtner B, Kuhl H, Struys EA, Wamelink M, Lehrach H, et al. Human iPSCs Harbor Homoplasmic and Heteroplasmic Mitochondrial DNA Mutations While Maintaining hESC-Like Metabolic Reprogramming. Stem Cells. 2011 doi: 10.1002/stem.683. [DOI] [PubMed] [Google Scholar]
- 37.Bustamante E, Pedersen PL. High aerobic glycolysis of rat hepatoma cells in culture: role of mitochondrial hexokinase. Proc Natl Acad Sci. 1977;74:3735–9. doi: 10.1073/pnas.74.9.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9:298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 39.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–40. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 40.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 41.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15:243–56. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perales-Clemente E, Folmes CD, Terzic A. Metabolic regulation of redox status in stem cells. Antioxid Redox Signal. 2014;21:1648–59. doi: 10.1089/ars.2014.6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 44.Miharada K, Karlsson G, Rehn M, Rorby E, Siva K, Cammenga J, et al. Cripto regulates hematopoietic stem cells as a hypoxic-niche-related factor through cell surface receptor GRP78. Cell Stem Cell. 2011;9:330–44. doi: 10.1016/j.stem.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 45.Miharada K, Karlsson G, Rehn M, Rorby E, Siva K, Cammenga J, et al. Hematopoietic stem cells are regulated by Cripto, as an intermediary of HIF-1alpha in the hypoxic bone marrow niche. Ann N Y Acad Sci. 2012;1266:55–62. doi: 10.1111/j.1749-6632.2012.06564.x. [DOI] [PubMed] [Google Scholar]
- 46.Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–39. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pattappa G, Heywood HK, de Bruijn JD, Lee DA. The metabolism of human mesenchymal stem cells during proliferation and differentiation. J Cell Physiol. 2011;226:2562–70. doi: 10.1002/jcp.22605. [DOI] [PubMed] [Google Scholar]
- 48.Pattappa G, Thorpe SD, Jegard NC, Heywood HK, de Bruijn JD, Lee DA. Continuous and uninterrupted oxygen tension influences the colony formation and oxidative metabolism of human mesenchymal stem cells. Tissue engineering Part C. Methods. 2013;19:68–79. doi: 10.1089/ten.TEC.2011.0734. [DOI] [PubMed] [Google Scholar]
- 49.Fischer B, Bavister BD. Oxygen tension in the oviduct and uterus of rhesus monkeys, hamsters and rabbits. J Reprod Fertil. 1993;99:673–9. doi: 10.1530/jrf.0.0990673. [DOI] [PubMed] [Google Scholar]
- 50.Forristal CE, Wright KL, Hanley NA, Oreffo RO, Houghton FD. Hypoxia inducible factors regulate pluripotency and proliferation in human embryonic stem cells cultured at reduced oxygen tensions. Reproduction. 2010;139:85–97. doi: 10.1530/REP-09-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Powers DE, Millman JR, Huang RB, Colton CK. Effects of oxygen on mouse embryonic stem cell growth, phenotype retention, and cellular energetics. Biotechnol Bioeng. 2008;101:241–54. doi: 10.1002/bit.21986. [DOI] [PubMed] [Google Scholar]
- 52.Westfall SD, Sachdev S, Das P, Hearne LB, Hannink M, Roberts RM, et al. Identification of oxygen-sensitive transcriptional programs in human embryonic stem cells. Stem Cells Dev. 2008;17:869–81. doi: 10.1089/scd.2007.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mathieu J, Zhang Z, Nelson A, Lamba DA, Reh TA, Ware C, et al. Hypoxia induces reentry of committed cells into pluripotency. Stem Cells. 2013;31:1737–48. doi: 10.1002/stem.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–64. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 56.Guppy M, Greiner E, Brand K. The role of the Crabtree effect and an endogenous fuel in the energy metabolism of resting and proliferating thymocytes. Eur J Biochem. 1993;212:95–9. doi: 10.1111/j.1432-1033.1993.tb17637.x. [DOI] [PubMed] [Google Scholar]
- 57.Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–7. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- 58.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 59.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 60.Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–9. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanover JA, Krause MW, Love DC. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol. 2012;13:312–21. doi: 10.1038/nrm3334. [DOI] [PubMed] [Google Scholar]
