Abstract
Class II transcription activators function by binding to a DNA site overlapping a core promoter and stimulating isomerization of an initial RNA polymerase (RNAP)-promoter closed complex into a catalytically competent RNAP-promoter open complex. Here we report a 4.4 Å crystal structure of an intact bacterial Class II transcription activation complex. The structure comprises Thermus thermophilus transcription activator protein TTHB099 (TAP; homolog of Escherichia coli catabolite activator protein, CAP), T. thermophilus RNAP σA holoenzyme, a Class II TAP-dependent promoter, and a ribotetranucleotide primer. The structure reveals the interactions between RNAP holoenzyme and DNA responsible for transcription initiation and reveals the interactions between TAP and RNAP holoenzyme responsible for transcription activation. The structure indicates that TAP stimulates isomerization through simple, adhesive, stabilizing protein-protein interactions with RNAP holoenzyme.
Simple bacterial transcription activators — those that stimulate transcription from a single DNA site without other factors — are divided into two classes (1-3). Class I transcription activators, typified by E. coli catabolite activator protein (CAP) at the lac promoter, stimulate transcription by binding to a specific DNA site upstream of a core promoter and facilitating binding of RNA polymerase (RNAP) holoenzyme to form an RNAP-promoter closed complex (RPc; 1-3). Class II transcription activators, typified by E. coli CAP at the gal promoter, stimulate transcription by binding to a specific DNA site overlapping a core promoter and facilitating conversion of RPc into a catalytically competent RNAP-promoter open complex (RPo) containing ∼13 bp of unwound DNA (“transcription bubble”; 1-3). A 20 Å resolution EM structure of a Class I transcription activation complex has been reported (4), but no structure of a Class II transcription activation complex previously has been reported. Here we determine the 4.4 Å resolution crystal structure of a Class II transcription activation complex comprising T. thermophilus transcription activator protein TTHB099 (TAP; a thermophilic sequence, structural, and functional homolog of E. coli CAP; 5), T. thermophilus RNAP σA holoenzyme, a Class II TAP-dependent promoter, and the ribotetranucleotide primer UpCpGpA (TAP-RPo; Table S1; Figs. 1, S1-S2).
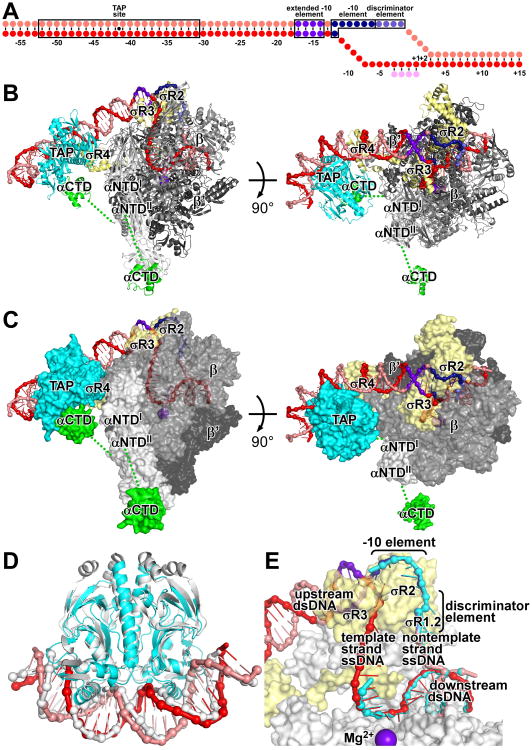
Fig. 1. Structure of TAP-RPo.
(A) Nucleic-acid scaffold. Pink, nontemplate strand; red, template strand; magenta, UpCpGpA; violet, extended -10 element; blue, -10 element; light blue, discriminator element.
(B-C) TAP-RPo (ribbons in B; surfaces in C; β′ nonconserved region omitted for clarity). Cyan, TAP; yellow, σ; white, green, gray, and dark gray, RNAP αNTD, αCTD, β, and β′. Other colors as in A. Dashed lines, αNTD-αCTD linkers.
(D) Comparison of TAP-DNA in TAP-RPo (colors as in B-C) to CAP-DNA (gray; 6).
(E) Comparison of transcription bubble and downstream dsDNA in TAP-RPo (colors as in B-C) to corresponding DNA segments in RPo (cyan; 7).
