Abstract
Genetically modified mice are extremely valuable tools for studying gene function and human diseases. Although the generation of mice with specific genetic modifications through traditional methods using homologous recombination in embryonic stem (ES) has been invaluable in the last two decades, it is an extremely costly, time-consuming and in some cases uncertain technology. The recently described CRISPR/Cas9 genome-editing technology significantly reduces the time and the cost that are required to generate genetically engineered mice, allowing scientist to test more precise and bold hypothesis in vivo. Using this revolutionary methodology we have generated more than one hundred novel genetically engineered mouse strains. In the current protocol, we describe in detail the optimal conditions to generate mice carrying point mutations, chromosomal deletions, conditional alleles, fusion tags or endogenous reporters.
PROTOCOL
MATERIALS
REAGENTS
Mice
C57BL/6, age 3-4 weeks or >8 weeks, for zygote collection (Jackson Laboratories) <R>.
ICR (CD-1) mice for pseudopregnant mother and vasectomized males (Charles Rivers Laboratories) <R>.
Plasmids
pMJ920-Cas9 (Addgene, cat. no. 42234), contains the Cas9 coding region <R>.
Circular DNA donor vector (if performing large fragment insertion) <R>.
OligoDNAs used for in vitro transcription (Sigma)
sgRNA template 5′NNNNNNNNNNNNNNNNNNNNGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTT3′ <R>
sgRNA_R 5′AAAAAAGCACCGACTCGGTG3′ <R>
T7-sgRNA_F 5′GAAATTAATACGACTCACTATAGGGAGANNNNNNNNNNNNNNNNNNNNGTTTTAGA3′<R>
Single strand DNA (ssDNA) used to insert small DNA fragments (Integrated DNA Technologies)
These oligoDNAs are ordered as a 4nm μltramer, desalted. Include phosphorothioate linkages to prevent degradation in the first and last three nucleotides of the oligoDNAs. <R>
Other reagents
1 M Tris pH 7.0 (Life Technologies, cat. no. AM9850G) <R>
1 M Tris pH 8.0 (Life Technologies, cat. no. AM9855G) <R>
Agarose (Life Technologies, cat. no. 17850) <R>
Agilent RNA 6000 Pico Kit (Agilent technologies, cat. no. 5067-1513) <R>
CutSmart buffer (New England Biolabs, cat. no. B7204S) <R>
Cytochalasin B (CB; Sigma, cat. no. C6762) <R>
DNeasy blood and tissue kit (Qiagen, cat. no. 69581) <R>
EDTA, 0.5M solution pH 8.0 (AmericanBio, cat. no. AB00502-01000) <R>
Gene specific primers for genotyping (Sigma) <R>
Human chorionic gonadotropin (hCG, Sigma, cat. no. C8554) <R>
Hyaluronidase (Sigma, cat. no. H4272) <R>
M2 medium (Sigma, cat. no. M7167) <R>
M16 medium (Sigma, cat. no. M7292) <R>
MEGAclear kit (Life Technologies, cat. no. AM1908) <R>
MEGAshortscript T7 kit (Life Technologies, cat. no. AM1354) <R>
Mineral oil light (Sigma, cat. no. 330779) <R>
mMESSAGE mMACHINE T7 μltra kit (Life Technologies, cat. no. AM1345) <R>
Nuclease-free water (Life Technologies, cat. no. AM9932) <R>
Phenol:Chloroform:Isoamyl Alcohol 25:24:1 (Sigma, cat. no. 2069) <R>
Platinum Taq DNA Polymerase High Fidelity (Life Technologies, cat. no. 11304-011) <R>
Polyvinylpyrrolidone (PVP; 360 kDa, Sigma, cat. no. PVP-360) <R>
Pregnant mare serum gonadotropin (PMSG; Sigma, cat. no. G4527) <R>
Qiagen endo free maxi kit (Qiagen, cat. no. 12362) <R>
QIAQuick PCR purification kit (Qiagen, cat. no. 28106) <R>
Restriction enzyme XbaI (New England Biolabs, cat. no. R0145S) <R>
TA Cloning kit (Invitrogen, cat. no. 450641) <R>
Tris-acetate-EDTA (TAE) buffer<R>
Tsg DNA polymerase (Lambda biotechnologies, cat. no. D101-200) <R>
ssDNA/RNA clean and concentrator kit (Zymo Research, cat. no. D7010) <R>
