Abstract
Acute respiratory infections caused by bacterial or viral pathogens are among the most common reasons for seeking medical care. Despite improvements in pathogen-based diagnostics, most patients receive inappropriate antibiotics. Host response biomarkers offer an alternative diagnostic approach to direct antimicrobial use. This observational, cohort study determined whether host gene expression patterns discriminate non-infectious from infectious illness, and bacterial from viral causes of acute respiratory infection in the acute care setting. Peripheral whole blood gene expression from 273 subjects with community-onset acute respiratory infection (ARI) or non-infectious illness as well as 44 healthy controls was measured using microarrays. Sparse logistic regression was used to develop classifiers for bacterial ARI (71 probes), viral ARI (33 probes), or a non-infectious cause of illness (26 probes). Overall accuracy was 87% (238/273 concordant with clinical adjudication), which was more accurate than procalcitonin (78%, p<0.03) and three published classifiers of bacterial vs. viral infection (78-83%). The classifiers developed here externally validated in five publicly available datasets (AUC 0.90-0.99). A sixth publically available dataset included twenty-five patients with co-identification of bacterial and viral pathogens. Applying the ARI classifiers defined four distinct groups: a host response to bacterial ARI; viral ARI; co-infection; and neither a bacterial nor viral response. These findings create an opportunity to develop and utilize host gene expression classifiers as diagnostic platforms to combat inappropriate antibiotic use and emerging antibiotic resistance.
Introduction
Respiratory tract infections caused 3.2 million deaths worldwide and 164 million disability-adjusted life years lost in 2011, more than any other cause.(1) Despite a viral etiology in the majority of cases, 73% of ambulatory care patients in the U.S. with acute respiratory infection (ARI) are prescribed an antibiotic, accounting for 41% of all antibiotics prescribed in this setting.(2, 3) Even when a viral pathogen is microbiologically confirmed, this does not exclude a possible concurrent bacterial infection leading to antimicrobial prescribing “just in case”. This empiricism drives antimicrobial resistance(4, 5), recognized as a national security priority.(6)
The host’s peripheral blood gene expression response to infection offers a diagnostic strategy complementary to those already in use. This strategy has successfully characterized the host response to viral (7-12) and bacterial ARI.(10, 13) Despite these advances, several issues preclude their use as diagnostics in patient care settings. An important consideration in the development of host-based molecular signatures is that they be developed in the intended use population.(14) However, nearly all published gene expression-based ARI classifiers used healthy individuals as controls and focused on small or homogeneous populations. Furthermore, the statistical methods used to identify gene-expression classifiers often include redundant genes based on clustering, univariate testing, or pathway association. These strategies identify relevant biology but do not maximize diagnostic performance. An alternative is to combine genes from potentially unrelated pathways to generate a more informative classifier.
We present evidence from a large observational cohort of Emergency Department patients that host responses to bacterial, viral, or non-infectious insults are unique and quantifiable. Therefore, the objective of this study is to show that the host response, as measured by peripheral blood gene expression changes, can accurately differentiate viral ARI, bacterial ARI, and non-infectious illness as an important step toward their routine use in clinical practice. Such an approach offers new opportunities to guide appropriate antibiotic use and combat emerging antibiotic resistance.
Results
Bacterial ARI, Viral ARI, and Non-Infectious Illness classifiers
In generating host gene expression-based classifiers that distinguish between clinical states, all relevant clinical phenotypes should be represented during the model training process. This imparts specificity, allowing the model to be applied to these included clinical groups but not to clinical phenotypes that were absent from model training.(14) The target population for an ARI diagnostic not only includes patients with viral and bacterial etiologies, but must also distinguish from the alternative – those without bacterial or viral ARI. Historically, healthy individuals have served as the uninfected control group. However, this fails to consider how patients with non-infectious illness, which can present with similar clinical symptoms, would be classified, serving as a potential source of diagnostic error. To our knowledge, no ARI gene-expression based classifier has included ill, uninfected controls in its derivation. We therefore enrolled a large, heterogeneous population of patients at initial clinical presentation with community-onset viral ARI (n=115), bacterial ARI (n=70), or non-infectious illness (n=88) (Table 1 and Table S1). We also included a healthy adult control cohort (n=44) to define the most appropriate control population for ARI classifier development. Clinical features of the subjects are summarized in Table S2.
Table 1. Demographic information for the enrolled cohort as well as independent datasets used for external validation.
| Cohort | Number of subjectsa |
Gender (M/F) |
Mean age, years (Range)b |
Ethnicity (B/W/O) |
Admitted | # Samples (Viral/ Bacterial/ Non- Infectious Illness) |
|---|---|---|---|---|---|---|
| Enrolled Derivation Cohort |
317 | 122/151 | 45 (6-88) | 135/116/22 | 61% | 115/70/88 |
| Viral | 115 | 44/71 | 45 (6-88) | 40/59/16 | 21% | |
| Bacterial | 70 | 35/35 | 49 (14-88) | 46/22/2 | 94% | |
| Non-infectious Illnessc | 88 | 43/45 | 49 (14-88) | 49/35/4 | 88% | |
| Healthy | 44 | 23/21 | 30 (20-59) | 8/27/6 d | 0% | |
| Validation Cohorts | ||||||
| Ramilo et al. (GSE6269) | 113e | 62/51 | 4 (0.04-36) | 22/37/54 | 100% | 28/85/0 |
| Hu et al. (GSE40396) |
43 | 25/18 | 14 (2-32) | 16/25/2 | N/A | 35/8/0 |
| Herberg et al. (GSE42026) |
59 | N/A | Pediatric | N/A | 100% | 18/41/0 |
| Parnell et al. 2011 (GSE20346)f |
10 | 4/6 | Adult | N/A | 100% | 19/26/0 |
| Bloom et al. (GSE42834) |
103 | N/A | Adult | N/A | N/A | 0/19/84 |
Only subjects with viral, bacterial, or non-infectious illness were included (when available) from each validation cohort.
When mean age was unavailable or could not be calculated, data is presented as either Adult or Pediatric.
Non-infectious illness was defined by the presence of SIRS criteria, which includes at least two of the following four features: Temperature <36° or >38°C; Heart rate >90 beats per minute; Respiratory rate >20 breaths per minute or arterial partial pressure of CO2 <32mmHg; and white blood cell count <4000 or >12,000 cells/mm3 or >10% band form neutrophils.
Three subjects did not report ethnicity.
In the case of GSE6269, subjects with Illumina Sentrix Hu6 expression data were excluded because array results could not be readily compared. Eight viral and 15 bacterial infections comprised the 24 excluded cases.
Subjects in the GSE20346 dataset include serial sampling. The number of samples exceeds the number of subjects because serial samples were treated as independent tests in the validation experiments.
M, Male. F, Female. B, Black. W, White. O, Other/Unknown. GSE numbers refer to NCBI Gene Expression Omnibus datasets. N/A, Not available based on published data.
