Abstract
Neuropeptide Y (NPY) is a physiological candidate gene for the regulation of body weight and has more recently been implicated in regulating bone mass. The current study sought to test if inherited variation in NPY might influence BMD in a population of African-ancestry men who have high bone mineral density (BMD). We genotyped 17 tagging single-nucleotide polymorphisms (SNPs) across the NPY gene region in 1,113 randomly selected men of African ancestry aged ≥40 years and tested for association with anthropometric characteristics and proximal femur BMD. The homozygous rare genotype of four SNPs was associated with a 0.92–1.59% decrease in stature (corrected P < 0.05). No SNP was associated with body mass index or body weight. Two SNPs in a 5-kb linkage disequilibrium block encompassing exons 3 and 4 were associated with proximal femur BMD, adjusted for age, body weight, and height (corrected P < 0.05). These results suggest that genetic variation at the NPY locus may contribute to bone density, independently of body weight.
Keywords: Neuropeptide Y, Bone mineral density, African, Polymorphism, Men
As with most common diseases, the pathogenesis of osteoporosis is due to a complex interplay between genetic and environmental factors. As much as 50–85% of the variation in adult bone mass is thought to be explained by genetic variation [1]. Identifying the genetic variation associated with osteoporosis-related phenotypes such as bone mineral density (BMD) may provide insight into the mechanisms underlying individual susceptibility to this common skeletal disorder.
The ventromedial hypothalamus, long known to regulate body composition through control of food intake and metabolism, has more recently been implicated as a neuroregulatory center for bone remodeling [2]. Neuropeptide Y (NPY), a peptide transmitter expressed in the arcuate nucleus of the hypothalamus, is a potent orexogen [3]. While NPY may act indirectly on bone through changes in energy balance and body composition, germline deletion of the Y2 receptor results in elevated bone mass and osteoblast activity in mice [4]. This phenotype is also observed with focal deletion of Y2 receptors in the arcuate, which implicates NPY as a central neuroregulator of bone homeostasis [4]. Nerve terminals containing NPY have also been identified in bone matrix, suggesting that NPY may also have a direct role in regulating bone remodeling in the periphery [5]. In the current study, we genotyped 17 tagging single-nucleotide polymorphisms (SNPs) across the NPY gene region in 1,113 men of African ancestry and tested for association with anthropometric characteristics and BMD.
Materials and Methods
Population Sample
Men were participants in the Tobago Bone Health Study, a study of 2,652 community-dwelling men aged 40 years and older who reside on the Caribbean island of Tobago. The majority of the population (94%) is of West African ancestry, with very little admixture compared to African Americans, as determined by ancestry-informative genetic markers [6, 7]. Men who were ambulatory and not terminally ill and who had not undergone a bilateral hip replacement were recruited between 1998 and 2004 by word-of-mouth and advertisements. Written informed consent was obtained from each participant using forms and procedures approved by the University of Pittsburgh Institutional Review Board, the U.S. Surgeon General’s Human Use Review Board, and the Tobago Division of Health and Social Services’ Institutional Review Board. We selected a random subset of 1,113 men for the current analysis who had four Afro-Caribbean grandparents, completed BMD measurements, and provided samples for genomic DNA extraction.
Anthropometry and Bone Density
Body weight was measured in kilograms using a calibrated balance beam scale, and participants wore indoor clothing with shoes removed. Standing height was measured in centimeters with a wall-mounted stadiometer. The average of two measurements was used in analysis. Body mass index (BMI) was calculated by dividing body weight (in kilograms) by height (in meters squared).
Participants completed a dual-energy X-ray absorptiometry (DXA) scan at the proximal femur on a QDR 4500W densitometer (Hologic, Bedford, MA). The same scanner was used for all participants. DXA scans were completed using the array beam mode. Standardized positioning and utilization of QDR software was based on the manufacturer’s recommended protocol. Scans were analyzed with QDR software version 8.26a. Coefficients of variation (CVs) were determined by repeating DXA measures on 12 participants; all CVs were ≤1.16%.
SNP Selection
In order to maximize our coverage of common polymorphisms in the NPY gene region, we used the International Haplotype Map Project Database (HapMap, www.hapmap.org, release 20) to identify validated SNPs from 5 kb upstream to 1 kb downstream of the NPY transcript [8]. A reference SNP set was generated using genotypes from the Yoruban population from Ibadan, Nigeria. Tagging SNPs were selected on the basis of linkage disequilibrium by a pairwise correlation method [9]. We required that the subset of tagging SNPs have a minor allele frequency ≥0.05 and predict the remaining SNPs with a minimum r2 of 0.80.