SURVEYOR Mutation Detection Kit - S100 (Transgenomics, cat. no. 706020) <R>
EQUIPMENT
2100 Bioanalyzer (Agilent technologies, cat. no. G2940CA)
26-G needle (Becton Dickinson, cat. no. 309625)
C1000 Touch termal cycler (Biorad)
CO2 incubator (Thermo, cat. no. BB15)
Falcon 6-cm (60 × 15 mm) dishes, used for longer culture (Becton Dickinson, cat. no. 351007)
Falcon 10-cm (100 × 20 mm) dishes; bottoms are suitable for oocyte/embryo collection and lids are suitable for micromanipulation (Becton Dickinson, cat. no. 353003)
Holding pipette (Origio, cat. no. MPH-SM-20)
Inverted microscope with Hoffman optics (Olympus, cat. no. IX73)
Microcentrifuge (Eppendorf, cat no. 5424)
Microinjection capillaries (Femtotip; 5242952.008; Eppendorf)
Microinjector (Cell tram vario; Eppendorf, cat. no. 5176.000.033)
Micromanipulator set (Narishige, cat. no. MMO-202ND)
Refrigerated microcentrifuge (Eppendorf, cat no. 5424R)
Stereomicroscope, such as the SZX7 (Olympus, cat. no. SZX7)
Transfer pipette (WPI, cat. no. TW100F-6)
PROCEDURE
Experimental design
sgRNA design
For sgRNA design, follow the protocol described by Zhang’s laboratory1.
For multiple-gene KO by indels, two or more sgRNAs are designed to target each gene.
For targeted chromosomal deletions, two sgRNAs are designed, each of which will generate a DSB at the start and end point of the sequence to be deleted.
To introduce a small DNA sequence (e.g. point mutation, fusion tag) a single sgRNA is designed. To generate a conditional allele by inserting two loxps in cis, two sgRNAs are designed at the insertion sites of interest.
To introduce a large DNA sequence (e.g. fluorescent protein) by HR a single sgRNA is designed at the insertion site of interest.
ssDNA donor oligo design
To introduce a short DNA sequence (e.g. point mutation, fusion tag), an ssDNA oligonucleotide that encodes the desired DNA insertion flanked on each side by 50-70 bases homologous to the sequence surrounding the sgRNA-mediated DSB is designed. The sequence to be inserted should be as close as possible to the generated DSB. We have been able to introduce up to 100 bases with homologous regions of 50 bases on each side.
To generate a conditional allele by introducing two loxp sites, two ssDNA oligonucleotides that encode the loxp sequence flanked on each side by 63 bases homologous to the sequence surrounding the sgRNA-mediated DSB are designed. The loxp sequence to be inserted should be as close as possible to the generated DSB.
Circular DNA donor vector design
To introduce a large DNA sequence (e.g. fluorescent protein), a plasmid that encodes the DNA sequence to be inserted flanked by >2kb bp of homology on each side should be designed and constructed through standard molecular biology methods. The DNA sequence of interest should be inserted as close as possible to the generated DSB. We have been able to introduce large fragments of DNA with smaller homology regions (~500 bp).
Critical consideration
When introducing a small or a large DNA fragment, a potential problem should be considered. After induction of the DSB and the DNA sequence has been inserted in the genome by HDR or HR, it is possible that the Cas9 can recut the modified allele if the sgRNA and PAM sequence have not been modified in the donor ssDNA or circular DNA vector. To prevent this, place the DNA sequence that encodes the desired insertion in a position in which the PAM sequence or the 6 nucleotides closest to the PAM sequence are disrupted.