To obtain a structure of TAP-RPo, we used a nucleic-acid scaffold corresponding to positions -57 to +15 of a Class II TAP-dependent promoter (positions numbered relative to transcription start site; Figs. 1A, S1). The scaffold contained a consensus DNA site for TAP centered between positions -41 and -42 (same position as DNA site for E. coli CAP in gal promoter; 1-2), a near-consensus extended -10 element (3), a consensus -10 element (3), a consensus discriminator element (3), a consensus core recognition element (3), a 13 bp transcription bubble (maintained in the unwound state by having non-complementary sequences on nontemplate and template strands), and UpCpGpA.
In the structure of TAP-RPo, TAP interacts with DNA, RNAP holoenzyme interacts with DNA, and TAP and RNAP holoenzyme make protein-protein interactions (Fig. 1B-C). The structure of TAP-DNA in TAP-RPo is superimposible on the structure of CAP-DNA (6), corroborating that TAP is a homolog of CAP (Fig. 1D). The structure of RPo in TAP-RPo is essentially superimposible on structures of RPo (7-10; neglecting RNAP α subunit C-terminal domain, αCTD, which was not resolved in previous structures), indicating that interactions between the Class II activator and RPo do not substantially alter the conformation of RPo (Fig. 1E).
RNAP contains two copies of αCTD, each of which is connected to the rest of RNAP through a flexible linker (1-3). In the structure of TAP-RPo, one αCTD (probably αCTDI; Fig. S3) interacts with TAP, and the other αCTD (probably αCTDII; Fig. S3) makes no interactions (Fig. 1B-C). In the crystal, the second αCTD is constrained by lattice contacts (i.e., contacts with TAP in an adjacent molecule of TAP-RPo in the lattice; Fig. S4). In solution, this αCTD would be free to adopt other positions.
The structure defines the interactions between RNAP holoenzyme and DNA that mediate promoter recognition and promoter unwinding in transcription initiation (Figs. 1-2) and the interactions between TAP and RNAP holoenzyme that mediate transcription activation (Figs. 1, 3-4).
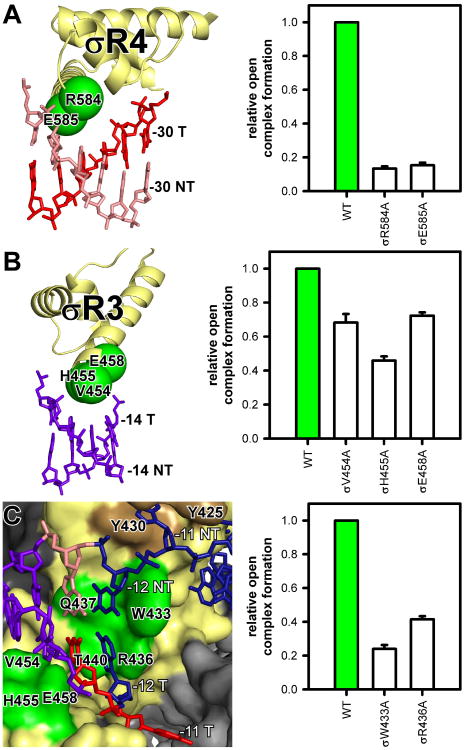
Fig. 2. Protein-DNA interactions that mediate promoter recognition.
Green, σ residues that contact DNA bases (numbered as in E. coli σ70); brown, σR2 residues that contact nontemplate-strand base -11. Other colors as in Fig. 1B-C. Graphs, effects on RPo formation of Ala substitutions of E. coli σ70 (mean±SEM; N ≥3).
(A) Interactions between σR4 and -35-region.
(B) Interactions between σR3 and extended -10 element.
(C) Interactions between σR2 and first (-12NT, -12T) and second (-11NT, -11T) positions of -10 element.