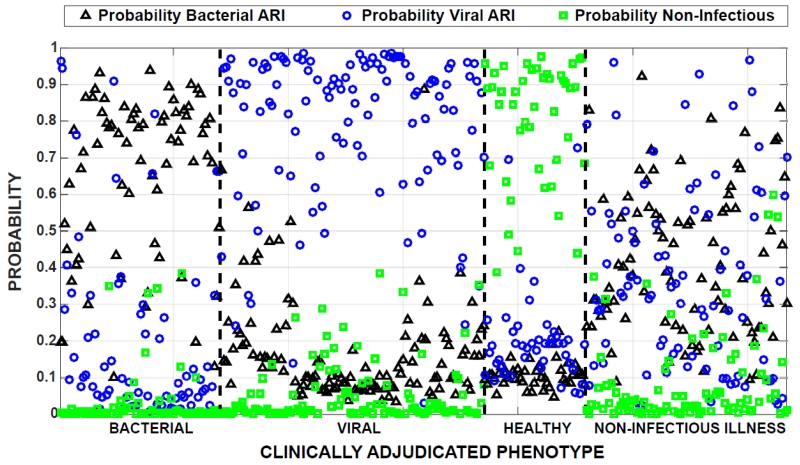
We first determined whether a gene expression classifier derived with healthy individuals as controls could accurately classify patients with non-infectious illness. Array data from patients with bacterial ARI, viral ARI, and healthy controls were used to generate gene expression classifiers for these conditions (Figure 1). Leave-one-out cross-validation revealed highly accurate discrimination between bacterial ARI (AUC 0.96), viral ARI (AUC 0.95), and healthy (AUC 1.0) subjects for a combined accuracy of 90% (Figure 2). However, when the classifier was applied to ill-uninfected patients, 48/88 were identified as bacterial, 35/88 as viral, and 5/88 as healthy. This highlighted that healthy individuals are a poor substitute for patients with non-infectious illness in the biomarker discovery process.
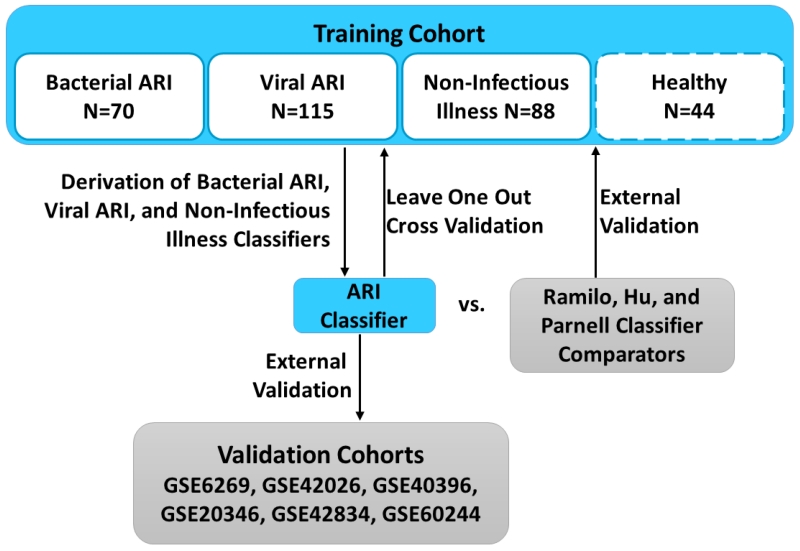
Fig. 1. Experimental flow.
A cohort of patients encompassing bacterial ARI, viral ARI, or non-infectious illness was used to develop classifiers of each condition. This combined ARI classifier was validated using leave-one-out cross-validation and compared to three published classifiers of bacterial vs. viral infection. The combined ARI classifier was also externally validated in six publically available datasets. In one experiment, healthy volunteers were included in the training set to determine their suitability as “no-infection” controls. All subsequent experiments were performed without the use of this healthy subject cohort.
Fig. 2. Evaluation of healthy adults as a no-infection control.
Classifiers of bacterial ARI, viral ARI, and no infection as represented by healthy controls were generated and applied using leave-one-out cross-validation. Each patient, represented along the horizontal axis, is assigned three distinct probabilities: bacterial ARI (black triangle), viral ARI (blue circle), and no infection (green square). The group on the right represents subjects with non-infectious illness.
Consequently, we re-derived an ARI classifier using a non-infectious illness control rather than healthy. Specifically, array data from these three groups was used to generate three gene-expression classifiers of host response to bacterial ARI, viral ARI, and non-infectious illness. Specifically, the Bacterial ARI classifier was tasked with positively identifying those with bacterial ARI vs. either viral ARI or non-infectious illnesses. The Viral ARI classifier was tasked with positively identifying those with viral ARI vs. bacterial ARI or non-infectious illnesses. The Non-Infectious Illness classifier was not generated with the intention of positively identifying all non-infectious illnesses, which would require an adequate representation of all such cases. Rather, it was generated as an alternative category, so that patients without bacterial or viral ARI could be assigned accordingly. Moreover, we hypothesized that such ill but non-infected patients were more clinically relevant controls because healthy people are unlikely to be the target for such a classification task.
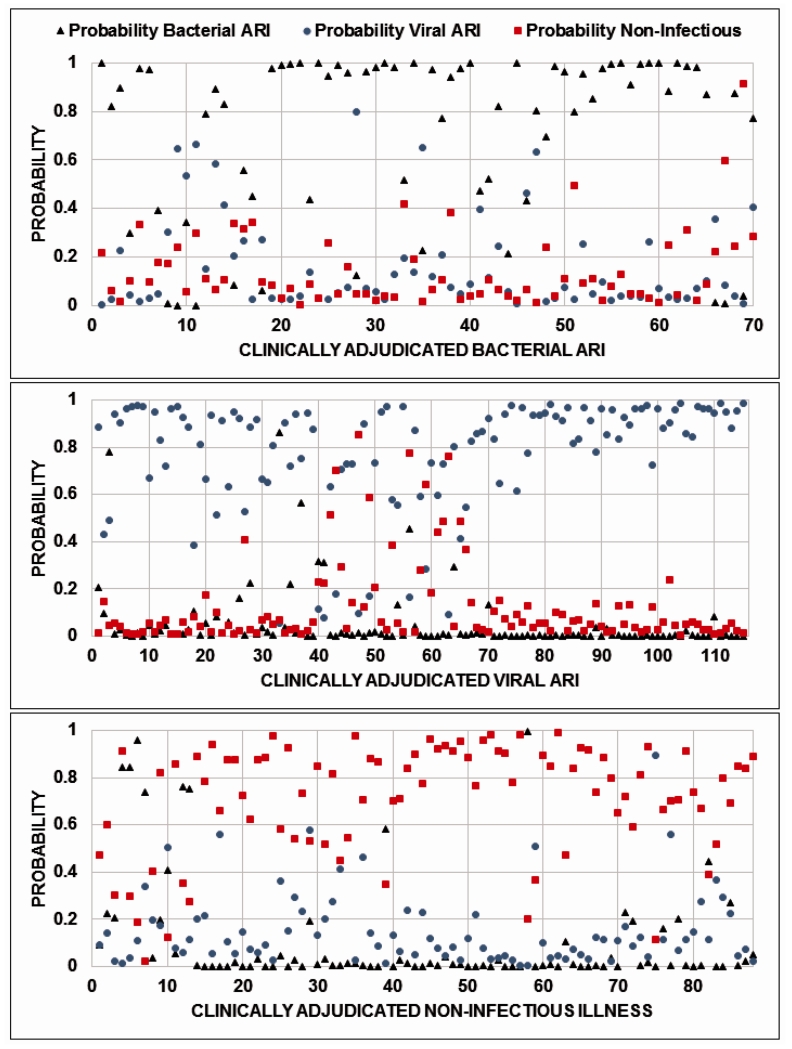
Six statistical strategies were employed to generate these gene-expression classifiers: linear support vector machines, supervised factor models, sparse multinomial logistic regression, elastic nets, K-nearest neighbor, and random forests. All performed similarly although sparse logistic regression required the fewest number of classifier genes and outperformed other strategies by a small but not significant margin (p-value>0.05 using McNemar’s tests between leave-one-out cross-validated predictions from sparse logistic regression vs. each alternative method). We also compared a strategy that generated three separate binary classifiers to a single multinomial classifier that would simultaneously assign a given subject to one of the three clinical categories. This latter approach required more genes and achieved an inferior accuracy. Consequently, we applied a sparse logistic regression model to define Bacterial ARI, Viral ARI, and Non-Infectious Illness classifiers containing 71, 33 and 26 probe signatures, respectively. Probe and classifier weights are shown in Table S3. Clinical decision making is infrequently binary, requiring the simultaneous distinction of multiple diagnostic possibilities. We applied all three classifiers, collectively defined as the ARI classifier, using leave-one-out cross-validation to assign probabilities of bacterial ARI, viral ARI, and non-infectious illness (Figure 3). These conditions are not mutually exclusive. For example, the presence of a bacterial ARI does not preclude a concurrent viral ARI or non-infectious disease. Moreover, the assigned probability represents the extent to which the patient’s gene expression response matches that condition’s canonical signature. Since each signature intentionally functions independently of the others, the probabilities are not expected to sum to one. To simplify classification, the highest predicted probability determined class assignment. Overall classification accuracy was 87% (238/273 concordant with adjudicated phenotype). Bacterial ARI was identified in 58/70 (83%) patients and excluded 179/191 (94%) without bacterial infection. Viral ARI was identified in 90% (104/115) and excluded in 92% (145/158) of cases. Using the non-infectious illness classifier, infection was excluded in 86% of cases (76/88). Sensitivity analyses was performed for positive and negative predictive values for all three classifiers given that prevalence can vary for numerous reasons including infection type, patient characteristics, or location (Figure S1).