Genotyping
Genomic DNA samples were genotyped using TaqMan SNP Genotyping Assays on the 7900 PCR system (Applied Biosystems, Foster City, CA). Two SNPs, rs16142 and rs3025120, were unable to be genotyped by TaqMan. A surrogate tag SNP (rs3025118) was in high linkage disequilibrium with rs3025120 (r2 = 1) and was genotyped instead. No surrogate existed for rs16142. As a quality-assurance measurement, approximately 5% of the DNA samples were included as blind duplicates. Genotypes were assigned by two independent callers, and only those samples assigned concordant calls were included in statistical analyses. All SNPs in the analysis had call rates >95% and replication rates >98% and conformed to the expectations of Hardy–Weinberg equilibrium (P > 0.05). One SNP (rs16122) failed to conform to any of these quality-control standards and was replaced with rs16123, a surrogate tag SNP (r2 = 1) that conformed to quality-control standards. The 17 tagging SNPs included in the analysis captured 33 of the 34 common variants in the NPY gene region (97%).
Statistical Analysis
Allele frequencies and Hardy–Weinberg equilibrium were assessed by gene counting and chi-squared analysis, respectively. Anthropometric characteristics (height, body weight, and BMI) and proximal femur BMD (total hip, femoral neck, trochanteric, and intertrochanteric) were analyzed for association with the 17 NPY variants. We tested for an additive association, using linear regression to test for association between the number of copies of the less common allele and the phenotype. All analyses were initially adjusted for age. Body weight was adjusted for age and height. BMD measurements were adjusted for age, body weight, and height. Version 9.1 of SAS statistics software (SAS Institute, Inc., Cary, NC) was used for all analyses.
Permutation testing was performed to account for testing multiple SNPs by regressing each of the 17 SNPs in NPY against the phenotype and randomly permuting the phenotype 1,000 times. The minimum P value across all NPY SNPs for each permutation was used to create the new empirical distribution, and the value observed in the single SNP analysis was corrected using this distribution (referred to as “corrected P” throughout). Permutation testing was conducted using the statistical package R (version 2.5.1, www.r-project.org).
Results
The 1,113 men analyzed in this study were aged 55 ± 11 years old, were 175 ± 7 cm tall, and had a mean BMI of 27 ± 4 kg/m2. BMD of the total hip and femoral neck was 1.153 ± 0.140 and 0.994 ± 0.140 g/cm2, respectively. The minor allele frequencies of the 17 tagging SNPs were comparable between our Afro-Caribbean cohort and the HapMap Yoruban sample (Table 1).
Table 1.
Characteristics of the 17 tagging SNPs in the NPY gene
| SNP | Base Change | Position in NPY | Minor allele frequency
|
|
|---|---|---|---|---|
| Afro-Caribbeans | HapMap YRIa | |||
| rs10951003 | G→A | Upstream | 0.087 | 0.083 |
| rs12700524 | T→C | Upstream | 0.100 | 0.117 |
| rs3905497 | C→G | Upstream | 0.129 | 0.142 |
| rs16479 | A→G | Upstream | 0.063 | 0.067 |
| rs16149 | G→A | Upstream | 0.340 | 0.322 |
| rs16148 | C→T | Upstream | 0.453 | 0.474 |
| rs3025123 | C→G | Intron 1 | 0.060 | 0.086 |
| rs16143 | C→T | Intron 1 | 0.399 | 0.333 |
| rs3025118 | G→T | Intron 2 | 0.063 | 0.068 |
| rs16477 | G→A | Intron 2 | 0.164 | 0.167 |
| rs16135 | A→G | Intron 2 | 0.055 | 0.075 |
| rs16131 | T→C | Intron 3 | 0.073 | 0.042 |
| rs16130 | C→T | Intron 3 | 0.164 | 0.150 |
| rs16476 | T→G | Intron 3 | 0.370 | 0.397 |
| rs5576 | A→G | 3′ UTR | 0.051 | 0.050 |
| rs2189495 | A→G | Downstream | 0.427 | 0.492 |
| rs16123 | A→G | Downstream | 0.056 | 0.060 |
YRI indicates HapMap Yoruba sample from Ibadan, Nigeria
Results from association tests for anthropometric phenotypes are summarized in Table 2. Nine of the 17 SNPs showed association with height; four of these SNPs remained significantly associated with height after correcting for multiple comparisons (Fig. 1). The homozygous rare genotype in these four SNPs was associated with a 0.92–1.59% decrease in height. The strongest SNP association was observed with rs16149, where men homozygous for the minor allele had 1.6 cm lower height compared to those homozygous for the major allele (corrected P = 0.018).