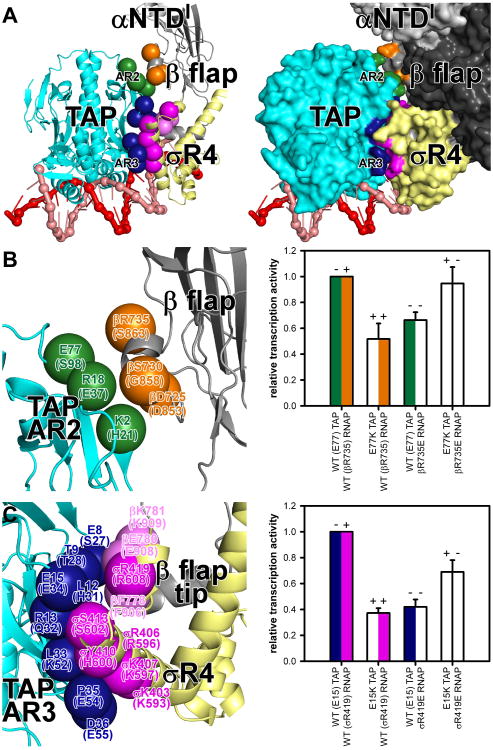
Fig. 3. Protein-protein interactions that mediate transcription activation: AR2 and AR3.
Green, TAP AR2; blue, TAP AR3; orange, RNAP β-flap residues that contact AR2; magenta and light magenta, σR4 and RNAP β-flap-tip residues that contact AR3 (numbered as in TAP and T. thermophilus RNAP holoenzyme and, in parentheses, as in CAP and E. coli RNAP holoenzyme). Other colors as in Fig. 1B-C. Graphs, effects on TAP-dependent transcription of single and double charge-reversal substitutions (mean±SEM; N≥3).
(A) Interactions between AR2, AR3, and RNAP holoenzyme (left, ribbons; right, surfaces).
(B) AR2 interactions.
(C) AR3 interactions.
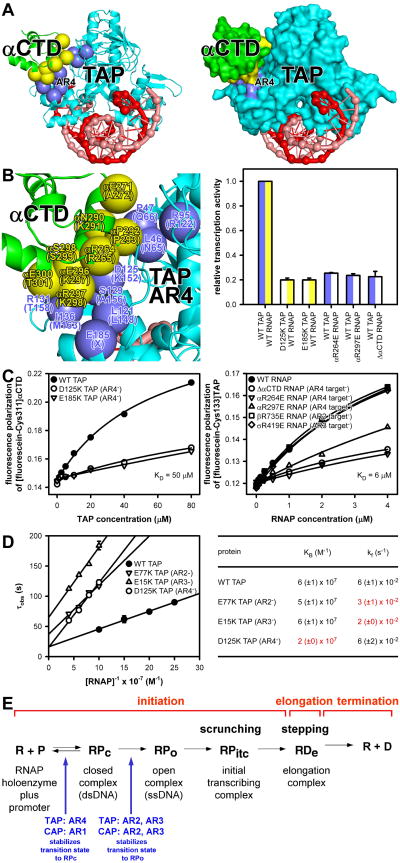
Fig. 4. Protein-protein interactions that mediate transcription activation: AR4 and kinetics.
(A-B) TAP AR4 interaction with αCTD (left, ribbons; right, surfaces; lower left, close-up). Violet, TAP AR4; yellow, αCTD residues that contact AR4. Other colors as in Fig. 1B-C. Graph, effects on TAP-dependent transcription of charge-reversal substitutions of AR4 and αCTD or truncation of αCTD (mean±SEM; N≥3).
(C) TAP-αCTD (left) and TAP-RNAP (right) interactions in the absence of DNA.
(D) Data (left) and parameters (right) for effects of substitutions of AR2, AR3, and AR4 on kinetics of transcription initiation.
(E) Summary of Class II activator-dependent transcription.
TAP and σ conserved region σR4 “co-recognize” the promoter -35-region, contacting the same DNA segment from different faces of the DNA helix (Figs. 1B-C, 2A). The general mode of interaction of σR4 with -35-region DNA in TAP-RPo — binding of the second α-helix of the σR4 helix-turn-helix motif in the DNA major groove — is the same as in RPo (Figs. 2A, S5; 8-10), but, due to DNA distortion by TAP, -35-region DNA is rotated ∼20° away from σR4 (Fig. S5). This rotation decreases the number of σR4 residues that contact DNA bases from 3 to 2 and decreases the number of contacted DNA bases from 4 to 2, providing a structural explanation for the observation that, although -35-region DNA sequences are recognized in Class II activator-dependent transcription, the recognition specificity is less and the number of recognized bases is smaller than in activator-independent transcription (11). Two σR4 residues are positioned to make contacts with DNA bases that potentially enable sequence readout (Fig. 2A). Substitution of these residues reduces RPo formation, verifying their importance (Fig. 2A).