Fig. 3. Leave-one-out cross-validation.
Classifiers of bacterial ARI, viral ARI, and no infection as represented by non-infectious illness were generated and applied using leave-one-out cross-validation. Each patient, represented along the horizontal axis, is assigned probabilities of having bacterial ARI (black triangle), viral ARI (blue circle), and non-infectious illness (red square). Patients clinically adjudicated as having bacterial ARI, viral ARI, or non-infectious illness are presented in the top, center, and bottom panels, respectively.
To determine if there was any effect of age, we included it as a covariate in the classification scheme. This resulted in two additional correct classifications, likely due to the over-representation of young people in the viral ARI cohort. However, we observed no statistically significant differences between correctly and incorrectly classified subjects due to age (Wilcoxon rank sum p=0.17). Likewise, patients with viral ARI tended to be less ill, as demonstrated by the lower rate of hospitalization. We therefore used hospitalization as a marker of disease severity and assessed its impact on classification performance, which revealed no statistical difference (Fisher’s exact test p-value of 1). As previously noted, the control cohort with systemic inflammatory response syndrome (SIRS) included subjects with both respiratory and non-respiratory etiologies. We assessed whether classification was statistically different in subjects with respiratory vs. non-respiratory SIRS and determined it was not (Fisher’s exact test p-value of 0.1305). Among the 47 subjects with respiratory SIRS, three were classified as having viral ARI and six were classified as having bacterial ARI. Among the 41 subjects with non-respiratory SIRS, one was classified as having viral ARI and two were classified as having bacterial ARI.
We compared this performance to procalcitonin, a widely used biomarker with some specificity for bacterial infection.(15) Procalcitonin concentrations were determined for the 238 subjects where samples were available and compared to ARI classifier performance for this subgroup. Procalcitonin concentrations >0.25μg/L assigned patients as having bacterial ARI, whereas values ≤0.25μg/L assigned patients as non-bacterial, which could be either viral ARI or non-infectious illness. Procalcitonin correctly classified 186 of 238 patients (78%) compared to 204/238 (86%) using the ARI classifier (p=0.03 by McNemar’s test). However, accuracy for the two strategies varied depending on the classification task. For example, performance was similar in discriminating viral from bacterial ARI. Procalcitonin correctly classified 136/155 (AUC 0.89) compared to 140/155 for the ARI classifier (p-value=0.65 using McNemar’s test). However, the ARI classifier was significantly better than procalcitonin in discriminating bacterial ARI from non-infectious illness [105/124 vs. 79/124 (AUC 0.72); p-value<0.001], and discriminating bacterial ARI from all other etiologies including viral and non-infectious etiologies [215/238 vs. 186/238 (AUC 0.82); p-value=0.02 by McNemar’s test].
We next compared the ARI classifier to three published gene expression classifiers of bacterial vs. viral infection, each of which was derived without uninfected ill controls. These included a 35-probe classifier (Ramilo) derived from children with influenza or bacterial sepsis(10); a 33-probe classifier (Hu) derived from children with febrile viral illness or bacterial infection(13); and a 29-probe classifier (Parnell) derived from adult ICU patients with community-acquired pneumonia or influenza(11). We hypothesized that classifiers generated using only patients with viral or bacterial infection would perform poorly when applied to a clinically relevant population that included ill but uninfected patients. Specifically, when presented with an individual with neither a bacterial nor a viral infection, the previously published classifiers would be unable to accurately assign that individual to a third, alternative category. We therefore applied the derived as well as published classifiers to our 273-patient cohort. Discrimination between bacterial ARI, viral ARI, and non-infectious illness was better with the derived ARI classifier (McNemar’s test, p=0.002 vs. Ramilo; p=0.0001 vs. Parnell; and p=0.08 vs. Hu) (Table 2).(16, 17) This underscores the importance of deriving gene-expression classifiers in a cohort representative of the intended use population, which in the case of ARI should include non-infectious illness.(14)
Table 2. Performance characteristics of the derived ARI classifier.
A combination of the Bacterial ARI, Viral ARI, and Non-Infectious Illness classifiers were validated using leave-one-out cross-validation in a population of bacterial ARI (n=70), viral ARI (n=115), or non-infectious illness (n=88). Three published bacterial vs. viral classifiers were identified and applied to this same population as comparators. Data are presented as number (%). Shaded cells indicate correct classifications.
| Clinical Assignment | |||||
|---|---|---|---|---|---|
| Bacterial | Viral | Non-infectious illness |
|||
| Ramilo et al. | Bacterial | 54 (77.1) | 4 (3.5) | 12 (13.6) | Classifier-Predicted Assignment |
| Viral | 6 (8.6) | 101 (87.8) | 12 (13.6) | ||
| Non-infectious illness | 12 (14.3) | 12 (8.7) | 64 (72.7) | ||
| Hu et al. | Bacterial | 53 (75.7) | 4 (3.5) | 9 (10.2) | |
| Viral | 9 (12.9) | 104 (90.4) | 9 (10.2) | ||
| Non-infectious illness | 8 (11.4) | 7 (6.1) | 70 (79.5) | ||
| Parnell et al. | Bacterial | 51 (72.8) | 8 (7.0) | 11 (12.5) | |
| Viral | 13 (18.6) | 94 (81.7) | 10 (11.4) | ||
| Non-infectious illness | 6 (8.6) | 13 (11.3) | 67 (76.1) | ||
| Derived ARI Classifier |
Bacterial | 58 (82.8) | 4 (3.4) | 8 (9.0) | |
| Viral | 9 (12.8) | 104 (90.4) | 4 (4.5) | ||
| Non-infectious illness | 3 (4.2) | 7 (6.0) | 76 (86.3) | ||
Discordant classifications
To better understand ARI classifier performance, we individually reviewed the 35 discordant cases (Table S4). Nine adjudicated bacterial infections were classified as viral and three as non-infectious illness. Four viral infections were classified as bacterial and seven as non-infectious. Eight non-infectious cases were classified as bacterial and four as viral. We did not observe a consistent pattern among discordant cases. However, notable examples included atypical bacterial infections. One patient with M. pneumoniae based on serological conversion, and one of three patients with Legionella pneumonia were classified as viral ARI. Of six patients with non-infectious illness due to autoimmune or inflammatory diseases, only one adjudicated as Still’s disease was classified as having bacterial infection.