Table 2.
NPY tagging SNPs significantly associated with height (cm)
| SNP | Adjusteda mean ± SE
|
P (corrected Pb) | ||
|---|---|---|---|---|
| 1/1 | 1/2 | 2/2 | ||
| rs16149 | 176.02 ± 0.29 | 174.79 ± 0.30 | 174.39 ± 0.56 | 0.001 (0.018) |
| rs16143 | 176.03 ± 0.31 | 175.19 ± 0.28 | 174.02 ± 0.46 | 0.0003 (0.038) |
| rs16477 | 175.65 ± 0.23 | 174.73 ± 0.37 | 172.96 ± 1.12 | 0.003 (0.038) |
| rs16130 | 175.61 ± 0.23 | 174.72 ± 0.36 | 172.83 ± 1.14 | 0.003 (0.038) |
Adjusted for age. 1 major allele, 2 minor allele
Corrected for multiple comparisons
Fig. 1.
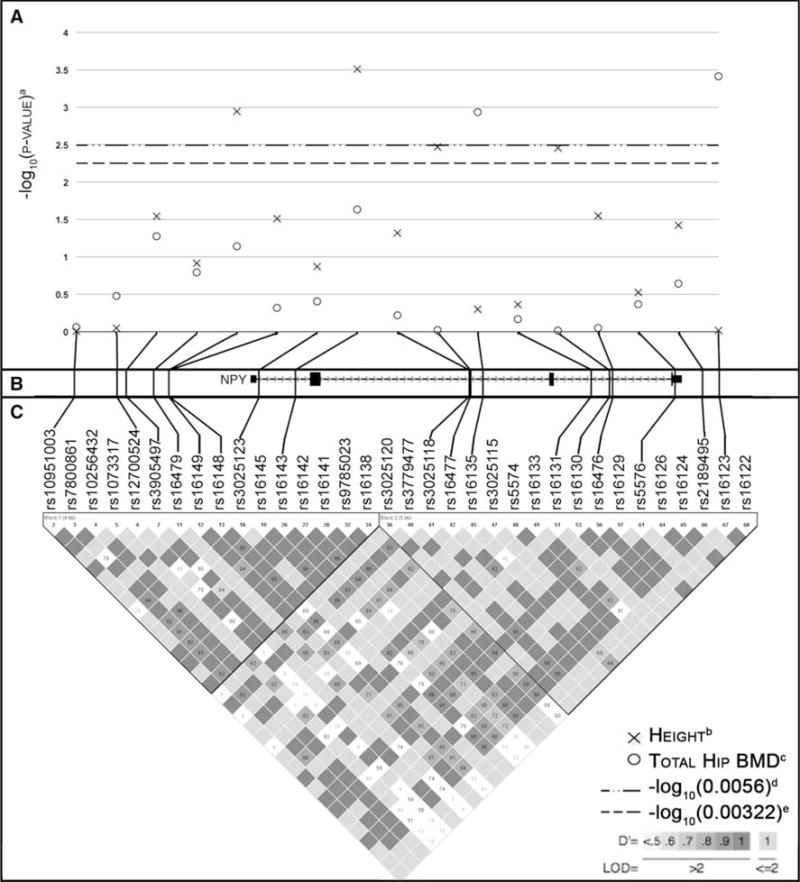
Genotyped SNPs, pairwise linkage disequilibrium, and genotype–phenotype association test results for NPY polymorphisms. a The −log10 of the uncorrected P value for the SNP associations with height and total-hip BMD is plotted above the gene region (x height, o total-hip BMD). Threshold lines corresponding to the corrected −log10(0.05) for height and total-hip BMD are included. Association tests for height were adjusted for age. Association tests for BMD were adjusted for age, weight, and height. b The 17 tagging SNPs that were genotyped are represented by vertical lines and are plotted relative to their position on chromosome 7 in the NPY gene region. c Linkage disequilibrium structure was determined using Haploview [28] and is presented below the gene region for all HapMap Yoruban SNPs with MAF > 0.05. Linkage disequilibrium values are shown as D′. a Uncorrected P value. b Adjusted for age. c Adjusted for age, weight, and height. d −log10(P = 0.0056); corresponds to corrected log10(0.05) for height. e −log10(P = 0.00322); corresponds to corrected log10(0.05) for total-hip BMD
Since several SNPs were associated with height, body weight measurements were adjusted for both age and height. None of the 17 NPY tagging SNPs was associated with BMI or body weight in these models (Table 2).