σ conserved region σR3 interacts with the promoter extended -10-region (Figs. 1B-C, 2B; 8-10). Three σR3 residues are positioned to make contacts with DNA bases (Figs. 2B, S6). Substitution of these residues reduces RPo formation, verifying their importance (Fig. 2B).
σ conserved region σR2 interacts with the promoter -10-element at the “upstream fork junction” where DNA unwinding occurs to form the transcription bubble (Figs. 1E, 2C). σR2 interacts with the first position of the -10-element (-12) as dsDNA and the second through sixth positions of the -10 element (-11 through -7) as nontemplate-strand ssDNA (Figs. 1E, 2C). σR2 Trp433 (numbered as in E. coli σ70) is positioned to stack on the nontemplate-strand base of base pair -12, forming a “wedge” that forces the nontemplate-stand -11 base to unstack and flip outside the DNA helix (9), where it is captured by binding within a pocket formed by residues of σR2 (Figs. 2C, S6; 7-10). σR2 Arg436 is positioned to stack on the template-strand base of base pair -12, forming an analogous “wedge” that forces the template-strand -11 base to unstack and flip outside the DNA helix, where it is captured within a channel formed by residues of RNAP, σR2, and σR3.2, that leads into the RNAP active-center cleft (Figs. 2C, S6-S7). Substitution of W433 or R436 results in defects in RPo formation, verifying their importance (Fig. 2C). A second pair of residues, Gln437 and Thr440, are positioned to make direct contacts with the nontemplate- and template-strand bases of the -12 base pair, providing a structural explanation for the observation that substitution of these residues alters specificity at -12 (Figs. 2C, S6; 12).
σ conserved region σR1.2 interacts with nontemplate-strand ssDNA of the discriminator element (Fig. 1E, 7-10). RNAP core interacts with the nontemplate-strand ssDNA of the core recognition element, template-strand ssDNA of the transcription bubble, and downstream dsDNA (Fig. 1E, 7-10).
Genetic and biochemical experiments indicate that Class II transcription activation by E. coli CAP involves three sets of protein-protein interactions: (i) activating region 1 (AR1) interacts with αCTD, (ii) activating region 2 (AR2) interacts with a species-specific insertion in αNTDI (162-165 determinant), and (iii) activating region 3 (AR3) interacts with αR4 (1-2).
In TAP-RPo, a surface of TAP corresponding to AR2 approaches αNTDI and contacts the RNAP β flap (Fig. 3A). Three residues of TAP AR2 are positioned to make direct contacts with three residues of RNAP β subunit (Fig. 3B). TAP Glu77 and RNAP β Arg735 are positioned to form a salt bridge in the AR2-RNAP interface (Fig. 3B). Charge-reversal substitution of either residue decreases TAP-dependent transcription, and charge-reversal substitution of both residues, which re-creates a salt bridge, restores TAP-dependent transcription, confirming the importance of the inferred interaction (Fig. 3B). Homology modeling of CAP-RPo based on TAP-RPo indicates that CAP AR2 is positioned to contact the αNTDI 162-165 determinant (a species-specific insertion present in E. coli RNAP but not in T. thermophilus RNAP; Fig. S8A-B), consistent with previous work (13). Homology modeling indicates that CAP AR2 also is positioned to contact the RNAP β flap (Fig. S8A-B). Substitution of the inferred interacting residues decreases CAP-dependent transcription, indicating the inferred interactions occur and are important (Fig. S8B).
In TAP-RPo, a surface of TAP corresponding to AR3 contacts σR4 α-helices 4 and 5 and the RNAP β flap-tip α-helix (Fig. 3A). Eight predominantly negatively charged residues of TAP AR3 are positioned to interact with six predominantly positively charged residues of σR4 and three predominantly positively charged residues of the β flap-tip α-helix (Fig. 3C). TAP Glu15 is positioned to form a salt bridge with a σR4 Arg residue at the center of the interface (Fig. 3C). Charge-reversal substitution of either residue decreases TAP-dependent transcription, and charge-reversal substitution of both residues, re-creating a salt bridge, restores TAP-dependent transcription, indicating the interactions occur and are important (Fig. 3C). Homology modeling of CAP-RPo based on TAP-RPo predicts equivalent interactions between seven predominantly negatively charged residues of CAP AR3 and five predominantly positively charged residues of σR4 and one residue of the β flap-tip (Fig. S8A,C), consistent with previous work (14-15).