External validation
Generating classifiers from high dimensional, gene expression data can result in over-fitting. We therefore validated the ARI classifier in silico using gene expression data from 328 individuals, represented in five available datasets (GSE6269, GSE42026, GSE40396, GSE20346, and GSE42834). These were chosen because they included at least two relevant clinical groups, varying in age, geographic distribution, and illness severity (Table 3). Applying the ARI classifier to four datasets with bacterial and viral ARI, AUC ranged from 0.90-0.99 (Figures S2-S5). Lastly, GSE42834 included patients with bacterial pneumonia (n=19), lung cancer (n=16), and sarcoidosis (n=68). Overall classification accuracy was 96% (99/103) corresponding to an AUC of 0.99 (Figure S6). GSE42834 included five subjects with bacterial pneumonia pre- and post-treatment. All demonstrated a treatment-dependent resolution of the bacterial response signature (Figure S7).
Table 3. External validation of the ARI classifier (combined bacterial ARI, viral ARI, and non-infectious classifiers).
Five Gene Expression Omnibus datasets were identified based on the inclusion of at least two of the relevant clinical groups: Viral ARI, Bacterial ARI, non-infectious illness (SIRS).
| Clinical Assignment | ||||||
|---|---|---|---|---|---|---|
| Bacterial | Viral | SIRS | AUC | |||
|
GSE6269: Hospitalized children with Influenza A or bacterial infection |
Classifier-Predicted Assignment | Bacterial | 84 | 1 | 0.95 | |
| Viral | 2 | 26 | ||||
|
GSE42026: Hospitalized children with Influenza H1N1/09, RSV, or bacterial infection |
Bacterial | 15 | 3 | 0.90 | ||
| Viral | 6 | 35 | ||||
|
GSE40396: Children with adenovirus, HHV-6, enterovirus, or bacterial infection |
Bacterial | 7 | 1 | 0.93 | ||
| Viral | 3 | 32 | ||||
|
GSE20346: Hospitalized adults with bacterial pneumonia or Influenza A |
Bacterial | 26 | 0 | 0.99 | ||
| Viral | 1 | 18 | ||||
|
GSE42834: Adults with bacterial pneumonia, lung cancer, or sarcoidosis |
Bacterial | 18 | 3 | 0.99 | ||
| SIRS | 1 | 81 | ||||
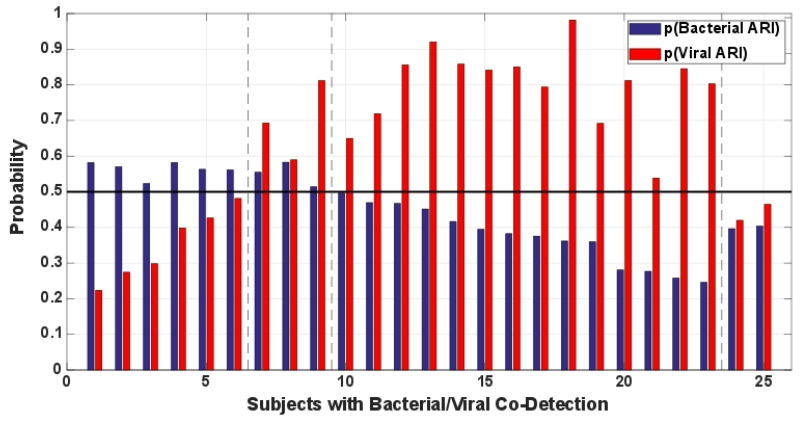
A subgroup of patients with ARI will have both bacterial and viral pathogens identified, often termed co-infection. However, it is unclear how the host responds in such situations. Illness may be driven by the bacteria, the virus, both, or neither at different times in the patient’s clinical course. In an exploratory analysis to determine whether co-infection could be identified with these methods, we applied the bacterial and viral ARI classifiers to patients with bacterial and viral co-identification. GSE60244 included bacterial pneumonia (n=22), viral respiratory tract infection (n=71), and bacterial/viral co-identification (n=25). The co-identification group was defined by the presence of both bacterial and viral pathogens without further information as to the likelihood of bacterial or viral disease.(18) We trained the ARI signatures in GSE60244 subjects with bacterial or viral infection and then validated in those with co-identification (Figure 4). The host response signature was deemed positive above a probability threshold of 0.5. We observed all four possible categories. Six of 25 subjects had a positive bacterial signature; 14/25 had a viral response; 3/25 had positive bacterial and viral signatures; and 2/25 had neither. These results suggest co-infection can be detected using the host response. Moreover, simply identifying bacterial and viral pathogens may not necessarily mean both are inducing a host response.
Fig. 4. Classifier performance in patients with co-infection defined by the identification of bacterial and viral pathogens.
Bacterial and Viral ARI classifiers were trained on subjects with bacterial (N=22) or viral (N=71) infection (GSE60244). This same dataset also included 25 subjects with bacterial/viral co-infection. Bacterial and viral classifier predictions were normalized to the same scale. Each subject receives two probabilities: that of a bacterial ARI host response and a viral ARI host response. A probability score of 0.5 or greater was considered positive. Subjects 1-6 have a bacterial host response. Subjects 7-9 have both bacterial and viral host responses. Subjects 10-23 have a viral host response. Subjects 24-25 do not have bacterial or viral host responses.
Biological pathways
The sparse logistic regression model that generated the classifiers penalizes selection of redundant (correlated) genes (e.g., if from the same pathway) if there is no additive diagnostic value. Consequently, conventional gene enrichment pathway analysis is not appropriate to perform. Moreover, such conventional gene enrichment analyses have been described.(8, 11, 13, 19, 20) Instead a literature review was performed for all classifier genes (Table S5). Overlap between Bacterial, Viral, and Non-infectious Illness Classifiers is shown in Figure S8.
The Viral classifier included known anti-viral response categories such as interferon response, T-cell signaling, and RNA processing. The Viral classifier had the greatest representation of RNA processing pathways such as KPNB1, which is involved in nuclear transport and is co-opted by viruses for transport of viral proteins and genomes.(21, 22) Its downregulation suggests it may play an antiviral role in the host response.
The Bacterial classifier encompassed the greatest breadth of cellular processes, notably cell cycle regulation, cell growth, and differentiation. The Bacterial classifier included genes important in T-, B-, and NK-cell signaling. Unique to the Bacterial classifier were genes involved in oxidative stress, and fatty acid and amino acid metabolism, consistent with sepsis-related metabolic perturbations.(23)
Discussion
Acute respiratory illness accounted for 71 million outpatient visits to U.S. providers in 2007.(24) Existing diagnostics fall short in their ability to differentiate bacterial, viral, and non-infectious etiologies contributing to the inappropriate prescription of antibiotics in 73% of such cases.(3) Created by President Obama in 2014, the Task Force for Combating Antibiotic-Resistant Bacteria has prioritized the development of new and next generation diagnostics.(6) One strategy to accurately define the infecting pathogen class is to use host-gene expression profiles. Using sparse logistic regression, we developed host gene expression profiles that accurately distinguished between bacterial and viral etiologies in patients with acute respiratory symptoms (external validation AUC 0.90-0.99). Deriving the ARI classifier with a non-infectious illness control group imparted a high negative predictive value across a wide range of prevalence estimates. These encouraging metrics offer an opportunity to provide clinically actionable results which can mitigate emerging antibiotic resistance.