- 62.Jang H, Kim TW, Yoon S, Choi SY, Kang TW, Kim SY, et al. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell. 2012;11:62–74. doi: 10.1016/j.stem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 63.Chou TY, Dang CV, Hart GW. Glycosylation of the c-Myc transactivation domain. Proc Natl Acad Sci U S A. 1995;92:4417–21. doi: 10.1073/pnas.92.10.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49:645–56. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 65.Shi FT, Kim H, Lu W, He Q, Liu D, Goodell MA, et al. Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J Biol Chem. 2013;288:20776–84. doi: 10.1074/jbc.M113.460386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, et al. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 2010;24:2784–99. doi: 10.1101/gad.1985910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lonergan T, Brenner C, Bavister B. Differentiation-related changes in mitochondrial properties as indicators of stem cell competence. J Cell Physiol. 2006;208:149–53. doi: 10.1002/jcp.20641. [DOI] [PubMed] [Google Scholar]
- 68.Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–33. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- 69.Suhr ST, Chang EA, Tjong J, Alcasid N, Perkins GA, Goissis MD, et al. Mitochondrial rejuvenation after induced pluripotency. PLoS One. 2010;5:e14095. doi: 10.1371/journal.pone.0014095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lonergan T, Bavister B, Brenner C. Mitochondria in stem cells. Mitochondrion. 2007;7:289–96. doi: 10.1016/j.mito.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Armstrong L, Tilgner K, Saretzki G, Atkinson SP, Stojkovic M, Moreno R, et al. Human induced pluripotent stem cell lines show similar stress defence mechanisms and mitochondrial regulation to human embryonic stem cells. Stem Cells. 2010;28:661–73. doi: 10.1002/stem.307. [DOI] [PubMed] [Google Scholar]
- 72.Facucho-Oliveira JM, St John JC. The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev Rep. 2009;5:140–58. doi: 10.1007/s12015-009-9058-0. [DOI] [PubMed] [Google Scholar]
- 73.St John JC, Ramalho-Santos J, Gray HL, Petrosko P, Rawe VY, Navara CS, et al. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells. 2005;7:141–53. doi: 10.1089/clo.2005.7.141. [DOI] [PubMed] [Google Scholar]
- 74.Yu WM, Liu X, Shen J, Jovanovic O, Pohl EE, Gerson SL, et al. Metabolic regulation by the mitochondrial phosphatase PTPMT1 is required for hematopoietic stem cell differentiation. Cell Stem Cell. 2013;12:62–74. doi: 10.1016/j.stem.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Zheng P. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205:2397–408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Norddahl GL, Pronk CJ, Wahlestedt M, Sten G, Nygren JM, Ugale A, et al. Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell. 2011;8:499–510. doi: 10.1016/j.stem.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 77.Rimmele P, Liang R, Bigarella CL, Kocabas F, Xie J, Serasinghe MN, et al. Mitochondrial metabolism in hematopoietic stem cells requires functional FOXO3. EMBO Rep. 2015;16:1164–76. doi: 10.15252/embr.201439704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.San Martin N, Cervera AM, Cordova C, Covarello D, McCreath KJ, Galvez BG. Mitochondria determine the differentiation potential of cardiac mesoangioblasts. Stem Cells. 2011;29:1064–74. doi: 10.1002/stem.654. [DOI] [PubMed] [Google Scholar]
- 79.Ma T, Li J, Xu Y, Yu C, Xu T, Wang H, et al. Atg5-independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. Nat Cell Biol. 2015;17:1379–87. doi: 10.1038/ncb3256. [DOI] [PubMed] [Google Scholar]
- 80.Facucho-Oliveira JM, Alderson J, Spikings EC, Egginton S, St John JC. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J Cell Sci. 2007;120:4025–34. doi: 10.1242/jcs.016972. [DOI] [PubMed] [Google Scholar]