In vitro transcription and purification of Cas9 mRNA
- Linearize the pMJ920-Cas9 plasmid that contains the Cas9 coding region under the T7 promoter with XbaI.
- Incubate the following reaction at 37°C for 16 hours.
- 5 μl of circular pMJ920-Cas9 plasmid (5ug of plasmid total)
- 0.5 μl of XbaI restriction enzyme
- 5 μl of 10X Cutsmart buffer
- 39.5 μl of Nuclease-free water
Add 1.5 ul of proteinase K (150 ng/μl) and 2.5 ul 20% SDS to the reaction mix. Incubate for 30 minutes at 50°C.
Run 5 μl of the digestion reaction on a 1% (wt/vol) agarose gel in TAE buffer to verify the successful linearization of the pMJ920-Cas9 plasmid.
Purify the linearized pMJ920-Cas9 plasmid with Phenol:Chloroform:Isoamyl Alcohol 25:24:1 according to the manufacturer’s instructions. Resuspend the purified linearized pMJ920-Cas9 plasmid to a concentration of 1ug/ul using nuclease-free water.
Use 1ug of the purified linearized pMJ920-Cas9 plasmid as the template for in vitro transcription to generate Cas9 mRNA using the mMESSAGE mMACHINE T7 kit according to the manufacturer’s instructions.
Purify the Cas9 mRNA using the MEGAclear kit according to the manufacturer’s instructions and elute it with 65 ul of elution buffer.
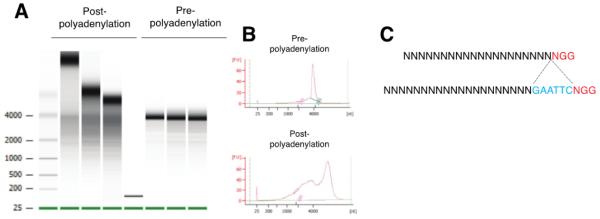
Critical step: After purification, determine the quality of the Cas9 mRNA by running 2.5 μl of the Cas9 mRNA prior poly-adenylation and post poly-adenylation in a bioanalyzer using the Agilent RNA 6000 Pico reagents and chips according to the manufacturer’s instructions. You should observe a discrete ~4 kb band in the sample prior poly-adenylation and an upward shift post poly-adenylation (Figure 1 A and B). Smeared bands indicate degradation. Samples should be discarded if degradation is observed. Calculate the concentration of your Cas9 mRNA according to the values reported in the Bioanalyzer.
Dilute the Cas9 mRNA to 400 ng/μl in nuclease-free microinjection buffer (5 mM Tris-HCl, pH 7.4, 0.1 mM EDTA, pH 8.0).
Figure 1. Quality control of Cas9 mRNA in a 2100 Bioanalyzer and ssDNA donor oligo design.
A. Electrophoresis of in vitro transcribed Cas9 mRNA prior poly-polyadenylation and post poly-adenylation. B. Electropherogram of in vitro transcribed Cas9 mRNA prior poly-polyadenylation and post poly-adenylation. C. Representation of a sequence that can be targeted with a specific sgRNA. Insertion of EcoRI (as an example) adjacent to the PAM sequence will preclude the Cas9 nuclease from cutting again after the genome has been repaired by HDR.
In vitro transcription and purification of sgRNAs
-
For sgRNA preparation, add the T7 promoter sequence to the sgRNA template by PCR amplification using T7-sgRNA_F and the sgRNA_R primers as follows:
PCR reactionComponent Amount Final sgRNA template (300 ng/μl) 4 μl 1200 ng 10X HiFi buffer 40 μl 1X MgSO4 (50mM) 12 μl 1.5mM dNTP (10mM) 8 μl 0.2 mM T7-sgRNA_F (10 μM) 8 μl 0.2 uM sgRNA_R (10 μM) 8 μl 0.2 uM High-fidelity polymerase 2.4 μl Nuclease-free water 317.6 μl Divide this PCR reaction into 8 PCR tubes (50 μl/each) and perform amplification using the following cycling conditions:Cycle number Denature Anneal Extend 1 94°C, 2 min 2-35 94°C, 20 sec 55°C, 30 sec 68°C, 40 sec 36 68°C, 7 min Run 5 μl of the PCR reaction on a 2.5% (wt/vol) agarose gel in 1XTAE buffer to verify that the product is unique and of the expected size. The size of the PCR product is ~120 bp for sgRNA templates.