Several studies made notable inroads in developing host-response diagnostics for ARI. This includes response to respiratory viruses (7, 9-11, 13), bacterial etiologies in an ICU population (11, 25), and tuberculosis (26-28). Many such studies define host response profiles compared to the healthy state, offering valuable insights into host biology.(29-31) However, these gene lists are suboptimal diagnostic targets because gene expression profiles should ideally be applied to similar populations from which they derive.(14) Since healthy individuals do not present with acute respiratory complaints, they should be excluded from host-response diagnostic development.
Including patients with bacterial and viral infections (10, 11, 13) allows for the distinction between these two states but does not address how to classify non-infectious illness. This phenotype is important to include because patients present in an undifferentiated manner whereby infectious and non-infectious etiologies are possible. This was the rationale for our approach, which was derived from, and can therefore be applied to, an undifferentiated clinical population where such a test is in greatest need. The cohort used to generate this classifier derived from the larger CAPSOD cohort, which includes patients with suspected sepsis of non-respiratory etiology as well. However, we only focused on patients with sepsis due to respiratory tract infection and therefore, we cannot assume these results would apply to a more general sepsis population.
In this study, we report three discrete host-response classifiers: Bacterial ARI, Viral ARI, and Non-Infectious Disease. However, the major clinical decision faced by clinicians is whether or not to prescribe antibacterials. A simpler diagnostic strategy might focus only on the probability of bacterial ARI. However, there is value in providing information about viral or non-infectious alternatives. For example, the confidence to withhold antibacterials in a patient with a low probability of bacterial ARI can be enhanced by a high probability of an alternative diagnosis. Second, a full diagnostic report could identify concurrent illness that a single classifier would miss. We observed this when validating in a population with bacterial-viral co-identification. Such patients are more commonly referred to as “co-infected”. To have infection, there must be a pathogen, a host, and a maladaptive interaction between the two. Simply identifying bacterial and viral pathogens should not imply co-infection. Although we cannot know the true infection status in these 25 subjects with bacterial/viral co-identification, our host response classifiers suggest the existence of multiple host-response states.
Discordant classifications may have arisen either from errors in classification or clinical phenotyping. Errors in clinical phenotyping can arise from a failure to identify causative pathogens due to limitations in current microbiological diagnostics. Alternatively, some non-infectious disease processes may in fact be infection-related through mechanisms that have yet to be discovered. Discordant cases were not clearly explained by a unifying variable such as pathogen type, syndrome, disease severity, or patient characteristic. As such, the gene expression classifiers presented herein are likely impacted by other factors including patient-specific variables (e.g., treatment, comorbidity, duration of illness); test-specific variables (e.g., sample processing, assay conditions, RNA quality and yield); or as-of-yet unidentified variables. These concerns are heightened when validating in publically available datasets where little to no information is made available about how such clinical labels are assigned. In the absence of phenotyping standards, errors in clinical diagnosis will propagate into poor performance of any classifier.
This study is limited in its ability to generalize to other special populations such as neonates, chronic viral infections, and the severely immunocompromised. Some of these patients were included in our cohort but not enough to draw definitive conclusions about classifier performance. In five patients (32), the host-response to bacterial infection resolved with treatment. However, a larger cohort is needed to answer whether ARI classifier kinetics can be used for treatment response monitoring. Moreover, the magnitude of gene expression changes may offer prognostic utility. Although we found no statistically significant difference in classification performance when comparing respiratory to non-respiratory SIRS, it is possible a true difference exists that we were underpowered to detect. We have undertaken a large, prospective collection of patients with acute respiratory complaints in order to directly address all of these limitations (supported by the Antibacterial Resistance Leadership Group, NIAID UM1AI104681).
These results define the necessary content to improve ARI diagnostics in a clinically relevant population. However, the technical hurdle to transfer these targets to a reliable, timely, affordable, and accessible platform remains. Doing so will directly answer the call for new diagnostics to combat antibiotic-resistant bacteria, a national security and public health priority.
Materials and Methods
Study Design
Studies were approved by relevant Institutional Review Boards, and in accord with the Declaration of Helsinki. All subjects or their legally authorized representatives provided written informed consent.
Patients with community-onset, suspected infection were enrolled in the Emergency Departments of Duke University Medical Center (DUMC; Durham, NC), the Durham VA Medical Center (DVAMC; Durham, NC), or Henry Ford Hospital (Detroit, MI) as part of the Community Acquired Pneumonia & Sepsis Outcome Diagnostics study (ClinicalTrials.gov NCT00258869).(23, 29, 33-35) Additional patients were enrolled through UNC Health Care Emergency Department (UNC; Chapel Hill, NC) as part of the Community Acquired Pneumonia and Sepsis Study. Patients were eligible if they had a known or suspected infection and if they exhibited two or more SIRS criteria.(36) The objective of these prospective, observational studies was to identify patients with suspected sepsis, collect clinical information, and bank samples for future research use. Upon adjudication and subject selection (described below), banked samples were accessed and analyzed. ARI cases included patients with upper or lower respiratory tract symptoms, as adjudicated by emergency medicine (SWG, EBQ) or infectious diseases (ELT) physicians. There is no currently accepted consensus criteria by which viral ARI or bacterial ARI can be defined. In this study, we performed retrospective adjudications based on manual chart reviews performed at least 28 days after enrollment and prior to any gene expression-based categorization as described in Langley et al. and in the text below.(23) Medical record information used to support adjudications included, but was not limited to, patient symptoms, physical examination findings, routine laboratory testing, and radiographic findings (when clinically indicated). In order to be categorized as having a viral or bacterial ARI, a subject must have had a compatible clinical syndrome and an identified, compatible pathogen. Seventy patients with microbiologically confirmed bacterial ARI were identified including four with pharyngitis and 66 with pneumonia. Bacterial pharyngitis was adjudicated based on patient-reported symptoms and examination such as tonsillar exudate or swelling, tender adenopathy, fever, and absence of cough along with the identification of Group A Streptococcus, either by antigen detection or culture. Bacterial pneumonia was adjudicated based on patient-reported symptoms and clinical evaluation such as productive cough, fever, leukocytosis/leukopenia, typical radiographic infiltrates (e.g., consolidation) along with the identification of bacterial pathogens known to cause pneumonia. Microbiological etiologies were determined using conventional culture of either blood or respiratory samples, urinary antigen testing (Streptococcus or Legionella), or with serological testing (Mycoplasma). There were 115 patients with viral ARI, including 48 students at Duke University enrolled through the DARPA Predicting Health and Disease study. Viral ARI was adjudicated based on patient-reported symptoms such as upper respiratory complaints (e.g., rhinorrhea, sneezing, post-nasal drip, sore throat); epidemiologic factors such as sick contacts; and clinical evaluation such as absence of radiological findings typical for bacterial infection. This was in conjunction with an identified viral etiology compatible with the clinical syndrome. Viral etiology testing was frequently performed as part of routine clinical care. Specimens were typically nasopharyngeal swabs or lower respiratory tract-derived. In addition, the ResPlex II v2.0 viral PCR multiplex assay (Qiagen) augmented clinical testing for viral etiology identification. This panel detects influenza A and B, adenovirus (B, E), parainfluenza 1-4, respiratory syncytial virus A and B, human metapneumovirus, human rhinovirus, coronavirus (229E, OC43, NL63, HKU1), coxsackie/echo virus, and bocavirus. Upon adjudication, a subset of enrolled patients was determined to have non-infectious illness (n=88) (Table S1). The determination of “non-infectious illness” was made only when an alternative diagnosis was established and results of any routinely ordered microbiological testing failed to support an infectious etiology. Inflammatory markers were not routinely measured for clinical purposes although we did measure procalcitonin concentrations for study purposes. However, because classification performance was compared to procalcitonin, this biomarker was intentionally excluded from the adjudication process. Through this adjudication process, subjects were assigned to one of five likelihoods of infection (23, 33): 1) Definite infection with an identified etiologic agent; 2) Definite infection without an identified etiologic agent; 3) Indeterminate, infection possible; 4) No evidence of infection without an identified non-infectious etiology; and 5) No evidence of infection with an alternative non-infectious etiology. In this study, we focused exclusively on Categories 1 and 5. Lastly, healthy controls (n=44; median age 30 years; range 23-59) were enrolled as part of a study on the effect of aspirin on platelet function among healthy volunteers without symptoms, where gene expression analyses was performed on pre-aspirin challenge time points.(37) The totality of information used to support these adjudications would not have been available to clinicians at the time of their evaluation.