In TAP-RPo, the surface of TAP corresponding to AR1 makes no interactions, and, instead, a different surface of TAP, here designated “activating region 4” (AR4), interacts with αCTD (Figs. 1B-C, 4A). The interface between TAP AR4 and αCTD is large (300 Å2; Fig. 4B). Nine residues of TAP AR4 are positioned to make direct contacts with eight residues of αCTD (Fig. 4B). Substitution of residues implicated in TAP AR4-αCTD interaction results in defects in TAP-dependent transcription (Fig. 4B). TAP-αCTD interactions differ from CAP-αCTD interactions not only in the identities of the activating regions (AR4 in TAP; AR1 in CAP), but also in the fact that TAP interacts with αCTD not bound to DNA, whereas CAP interacts with αCTD bound to DNA immediately upstream of CAP (Figs. 1B-C, 4A; 1-2). Hydroxyl-radical DNA footprinting confirms that αCTD functions differently in T. thermophilus than in E. coli. Thus, T. thermophilus αCTD does not footprint DNA at a Class II TAP-dependent promoter or a ribosomal RNA promoter (Figs. S9-S11), in contrast to E. coli αCTD, which footprints DNA immediately upstream of CAP at a Class II CAP-dependent promoter and A/T-rich UP-element DNA immediately upstream of the -35 element at a ribosomal RNA promoter (Figs. S9-S11). Consistent with the structure of TAP-RPo, fluorescence-polarization assays show that TAP is able to bind to αCTD in the absence of DNA and that the binding requires AR4 (Fig. 4C, left). Further consistent with the structure, fluorescence-polarization assays show that TAP is able to bind to RNAP holoenzyme in the absence of DNA and that the binding requires AR4 interactions and does not require AR2 and AR3 interactions (Fig. 4C, right).
The finding that TAP is able to bind to RNAP holoenzyme in the absence of DNA raises the possibility that TAP, in contrast to CAP, can access not only a “recruitment” pathway, in which the activator interacts first with DNA and then with RNAP holoenzyme, but also a “pre-recruitment” pathway, in which the activator interacts first with RNAP holoenzyme and then with DNA (Fig. S12; 16). Based on the KD for TAP-RNAP holoenzyme interaction (6 μM; Fig. 4C, right) and the concentration of non-transcribing RNAP in bacteria in vivo (5 μM; 17), it appears likely that a pre-recruitment pathway contributes to TAP-dependent transcription in T. thermophilus in vivo.
Measurements of effects of substitution of TAP activating regions on the kinetics of transcription initiation indicate that TAP AR2 and AR3 promote isomerization of RPc to RPo (kf), and TAP AR4 promotes formation of RPc (KB; Fig. 4D). This pattern is reminiscent of the pattern for E. coli CAP, for which AR2 and AR3 promote isomerization of RPc to RPo (11,13,15), and AR1, through interaction with αCTD, promotes formation of RPc (11,13).
A long-standing question has been how a Class II activator promotes isomerization of RPc to RPo, which entails loading of DNA into the RNAP active-center cleft, unwinding of DNA to form the transcription bubble, and closure of the RNAP clamp (1-3,13,18-20). The structure of TAP-RPo reveals that TAP does not interact with, and does not alter the conformation or interactions of, the RNAP active-center cleft, the transcription bubble, or the RNAP clamp. The structure further reveals that the interactions that promote isomerization — AR2 and AR3 interactions — are simple, adhesive, stabilizing protein-protein interactions between exposed surfaces of TAP and exposed surfaces of RNAP holoenzyme (Figs. 3, S8). We infer that interactions between a Class II activator and RNAP holoenzyme that promote formation of RPc (AR4 interactions for TAP; AR1 interactions for CAP) and interactions between Class II activator and RNAP holoenzyme that promote isomerization (AR2 and AR3 interactions) do not differ in character, but, instead, differ only in timing (13,18-20). The former first occur in the transition state for formation of RPc and stabilize both RPc and RPo, whereas the later first occur in the transition state for isomerization of RPc to RPo, and stabilize RPo (Fig. 4E).