Purify the T7-sgRNA PCR product using the QIAQuick PCR purification kit according to the manufacturer’s instructions. Elute in 30 μl of nuclease free water and resuspend the purified PCR product to a concentration of 120.5 ng/μl.
Use 1 ul of the purified T7-sgRNA PCR product as the template for in vitro transcription of sgRNA using the MEGAshortscript T7 kit according to the manufacturer’s instructions.
Purify the sgRNA using the MEGAclear kit and elute with elution buffer according to the manufacturer’s instructions.
Dilute the sgRNA to 500 ng/μl in nuclease-free microinjection buffer (5 mM Tris-HCl, pH 7.4, 0.1 mM EDTA, pH 8.0) and check its quality on a 2% (wt/vol) agarose gel in TAE buffer. Smeared bands indicate degradation. Samples should be discarded if degradation is observed.
Freeze 30-40 ul aliquots at −80°C.
Purification of ssDNA used to insert small DNA fragments
Purify the donor ssDNA using the ssDNA/RNA clean and concentrator kit according to the manufacturer’s instructions and elute with 100 μl of elution buffer.
Dilute the donor ssDNA to 500 ng/μl in nuclease-free microinjection buffer (5 mM Tris-HCl, pH 7.4, 0.1 mM EDTA, pH 8.0).
Freeze 30-40 ul aliquots at −80°C.
Purification of circular DNA donor used to insert large DNA fragments
Prepare the circular DNA donor vector using the Qiagen endo free maxi kit according to the manufacturer’s instructions.
Purify the circular DNA donor vector using the using the QIAQuick PCR purification kit according to the manufacturer’s instructions. Elute in 30 μl of nuclease-free microinjection buffer (5 mM Tris-HCl, pH 7.4, 0.1 mM EDTA, pH 8.0).
Preparation of samples for microinjection
- Prepare the appropriate injection mix depending on the aim of the experiment as outlined below.
- Gene disruption by NHEJ or small/large chromosomal deletions: Add 10μl of Cas9 mRNA stock, 4 μl of each sgRNA stock and nuclease-free microinjection buffer up to 40 μl. The final concentrations are 100 ng/ul of Cas9 mRNA, 50-100 ng/ul of each sgRNA.
- Point mutation, small tag insertion or conditional allele generation by HDR: Add 10μl of Cas9 mRNA stock, 4 μl of each sgRNA stock, 8 ul of donor ssDNA stock and nuclease-free microinjection buffer up to 40 μl. The final concentrations are 100 ng/ul of Cas9 mRNA, 50-100 ng/ul of each sgRNA and 100 ng/ul of each donor ssDNA.
- Large fragment insertion by HR: Add 10μl of Cas9 mRNA stock, 4μl of each sgRNA stock, the circular DNA donor vector and nuclease-free microinjection buffer up to 40 μl. The final concentrations are 100 ng/ul of Cas9 mRNA, 50-100 ng/ul of each sgRNA and and 200 ng/ul of the circular DNA donor vector.
Critical step: Centrifuge the stock aliquots of the Cas9 mRNA, sgRNAs and ssDNA donor oligos at 13200 rpm/4°C for 10 minutes.
Mix the correct amount for each component as described in step 1a-c in an Rnase free microcentrifuge tube.
Critical step: Centrifuge the microinjection mixture at 13200 rpm/4°C for 6 minutes. Remove 30 ul and transfer to a new Rnase free microcentrifuge tube.