Procalcitonin Measurement
Concentrations were measured at different stages during the study and as a result, different platforms were utilized based on availability. Some serum measurements were made on a Roche Elecsys 2010 analyzer (Roche Diagnostics) by electrochemiluminescent immunoassay. Additional serum measurements were made using the miniVIDAS immunoassay (bioMerieux). When serum was unavailable, measurements were made by the Phadia Immunology Reference Laboratory in plasma-EDTA by immunofluorescence using the B·R·A·H·M·S PCT sensitive KRYPTOR (Thermo Fisher Scientific). Replicates were performed for some paired serum and plasma samples, revealing equivalence in concentrations. Therefore, all procalcitonin measurements were treated equivalently, regardless of testing platform.
Microarray Generation
At initial clinical presentation, patients were enrolled and samples collected for analysis. After adjudications were performed as described above, 317 subjects with clear clinical phenotypes were selected for gene expression analysis. Total RNA was extracted from human blood using the Qiagen PAXgene Blood RNA Kit according to the manufacturer’s protocol. RNA quantity and quality were assessed using the Nanodrop spectrophotometer (Thermo Scientific) and Agilent 2100 Bioanalyzer, respectively. Microarrays were RMA-normalized. Hybridization and data collection were performed at Expression Analysis using the GeneChip Human Genome U133A 2.0 Array according to the Affymetrix Technical Manual.
Validation
The ARI classifier was validated using leave-one-out cross-validation in the same population from which it was derived. Independent, external validation occurred using publically available human gene expression datasets from 328 individuals (GSE6269, GSE42026, GSE40396, GSE20346, and GSE42834). Datasets were chosen if they included at least two clinical groups (bacterial ARI, viral ARI, or non-infectious illness). We also used GSE60244 to specifically validate classifier performance in 25 subjects with bacterial/viral co-identification. To match probes across different microarray platforms, each ARI classifier probe was converted to gene symbols, which were used to identify corresponding target microarray probes. Batch differences across these independent datasets precluded the direct application of the ARI classifier. Consequently, the signatures in the ARI classifier were tuned to each dataset in order to assess classification performance.
Statistical Analysis
The transcriptomes of 317 subjects (273 ill patients and 44 healthy volunteers) were measured in two microarray batches with seven overlapping samples (GSE63990). Exploratory principal component analysis and hierarchical clustering revealed substantial batch differences. These were corrected by first estimating and removing probe-wise mean batch effects using a Bayesian fixed effects model. Next, we fitted a robust linear regression model with Huber loss function using seven overlapping samples, which was used to adjust the remaining expression values.
Sparse classification methods such as sparse logistic regression perform classification and variable selection simultaneously while reducing over-fitting risk.(38) Therefore, separate gene selection strategies such as univariate testing or sparse factor models are unnecessary. Here, a sparse logistic regression model was fitted independently to each of the binary tasks using the 40% of probes with the largest variance after batch correction.(39) Specifically, we used a LASSO regularized generalized linear model with binomial likelihood with nested cross-validation to select for the regularization parameters. Scripts were written in Matlab using the Glmnet toolbox (http://www.stanford.edu/~hastie/glmnet_matlab/) and can be located at https://bitbucket.org/rhenao/ari_stm. This generated Bacterial ARI, Viral ARI, and Non-Infectious Illness classifiers. Provided that each binary classifier estimates class membership probabilities (e.g., probability of bacterial vs. either viral or non-infectious in the case of the Bacterial ARI classifier), we can combine the three classifiers into a single decision model (termed the ARI classifier) by following a one-versus-all scheme whereby largest membership probability assigns class label.(38, 40) Classification performance metrics included area-under-the-receiving-operating-characteristic-curve (AUC) for binary outcomes and confusion matrices for ternary outcomes.(41) Determinations of significance included Wilcoxon rank sum, Fisher’s exact test, and McNemar’s test with Yates correction. Corrections for multiple testing and significance cutoffs are noted in the Results.
Supplementary Material
Acknowledgments
Funding: Supported by the US Defense Advanced Research Projects Agency (DARPA) through contracts N66001-07-C-2024 and N66001-09-C-2082; and by grants from the NIH (U01AI066569, P20RR016480, and HHSN266200400064C). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. ELT was supported by a National Research Service Award training grant provided by the Agency for Healthcare Research and Quality. ELT and MTM were also supported by Award Number 1IK2CX000530 and 1IK2CX000611, respectively, from the Clinical Science Research and Development Service of the VA Office of Research and Development. The views expressed in this article are those of the authors and do not necessarily represent the views of the Department of Veterans Affairs. VGF was supported by a Mid-Career Mentoring Award K24-AI093969 from the National Institutes of Health. The authors acknowledge bioMérieux, Inc. for kind contribution of reagents used to measure procalcitonin concentrations. The data contained in this manuscript has not previously been presented.
Footnotes
Fig. S1: Positive and negative predictive values for A) Bacterial and B) Viral ARI classification as a function of prevalence.
Fig. S2: Validation of Bacterial and Viral ARI Classifiers in GSE6269.
Fig. S3: Validation of Bacterial and Viral ARI Classifiers in GSE42026.
Fig. S4: Validation of Bacterial and Viral ARI Classifiers in GSE40396.
Fig. S5: Validation of Bacterial and Viral ARI Classifiers in GSE20346.
Fig. S6: Validation of Bacterial ARI and Non-Infectious Illness Classifiers in GSE42834.
Fig. S7: Treatment effect on bacterial ARI classification.
Fig. S8: Venn diagram representing overlap in the Bacterial ARI, Viral ARI, and Non-infectious Illness Classifiers.
Table S1: Etiological causes of illness for subjects with viral ARI, bacterial ARI, and non-infectious illness.
Table S2: Summary of clinical features for the derivation cohort.
Table S3: Probes selected for the Bacterial ARI, Viral ARI, and Non-infectious Illness Classifiers.
Table S4: Subjects with discordant predictions compared to clinical assignments.
Table S5: Genes in the Bacterial ARI, Viral ARI, and Non-infectious Illness Classifiers, grouped by biologic process.
Author contributions: ELT, MN, TB, MTM, AKZ, LC, GSG, and CWW helped conceive the study. All authors helped acquire, analyze, or interpret data. ELT, RH, and ERK drafted the manuscript, which was critically revised by all remaining authors. Statistical analysis was specifically performed by RH, JL, and LC. Funding was obtained by CBC, EPR, AKZ, SFK, CGF, GSG, and CWW. All authors had full access to all data in this study.