Supplementary Material
Acknowledgments
We thank the National Synchrotron Light Source for beamline access and E. Arnold for discussion. This work was funded by NIH grant GM041376 to RHE. The Protein Data Bank accession code is 5I2D.
References
- 1.Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 2.Lawson C, Swigon D, Murakami K, Darst S, Berman H, Ebright RH. Catabolite activator protein (CAP): DNA binding and transcription activation. Curr Opin Structl Biol. 2004;14:10. doi: 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decker K, Hinton D. Transcription regulation at the core: similarities among bacterial, archaeal, and eukaryotic RNA polymerases. Annu Rev Microbiol. 2013 Jun 13;67:113. doi: 10.1146/annurev-micro-092412-155756. [DOI] [PubMed] [Google Scholar]
- 4.Hudson B, Quispe J, Lara-González S, Kim Y, Berman H, Arnold E, et al. Three-dimensional EM structure of an intact activator-dependent transcription initiation complex. Proc Natl Acad Sci USA. 2009;106:19830. doi: 10.1073/pnas.0908782106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agari Y, Kuramitsu S, Shinkai A. X-ray crystal structure of TTHB099, a CRP/FNR superfamily transcriptional regulator from Thermus thermophilus HB8, reveals a DNA-binding protein with no required allosteric effector molecule. Proteins. 2012;80:1490. doi: 10.1002/prot.24049. [DOI] [PubMed] [Google Scholar]
- 6.Schultz S, Shields G, Steitz T. Crystal structure of a CAP-DNA complex: the DNA is bent by 90°. Science. 1991;253:1001. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho M, Arnold E, et al. Structural basis of transcription initiation. Science. 2012;338:1076. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuo Y, Steitz T. Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol Cell. 2015;58:534. doi: 10.1016/j.molcel.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bae B, Feklistov A, Lass-Napiorkowska A, Landick R, Darst S. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife. 2015;4:4:e08504. doi: 10.7554/eLife.08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bae B, Chen J, Davis E, Leon K, Darst S, Campbell E. CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex. eLife. 2015;4:e08505. doi: 10.7554/eLife.08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhodius V, West D, Webster C, Busby S, Savery N. Transcription activation at Class II CRP- dependent promoters: the role of different activating regions. Nucl Acids Res. 1997;25:326. doi: 10.1093/nar/25.2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan C, Lonetto M, Gross C. Sigma domain structure. Structure. 1996;4:1235. doi: 10.1016/s0969-2126(96)00131-1. [DOI] [PubMed] [Google Scholar]
- 13.Niu W, Kim Y, Tau G, Heyduk T, Ebright RH. Transcription activation at Class II CAP- dependent promoters: two interactions between CAP and RNA polymerase. Cell. 1996;87:1123. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lonetto M, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase σ70 subunit. J Mol Biol. 1998;284:1353. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- 15.Rhodius V, Busby J. Interactions between activating region 3 of the Escherichia coli cyclic AMP receptor protein and region 4 of the RNA polymerase σ70 subunit: application of suppression genetics. J Mol Biol. 2000;299:311. doi: 10.1006/jmbi.2000.3737. [DOI] [PubMed] [Google Scholar]
- 16.Zafar M, Shah I, Wolf R. Protein-protein interactions between σ70 region 4 of RNA polymerase and Escherichia coli SoxS, a transcription activator that functions by the prerecruitment mechanism. J Mol Biol. 2010;401:13. doi: 10.1016/j.jmb.2010.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patrick M, Dennis P, Ehrenberg M, Bremer H. Free RNA polymerase in Escherichia coli. Biochimie. 2015;119:80. doi: 10.1016/j.biochi.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 19.Roy S, Garges S, Adhya S. Activation and repression of transcription by differential contact: two sides of a coin. J Biol Chem. 1998;273:14059. doi: 10.1074/jbc.273.23.14059. [DOI] [PubMed] [Google Scholar]
- 20.Dove S, Huang F, Hochschild A. Mechanism for a transcriptional activator that works at the isomerization step. Proc Natl Acad Sci USA. 2000;97:13215. doi: 10.1073/pnas.97.24.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.