Critical step: Centrifuge the microinjection mixture at 13200 rpm/4°C for 6 minutes. Place the mixture on ice and begin microinjection.
Zygote preparation
Inject 15 female C57BL/6 (3–4 weeks old) mice with PMSG (5 IU) on day 1.
After 48 h (i.e., on day 3), inject female mice with hCG (5 IU). After the hCG injection, house female mice with proven stud C57BL/6 male mice overnight.
On the morning of day 4, collect female mice with plugs for zygote preparation and euthanize the females mice without plugs.
Prepare the medium for embryo culture. Place several drops (30–50 μl for each drop) of M16 medium on a 6-cm dish, cover it with light mineral oil, and then place the dish into a 37 °C incubator for at least 30 min before use.
At 20–21 h after hCG injection, euthanize the mice and collect zygote-cumulus complexes from the oviduct.
Prepare the medium for embryo collection by adding hyaluronidase to M2 media to obtain a working concentration of 300 μg/ml (M2 + Hy medium). Move the zygote-cumulus complexes into M2 + Hy medium, pipette up and down several times, pick them up with a transfer pipette, wash several times in M2 medium and place the embryos into M16 medium at 37 °C in a 5% CO2 incubator.
Microinjection of zygotes
Use an embryo transfer pipette to transfer a group of fertilized oocytes into the injection chamber containing M16 medium. The number of zygotes to be moved into the microinjection drop should be determined by the skills of the injector and quality of the setup. Wait for at least 5 min before starting the injection.
Examine the zygotes under high power, making sure that two pronuclei are visible and that the morphology is good. Discard all zygotes that appear abnormal.
Aspirate the injection mix into microinjection capillary.
To determine whether the microinjection capillary is open and unclogged, place the tip of the microinjection capillary close to a zygote in the same horizontal plane under a continuous flow stream. If the microinjection capillary is open, a stream of DNA will move the zygote away from the tip of the microinjection capillary. If the pipette is closed or clogged, flush DNA with high power through the microinjection capillary by using the “Clear” function on the microinjector. Repeat the test. If the tip is still not open, tip it carefully on the holding pipette and so break up to a larger tip diameter. If the diameter becomes too large, or the tip is still not open, discard the pipette and use a new one.
To prepare a zygote for injection, place the tip of the holding pipette next to the zygote and apply a negative pressure to the pressure control unit. Focus the microscope to locate the pronuclei. The pronucleus should also be as close as possible to the central axis of the holding pipette. Refocus on the pronucleus to be injected, bring the tip of the microinjection capillary into the same focal plane as the mid-plane of the pronucleus. Move the injection pipette to the same y-axis position as the targeted pronucleus and adjust the height of the pipette so that the tip of the microinjection capillary appears completely sharp.
Move the injection pipette to a 3 o’clock position without changing its height. Push the microinjection pipette through the zona pellucida, into the cytoplasm, and toward the pronucleus. When the tip of microinjection capillary appears to be inside the cytoplasm apply injection pressure through the injector.
If the cytoplasm swells visibly, it has been successfully injected. Quickly pull the pipette out of the zygote. If the cytoplasm does not swell, the pipette has become clogged or has not punctured the oocyte plasma membrane. If a small round “bubble” forms around the tip of the pipette, then the pipette has not punctured the plasma membrane. Cytoplasmic granules flowing out of the oocyte after removal of the injection pipette are a clear sign that the zygote will soon lyse. In this case, the oocyte should be discarded. If the zygote appears to be intact and successfully injected, it should be sorted and another zygote should be picked up for injection.
When all the zygotes in the chamber have been injected, they should immediately be moved back into M16 medium and incubated at 37 °C in a 5% CO2 incubator 37°C.
Some injected zygotes will inevitably lyse due to the mechanical damage caused by the injection procedure. The lysed zygotes can be from healthy ones by the appearance of the zona pellucida as it is more translucid. Typically, about 75% of the zygotes survive the microinjection.