Competing interests: All authors report no competing interests as it pertains to this manuscript. The following individuals report additional activities, but not as competing interests to this manuscript: CWW served as a scientific consultant to bioMerieux, Becton Dickinson, and Verigene. He received research support from the NIH, the Defense Advanced Research Projects Agency (DARPA), the Defense Threat Reduction Agency (DTRA), the Bill and Melinda Gates Foundation (BMGF), the Veterans Administration (VHA), the Centers for Disease Control and Prevention, Novartis Pharmaceuticals, Roche Molecular, bioMerieux, and Qiagen. SFK served as a scientific advisor to Parabase Genomics Inc. and Edico Genomics Inc. He received research support from the NIH. ELT has received research support from DARPA, DTRA, BMGF, VHA, and Novartis Pharmaceuticals and has served as a scientific consultant to Immunexpress. VGF has grants from the NIH, MedImmune, Forest/Cerexa, Pfizer, Merck, Advanced Liquid Logics, Theravance, Novartis, and Cubist. He served as the Chair of the Merck scientific advisory board for the V710 S. aureus vaccine. He has been a consultant for Pfizer, Novartis, Galderma, Novadigm, Durata, Debiopharm, Genentech, Achaogen, Affinium, Medicines Co., Cerexa, Tetraphase, Trius, MedImmune, Bayer, Theravance, Cubist, Basilea, Affinergy, and Contrafect. He also received royalties from UpToDate and has been paid for the development of educational presentations for Green Cross, Cubist, Cerexa, Durata, and Theravance. GSG has consulted for US Diagnostic Standards and has served on the Scientific Advisory Board for Pappas Ventures. He has received grants from the US Defense Advanced Research Projects Agency, the Gates Foundation, and Novartis Vaccines and Diagnostics. GSG, ELT, VGF, and CWW have a patent pending for host gene expression signatures of Staphylococcus aureus and Escherichia coli infections. GSG, ELT, RH, TB, MTM, LC, and CWW have filed a patent for methods of identifying infectious disease and assays for identifying infectious disease, as well as for molecular predictors of fungal infection. RJL and SFK have a patent pending for sepsis prognosis biomarkers.
Data and materials availability: Gene expression data generated in this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GSE63990). This study also utilized gene expression data from existing datasets (GSE6269, GSE42026, GSE40396, GSE20346, GSE42834, and GSE60244).
References and Notes
- 1.WHO [accessed on Nov 8, 2011];Mortality and burden of disease: Cause-specific mortality, 2008: WHO regions. Global Health Observatory Data Repository. 2011 Available at: http://apps.who.int/ghodata/
- 2.Shapiro DJ, Hicks LA, Pavia AT, Hersh AL. Antibiotic prescribing for adults in ambulatory care in the USA, 2007-09. The Journal of antimicrobial chemotherapy. 2014;69:234–240. doi: 10.1093/jac/dkt301. [DOI] [PubMed] [Google Scholar]
- 3.Lee GC, Reveles KR, Attridge RT, Lawson KA, Mansi IA, Lewis JS, 2nd, Frei CR. Outpatient antibiotic prescribing in the United States: 2000 to 2010. BMC medicine. 2014;12:96. doi: 10.1186/1741-7015-12-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gould IM. Antibiotic resistance: the perfect storm. International Journal of Antimicrobial Agents. 2009;34(Supplement 3):S2–S5. doi: 10.1016/S0924-8579(09)70549-7. [DOI] [PubMed] [Google Scholar]
- 5.Kim JH, Gallis HA. Observations on spiraling empiricism: its causes, allure, and perils, with particular reference to antibiotic therapy. Am J Med. 1989;87:201–206. doi: 10.1016/s0002-9343(89)80697-7. [DOI] [PubMed] [Google Scholar]
- 6.Executive Order - Combating Antibiotic-Resistant Bacteria. 2014 http://www.whitehouse.gov/the-press-office/2014/09/18/executive-order-combating-antibiotic-resistant-bacteria.
- 7.Zaas AK, Chen M, Varkey J, Veldman T, Hero AO, 3rd, Lucas J, Huang Y, Turner R, Gilbert A, Lambkin-Williams R, Oien NC, Nicholson B, Kingsmore S, Carin L, Woods CW, Ginsburg GS. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe. 2009;6:207–217. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woods CW, McClain MT, Chen M, Zaas AK, Nicholson BP, Varkey J, Veldman T, Kingsmore SF, Huang Y, Lambkin-Williams R, Gilbert AG, Hero AO, III, Ramsburg E, Glickman S, Lucas JE, Carin L, Ginsburg GS. A Host Transcriptional Signature for Presymptomatic Detection of Infection in Humans Exposed to Influenza H1N1 or H3N2. PLoS ONE. 2013;8:e52198. doi: 10.1371/journal.pone.0052198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mejias A, Dimo B, Suarez NM, Garcia C, Suarez-Arrabal MC, Jartti T, Blankenship D, Jordan-Villegas A, Ardura MI, Xu Z, Banchereau J, Chaussabel D, Ramilo O. Whole Blood Gene Expression Profiles to Assess Pathogenesis and Disease Severity in Infants with Respiratory Syncytial Virus Infection. PLoS medicine. 2013;10:e1001549. doi: 10.1371/journal.pmed.1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, Chaussabel D. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parnell GP, McLean AS, Booth DR, Armstrong NJ, Nalos M, Huang SJ, Manak J, Tang W, Tam OY, Chan S, Tang BM. A distinct influenza infection signature in the blood transcriptome of patients who presented with severe community acquired pneumonia. Crit Care. 2012;16:R157. doi: 10.1186/cc11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaas AK, Burke T, Chen M, McClain M, Nicholson B, Veldman T, Tsalik EL, Fowler V, Rivers EP, Otero R, Kingsmore SF, Voora D, Lucas J, Hero AO, Carin L, Woods CW, Ginsburg GS. A host-based RT-PCR gene expression signature to identify acute respiratory viral infection. Sci Transl Med. 2013;5:203ra126. doi: 10.1126/scitranslmed.3006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu X, Yu J, Crosby SD, Storch GA. Gene expression profiles in febrile children with defined viral and bacterial infection. Proceedings of the National Academy of Sciences. 2013 doi: 10.1073/pnas.1302968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lytkin NI, McVoy L, Weitkamp JH, Aliferis CF, Statnikov A. Expanding the understanding of biases in development of clinical-grade molecular signatures: a case study in acute respiratory viral infections. PLoS One. 2011;6:e20662. doi: 10.1371/journal.pone.0020662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon L, Gauvin F, Amre DK, Saint-Louis P, Lacroix J. Serum procalcitonin and C-reactive protein levels as markers of bacterial infection: a systematic review and meta-analysis. Clin Infect Dis. 2004;39:206–217. doi: 10.1086/421997. [DOI] [PubMed] [Google Scholar]
- 16.McNemar Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika. 1947;12:153–157. doi: 10.1007/BF02295996. [DOI] [PubMed] [Google Scholar]
- 17.Yates F. Contingency Tables Involving Small Numbers and the χ2 Test. Supplement to the Journal of the Royal Statistical Society. 1934;1:217–235. [Google Scholar]