Although neither body weight nor BMI was associated with the NPY tagging SNPs, we adjusted BMD measurements for body weight, in addition to age and height, to minimize the potential effects of even small genotype-related differences on BMD. Two SNPs were associated with total-hip BMD (rs16135 and rs16123) after correction for multiple comparison testing (corrected P = 0.017 and P = 0.0007) (Table 3). These two SNPs were also associated with intertrochanteric and trochanteric BMD after correcting for multiple comparisons. Presence of one copy of the rare allele in rs16135 or rs16123 was associated with a 3.5% higher total-hip BMD compared with the homozygous major genotype. None of the SNPs in this study was associated with femoral neck cross-sectional area (data not shown).
Table 3.
NPY tagging SNPs significantly associated with proximal femur BMD (g/cm2)
| SNP | Adjusteda mean ± SE (unadjusted mean)
|
P (corrected Pb) | ||
|---|---|---|---|---|
| 1/1 | 1/2 | 2/2 | ||
| rs16135 | ||||
| Total hip | 1.15 ± 0.004 (1.15) |
1.19 ± 0.01 (1.19) |
1.21 ± 0.09 (1.21) |
0.001 (0.017) .005 |
| Trochanteric | 0.89 ± 0.004 (0.89) |
0.92 ± 0.01 (0.92) |
0.90 ± 0.08 (0.91) |
0.003 (0.042) 0.011 |
| Intertrochanteric | 1.33 ± 0.005 (1.33) |
1.38 ± 0.01 (1.37) |
1.43 ± 0.11 (1.43) |
0.001 (0.010) 0.005 |
| rs16123 | ||||
| Total hip | 1.15 ± 0.004 (1.15) |
1.19 ± 0.01 (1.19) |
1.31 ± 0.13 (1.18) |
0.0004 (0.0007) 0.003 |
| Trochanteric | 0.89 ± 0.004 (0.89) |
0.93 ± 0.01 (0.93) |
1.08 ± 0.12 (0.94) |
0.0003 (0.042) 0.005 |
| Intertrochanteric | 1.33 ± 0.005 (1.33) |
1.38 ± 0.01 (1.38) |
1.49 ± 0.15 (1.34) |
0.001 (0.010) 0.003 |
Adjusted for age, weight, and height. 1 major allele, 2 minor allele
Corrected for multiple comparisons
Discussion
An important and novel role for NPY signaling in the control of bone homeostasis has recently emerged. Thus, a primary aim of our analysis was to test if inherited variation in NPY might influence BMD in a population of African-ancestry men who have high BMD [10]. To address this aim, we examined the association of 17 tagging SNPs across the NPY gene region and proximal femur BMD but not bone area measurements. Our results suggest that common polymorphisms in the NPY gene region may be associated with proximal femur BMD in this population independently of anthropometric characteristics. Two polymorphisms in a single linkage disequilibrium block spanning exons 3 and 4 of NPY were associated with BMD, even after correcting for multiple comparisons. Although these specific polymorphisms have not been previously associated with BMD, a study of 316 postmenopausal Finnish women identified an association of a Leu7Pro polymorphism in NPY with femoral neck BMD [11]. This Leu7Pro polymorphism is not present in African-ancestry populations (www.ncbi.nlm.nih.gov/snp) and, thus, was not genotyped in the current study.
NPY may regulate bone remodeling through a central mechanism [12]. Both germline and focal hypothalamic deletions of Y2 receptors yield increased bone mass in mice [4]. There is also evidence that NPY may regulate bone remodeling through direct, peripheral mechanisms [13]. Further, in vitro treatment of marrow stromal cells with NPY reduces their proliferation, and NPY-mediated signaling may render stromal cells less pluripotent [14]. Additionally, Y1 receptors are expressed in stromal cells and osteoblasts, and deletion of these receptors yields an anabolic bone phenotype [15]. The Y1 receptor has also emerged as a potential drug target for prevention and treatment of bone loss [16].