Embryo transfer and production of mice
Prepare pseudopregnant foster mothers by mating estrous CD1 female mice with vasectomized male mice on the same day as injection.
Transfer ~25 microinjected zygotes into oviducts of 0.5 days post coitum (d.p.c.) recipients. Alternatively, injected zygotes can be cultured in M16 medium at 37°C in a 5% CO2 incubator until they reach the two-cell stage 24 h later and then be transferred into oviducts of 0.5 days post coitum (d.p.c.) recipients.
Recipient mothers will deliver pups at 19.5 d.p.c.
Separate male and female offspring into individual cages at 3 weeks after birth.
Genotyping
Extract genomic DNA from tail biopsies of the 10 day-old mice by using a DNeasy blood and tissue kit according to the manufacturer’s protocol.
-
To genotype small genomic modifications such as point mutations, gene disruptions, small sequence insertions or conditional alleles perform PCR amplification using gene-specific primers under the following conditions:
PCR reactionComponent Amount Final Genomic DNA 2.5 μl 10X Tsg buffer 2.5 μl 1X MgCl2 1.5 μl 1.5mM dNTP (10mM) 0.5 μl 0.2 mM Primer_F (10 μM) 0.5 μl 0.2 uM Primer_R (10 μM) 0.5 μl 0.2 uM Tsg Polymerase 0.3 μl Nuclease-free water 16.7 μl Amplification conditionsCycle number Denature Anneal Extend 1 95°C, 2 min 2-35 94°C, 30 sec 72°C, 30 sec, −0.5 °C/Cycle 72°C, 45 sec 36 72°C, 7 min To check for indels or small mutations perform a surveyor assay directly on the PCR product according to the manufacture’s instructions.
Alternatively, purify the PCR products by using the QIAQuick PCR purification kit according to the manufacturer’s instructions. If a restriction enzyme has been inserted or removed, perform restriction enzyme digestion of the PCR product. To verify mutations clone the PCR products using the TA Cloning kit, and sequence according to the manufacturer’s instructions.
-
To genotype for insertions of tags, loxps or deletions perform PCR amplification using gene-specific primers under the following conditions:
PCR reactionComponent Amount Final Genomic DNA 2.5 μl 10X Tsg buffer 2.5 μl 1X MgCl2 1.5 μl 1.5mM dNTP (10mM) 0.5 μl 0.2 mM Primer_F (10 μM) 0.5 μl 0.2 uM Primer_R (10 μM) 0.5 μl 0.2 uM Tsg Polymerase 0.3 μl Nuclease-free water 16.7 μl Amplification conditionsCycle number Denature Anneal Extend 1 95°C, 2 min 2-35 94°C, 30 sec 72°C, 30 sec, −0.5 °C/Cycle 72°C, 45 sec 36 72°C, 7 min Run the PCR product on a 2% (wt/vol) agarose gel in 1XTAE buffer to verify that the product is unique and of the expected size.
To genotype for large insertions perform PCR amplification for large fragments using gene-specific primers that allows detecting the insertion of the fragment at the specific genomic location. PCR conditions should be optimized in each case.
EXPECTED RESULTS
On the basis of the targeting experiments that we have done, which accounts to over four hundred different microinjections and more than one hundred new mouse strains generated, the great majority of microinjections with only Cas9 mRNA and sgRNAs resulted in mutant alleles containing indels with high efficiency (>80% of pups were targeted for one allele). The efficiency of targeted chromosomal deletions greatly depends on the size of the deletion, we have been successful in deleting chromosomal regions up to ~200 kb, we observe efficiencies for targeted deletions that range from 10-40% of born pups being targeted. With co-injection of donor ssDNA, the efficiency of homology-directed repair (HDR) varies from 10-60% depending on the size of the fragment that is being inserted/modified. By using double-stranded plasmid donor DNA, the efficiency of HR varies greatly and requires significant optimization.
References
- 1.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]