- 18.Suarez NM, Bunsow E, Falsey AR, Walsh EE, Mejias A, Ramilo O. Transcriptional Profiling is Superior to Procalcitonin to Discriminate Bacterial vs Viral Lower Respiratory Tract Infections in Hospitalized Adults. Journal of Infectious Diseases. 2015 doi: 10.1093/infdis/jiv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Zaas AK, Rao A, Dobigeon N, Woolf PJ, Veldman T, Oien NC, McClain MT, Varkey JB, Nicholson B, Carin L, Kingsmore S, Woods CW, Ginsburg GS, Hero AO., 3rd Temporal dynamics of host molecular responses differentiate symptomatic and asymptomatic influenza a infection. PLoS genetics. 2011;7:e1002234. doi: 10.1371/journal.pgen.1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsalik EL, Langley RJ, Dinwiddie DL, Miller NA, Yoo B, van Velkinburgh JC, Smith LD, Thiffault I, Jaehne AK, Valente AM, Henao R, Yuan X, Glickman SW, Rice BJ, McClain MT, Carin L, Corey GR, Ginsburg GS, Cairns CB, Otero RM, Fowler VG, Jr., Rivers EP, Woods CW, Kingsmore SF. An integrated transcriptome and expressed variant analysis of sepsis survival and death. Genome medicine. 2014;6:111. doi: 10.1186/s13073-014-0111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bukrinsky MI, Sharova N, Dempsey MP, Stanwick TL, Bukrinskaya AG, Haggerty S, Stevenson M. Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc Natl Acad Sci U S A. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghildyal R, Ho A, Wagstaff KM, Dias MM, Barton CL, Jans P, Bardin P, Jans DA. Nuclear import of the respiratory syncytial virus matrix protein is mediated by importin beta1 independent of importin alpha. Biochemistry. 2005;44:12887–12895. doi: 10.1021/bi050701e. [DOI] [PubMed] [Google Scholar]
- 23.Langley RJ, Tsalik EL, Velkinburgh J. C. v., Glickman SW, Rice BJ, Wang C, Chen B, Carin L, Suarez A, Mohney RP, Freeman DH, Wang M, You J, Wulff J, Thompson JW, Moseley MA, Reisinger S, Edmonds BT, Grinnell B, Nelson DR, Dinwiddie DL, Miller NA, Saunders CJ, Soden SS, Rogers AJ, Gazourian L, Fredenburgh LE, Massaro AF, Baron RM, Choi AMK, Corey GR, Ginsburg GS, Cairns CB, Otero RM, Fowler VG, Rivers EP, Woods CW, Kingsmore SF. An Integrated Clinico-Metabolomic Model Improves Prediction of Death in Sepsis. Science Translational Medicine. 2013;5:195ra195. doi: 10.1126/scitranslmed.3005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Center for Health Statistics . Ambulatory medical care utilization estimates for 2007. 2011. [PubMed] [Google Scholar]
- 25.Severino P, Silva E, Baggio-Zappia GL, Brunialti MKC, Nucci LA, Rigato O, Jr, da Silva IDCG, Machado FR, Salomao R. Patterns of Gene Expression in Peripheral Blood Mononuclear Cells and Outcomes from Patients with Sepsis Secondary to Community Acquired Pneumonia. PLoS ONE. 2014;9:e91886. doi: 10.1371/journal.pone.0091886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson ST, Kaforou M, Brent AJ, Wright VJ, Banwell CM, Chagaluka G, Crampin AC, Dockrell HM, French N, Hamilton MS, Hibberd ML, Kern F, Langford PR, Ling L, Mlotha R, Ottenhoff THM, Pienaar S, Pillay V, Scott JAG, Twahir H, Wilkinson RJ, Coin LJ, Heyderman RS, Levin M, Eley B. Diagnosis of Childhood Tuberculosis and Host RNA Expression in Africa. New England Journal of Medicine. 2014;370:1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, Quinn C, Blankenship D, Dhawan R, Cush JJ, Mejias A, Ramilo O, Kon OM, Pascual V, Banchereau J, Chaussabel D, O’Garra A. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaforou M, Wright VJ, Oni T, French N, Anderson ST, Bangani N, Banwell CM, Brent AJ, Crampin AC, Dockrell HM, Eley B, Heyderman RS, Hibberd ML, Kern F, Langford PR, Ling L, Mendelson M, Ottenhoff TH, Zgambo F, Wilkinson RJ, Coin LJ, Levin M. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS medicine. 2013;10:e1001538. doi: 10.1371/journal.pmed.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahn SH, Tsalik EL, Cyr DD, Zhang Y, van Velkinburgh JC, Langley RJ, Glickman SW, Cairns CB, Zaas AK, Rivers EP, Otero RM, Veldman T, Kingsmore SF, Lucas J, Woods CW, Ginsburg GS, Fowler VG., Jr. Gene Expression-Based Classifiers Identify Staphylococcus aureus Infection in Mice and Humans. PLoS ONE. 2013;8:e48979. doi: 10.1371/journal.pone.0048979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banchereau R, Jordan-Villegas A, Ardura M, Mejias A, Baldwin N, Xu H, Saye E, Rossello-Urgell J, Nguyen P, Blankenship D, Creech CB, Pascual V, Banchereau J, Chaussabel D, Ramilo O. Host immune transcriptional profiles reflect the variability in clinical disease manifestations in patients with Staphylococcus aureus infections. PLoS One. 2012;7:e34390. doi: 10.1371/journal.pone.0034390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herberg JA, Kaforou M, Gormley S, Sumner ER, Patel S, Jones KDJ, Paulus S, Fink C, Martinon-Torres F, Montana G, Wright VJ, Levin M. Transcriptomic Profiling in Childhood H1N1/09 Influenza Reveals Reduced Expression of Protein Synthesis Genes. Journal of Infectious Diseases. 2013;208:1664–1668. doi: 10.1093/infdis/jit348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bloom CI, Graham CM, Berry MP, Rozakeas F, Redford PS, Wang Y, Xu Z, Wilkinson KA, Wilkinson RJ, Kendrick Y, Devouassoux G, Ferry T, Miyara M, Bouvry D, Valeyre D, Gorochov G, Blankenship D, Saadatian M, Vanhems P, Beynon H, Vancheeswaran R, Wickremasinghe M, Chaussabel D, Banchereau J, Pascual V, Ho LP, Lipman M, O’Garra A. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One. 2013;8:e70630. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glickman SW, Cairns CB, Otero RM, Woods CW, Tsalik EL, Langley RJ, van Velkinburgh JC, Park LP, Glickman LT, Fowler VG, Jr., Kingsmore SF, Rivers EP. Disease progression in hemodynamically stable patients presenting to the emergency department with sepsis. Acad Emerg Med. 2010;17:383–390. doi: 10.1111/j.1553-2712.2010.00664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsalik EL, Jaggers LB, Glickman SW, Langley RJ, van Velkinburgh JC, Park LP, Fowler VG, Cairns CB, Kingsmore SF, Woods CW. Discriminative value of inflammatory biomarkers for suspected sepsis. J Emerg Med. 2012;43:97–106. doi: 10.1016/j.jemermed.2011.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsalik EL, Jones D, Nicholson B, Waring L, Liesenfeld O, Park LP, Glickman SW, Caram LB, Langley RJ, van Velkinburgh JC, Cairns CB, Rivers EP, Otero RM, Kingsmore SF, Lalani T, Fowler VG, Woods CW. Multiplex PCR to diagnose bloodstream infections in patients admitted from the emergency department with sepsis. Journal of clinical microbiology. 2010;48:26–33. doi: 10.1128/JCM.01447-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 37.Voora D, Ortel TL, Lucas JE, Chi JT, Becker RC, Ginsburg GS. Abstract 16293: A Whole Blood RNA Signature Accurately Classifies Multiple Measures of Platelet Function on Aspirin in Healthy Volunteers and Highlights a Common Underlying Pathway. Circulation. 2010;122:A16293. [Google Scholar]
- 38.Bishop CM. Pattern Recognition and Machine Learning (Information Science and Statistics) Springer-Verlag; New York, Inc.: 2006. [Google Scholar]
- 39.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of statistical software. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 40.Rifkin R, Klautau A. In Defense of One-Vs-All Classification. J. Mach. Learn. Res. 2004;5:101–141. [Google Scholar]
- 41.Fawcett T. An introduction to ROC analysis. Pattern Recognition Letters. 2006;27:861–874. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.