- 81.Todd LR, Damin MN, Gomathinayagam R, Horn SR, Means AR, Sankar U. Growth factor erv1-like modulates Drp1 to preserve mitochondrial dynamics and function in mouse embryonic stem cells. Mol Biol Cell. 2010;21:1225–36. doi: 10.1091/mbc.E09-11-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katajisto P, Dohla J, Chaffer CL, Pentinmikko N, Marjanovic N, Iqbal S, et al. Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science. 2015;348:340–3. doi: 10.1126/science.1260384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prigione A, Hossini AM, Lichtner B, Serin A, Fauler B, Megges M, et al. Mitochondrial-associated cell death mechanisms are reset to an embryonic-like state in aged donor-derived iPS cells harboring chromosomal aberrations. PLoS One. 2011;6:e27352. doi: 10.1371/journal.pone.0027352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mah N, Wang Y, Liao MC, Prigione A, Jozefczuk J, Lichtner B, et al. Molecular insights into reprogramming-initiation events mediated by the OSKM gene regulatory network. PLoS One. 2011;6:e24351. doi: 10.1371/journal.pone.0024351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 2011;30:4860–73. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mattenberger Y, James DI, Martinou JC. Fusion of mitochondria in mammalian cells is dependent on the mitochondrial inner membrane potential and independent of microtubules or actin. FEBS Lett. 2003;538:53–9. doi: 10.1016/s0014-5793(03)00124-8. [DOI] [PubMed] [Google Scholar]
- 87.Teslaa T, Teitell MA. Pluripotent stem cell energy metabolism: an update. EMBO J. 2015;34:138–53. doi: 10.15252/embj.201490446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shyh-Chang N, Zheng Y, Locasale JW, Cantley LC. Human pluripotent stem cells decouple respiration from energy production. EMBO J. 2011;30:4851–2. doi: 10.1038/emboj.2011.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sukumar M, Liu J, Mehta GU, Patel SJ, Roychoudhuri R, Crompton JG, et al. Mitochondrial Membrane Potential Identifies Cells with Enhanced Stemness for Cellular Therapy. Cell Metab. 2015 doi: 10.1016/j.cmet.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schieke SM, Ma M, Cao L, McCoy JP, Jr, Liu C, Hensel NF, et al. Mitochondrial metabolism modulates differentiation and teratoma formation capacity in mouse embryonic stem cells. J Biol Chem. 2008;283:28506–12. doi: 10.1074/jbc.M802763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol. 2012;13:270–76. doi: 10.1038/nrm3305. [DOI] [PubMed] [Google Scholar]
- 92.Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402. doi: 10.1016/j.cmet.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 93.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–80. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bracha AL, Ramanathan A, Huang S, Ingber DE, Schreiber SL. Carbon metabolism-mediated myogenic differentiation. Nat Chem Biol. 2010;6:202–4. doi: 10.1038/nchembio.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Oburoglu L, Tardito S, Fritz V, de Barros SC, Merida P, Craveiro M, et al. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell. 2014;15:169–84. doi: 10.1016/j.stem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 96.Folmes CD, Terzic A. Stem cell lineage specification: you become what you eat. Cell Metab. 2014;20:389–91. doi: 10.1016/j.cmet.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–6. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–9. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alexander PB, Wang J, McKnight SL. Targeted killing of a mammalian cell based upon its specialized metabolic state. Proc Natl Acad Sci U S A. 2011;108:15828–33. doi: 10.1073/pnas.1111312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Han C, Gu H, Wang J, Lu W, Mei Y, Wu M. Regulation of L-threonine dehydrogenase in somatic cell reprogramming. Stem Cells. 2013;31:953–65. doi: 10.1002/stem.1335. [DOI] [PubMed] [Google Scholar]
- 101.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–6. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shiraki N, Shiraki Y, Tsuyama T, Obata F, Miura M, Nagae G, et al. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19:780–94. doi: 10.1016/j.cmet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 103.Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, et al. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18:1350–8. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Knobloch M, Braun SM, Zurkirchen L, von Schoultz C, Zamboni N, Arauzo-Bravo MJ, et al. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature. 2013;493:226–30. doi: 10.1038/nature11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Folmes CD, Park S, Terzic A. Lipid metabolism greases the stem cell engine. Cell Metab. 2013;17:153–5. doi: 10.1016/j.cmet.2013.01.010. [DOI] [PubMed] [Google Scholar]