We also observed significant associations between NPY polymorphisms and height after controlling for multiple comparisons. The associated SNPs were located across the NPY gene region in two distinct linkage disequilibrium blocks. We are unaware of other studies demonstrating an association between NPY polymorphisms and height, including several recent genomewide association studies conducted largely in Caucasians [17–19]. Nonetheless, this association is biologically plausible as NPY may participate in the autofeedback regulation of growth hormone secretion [20]. None of the SNPs associated with height was associated with BMD, raising the possibility that different mechanisms may underlie these genetic associations.
We were unable to document an association between NPY SNPs and body weight or BMI. Our results are consistent with a study of 26 tagging SNPs in the NPY gene region and BMI among 2,800 Caucasians [21]. Our data, along with the data from the aforementioned study, suggest that common SNPs in NPY are not likely to be associated with susceptibility to obesity in the general population. However, we cannot exclude the possibility that rare variation in NPY is associated with obesity phenotypes in our cohort.
Our study examined the association of NPY variants and BMD in men of African ancestry. Studies of African-ancestry populations are important for understanding the genetic basis for complex disease. Although African-derived populations have higher levels of genetic diversity and lower levels of linkage disequilibrium than non-African populations, they are often underrepresented in population genetic studies [22]. Nonetheless, our results may not be generalizable to populations of other ethnic backgrounds or to women. For example, recent genomewide association studies have not documented an association between NPY variants and BMD in Caucasians or Asians [23–27]. Replication of our results in other African-ancestry populations will be needed to support the possible role of NPY variants in BMD regulation in this population group. In addition, the physiological mechanisms by which these SNPs are associated with height and BMD will need to be examined as the SNPs identified in this study may be in linkage disequilibrium with the causative variants. In particular, we only studied SNPs located 5 kb upstream of NPY and, thus, may have missed more distant regulatory SNPs in our analysis.
In conclusion, NPY signaling has emerged as having a potentially important regulatory role in skeletal homeostasis. Our results suggest that common genetic variation at the NPY locus may contribute in part to height and proximal femur bone density independently of anthropometric characteristics among men of African ancestry.
Acknowledgments
This study was supported by grant R01-AR049747 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases.
Contributor Information
Louis J. Goodrich, Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, 130 DeSoto St, Pittsburgh, PA 15261, USA
Laura M. Yerges-Armstrong, Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, 130 DeSoto St, Pittsburgh, PA 15261, USA
Iva Miljkovic, Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, 130 DeSoto St, Pittsburgh, PA 15261, USA.
Cara S. Nestlerode, Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, 130 DeSoto St, Pittsburgh, PA 15261, USA
Allison L. Kuipers, Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, 130 DeSoto St, Pittsburgh, PA 15261, USA
Clareann H. Bunker, Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, 130 DeSoto St, Pittsburgh, PA 15261, USA
Alan L. Patrick, The Tobago Health Studies Office, Scarborough, Tobago, Trinidad and Tobago
Victor W. Wheeler, The Tobago Health Studies Office, Scarborough, Tobago, Trinidad and Tobago
Joseph M. Zmuda, Email: zmudaj@edc.pitt.edu, Department of Human Genetics, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, USA; Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, 130 DeSoto St, Pittsburgh, PA 15261, USA.
References
- 1.Ralston SH, de Crombrugghe B. Genetic regulation of bone mass and susceptibility to osteoporosis. Genes Dev. 2006;20:2492–2506. doi: 10.1101/gad.1449506. [DOI] [PubMed] [Google Scholar]
- 2.Baldock PA, Sainsbury A, Allison S, Lin EJ, Couzens M, Boey D, Enriquez R, During M, Herzog H, Gardiner EM. Hypothalamic control of bone formation: distinct actions of leptin and Y2 receptor pathways. J Bone Miner Res. 2005;20:1851–1857. doi: 10.1359/JBMR.050523. [DOI] [PubMed] [Google Scholar]
- 3.Hillebrand JJ, de Wied D, Adan RA. Neuropeptides, food intake and body weight regulation: a hypothalamic focus. Peptides. 2002;23:2283–2306. doi: 10.1016/s0196-9781(02)00269-3. [DOI] [PubMed] [Google Scholar]
- 4.Baldock PA, Sainsbury A, Couzens M, Enriquez RF, Thomas GP, Gardiner EM, Herzog H. Hypothalamic Y2 receptors regulate bone formation. J Clin Invest. 2002;109:915–921. doi: 10.1172/JCI14588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjurholm A, Kreicbergs A, Terenius L, Goldstein M, Schultzberg M. Neuropeptide Y-, tyrosine hydroxylase- and vasoactive intestinal polypeptide-immunoreactive nerves in bone and surrounding tissues. J Auton Nerv Syst. 1988;25:119–125. doi: 10.1016/0165-1838(88)90016-1. [DOI] [PubMed] [Google Scholar]
- 6.Miljkovic-Gacic I, Ferrell RE, Patrick AL, Kammerer CM, Bunker CH. Estimates of African, European and Native American ancestry in Afro-Caribbean men on the island of Tobago. Hum Hered. 2005;60:129–133. doi: 10.1159/000089553. [DOI] [PubMed] [Google Scholar]
- 7.Chakraborty R, Kamboh MI, Nwankwo M, Ferrell RE. Caucasian genes in American blacks: new data. Am J Hum Genet. 1992;50:145–155. [PMC free article] [PubMed] [Google Scholar]
- 8.HapMap Project. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 9.Roeder K, Bacanu SA, Sonpar V, Zhang X, Devlin B. Analysis of single-locus tests to detect gene/disease associations. Genet Epidemiol. 2005;28:207–219. doi: 10.1002/gepi.20050. [DOI] [PubMed] [Google Scholar]
- 10.Hill DD, Cauley JA, Sheu Y, Bunker CH, Patrick AL, Baker CE, Beckles GL, Wheeler VW, Zmuda JM. Correlates of bone mineral density in men of African ancestry: the Tobago bone health study. Osteoporos Int. 2008;19:227–234. doi: 10.1007/s00198-007-0450-9. [DOI] [PubMed] [Google Scholar]
- 11.Heikkinen AM, Niskanen LK, Salmi JA, Koulu M, Pesonen U, Uusitupa MI, Komulainen MH, Tuppurainen MT, Kroger H, Jurvelin J, Saarikoski S. Leucine7 to proline7 polymorphism in prepro-NPY gene and femoral neck bone mineral density in postmenopausal women. Bone. 2004;35:589–594. doi: 10.1016/j.bone.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Patel MS, Elefteriou F. The new field of neuroskeletal biology. Calcif Tissue Int. 2007;80:337–347. doi: 10.1007/s00223-007-9015-3. [DOI] [PubMed] [Google Scholar]
- 13.Denes A, Boldogkoi Z, Uhereczky G, Hornyak A, Rusvai M, Palkovits M, Kovacs KJ. Central autonomic control of the bone marrow: multisynaptic tract tracing by recombinant pseudorabies virus. Neuroscience. 2005;134:947–963. doi: 10.1016/j.neuroscience.2005.03.060. [DOI] [PubMed] [Google Scholar]
- 14.Lundberg P, Allison SJ, Lee NJ, Baldock PA, Brouard N, Rost S, Enriquez RF, Sainsbury A, Lamghari M, Simmons P, Eisman JA, Gardiner EM, Herzog H. Greater bone formation of Y2 knockout mice is associated with increased osteoprogenitor numbers and altered Y1 receptor expression. J Biol Chem. 2007;282:19082–19091. doi: 10.1074/jbc.M609629200. [DOI] [PubMed] [Google Scholar]
- 15.Baldock PA, Allison SJ, Lundberg P, Lee NJ, Slack K, Lin EJ, Enriquez RF, McDonald MM, Zhang L, During MJ, Little DG, Eisman JA, Gardiner EM, Yulyaningsih E, Lin S, Sainsbury A, Herzog H. Novel role of Y1 receptors in the coordinated regulation of bone and energy homeostasis. J Biol Chem. 2007;282:19092–19102. doi: 10.1074/jbc.M700644200. [DOI] [PubMed] [Google Scholar]
- 16.Sousa DM, Herzog H, Lamghari M. NPY signalling pathway in bone homeostasis: Y1 receptor as a potential drug target. Curr Drug Targets. 2009;10:9–19. doi: 10.2174/138945009787122888. [DOI] [PubMed] [Google Scholar]
- 17.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, Sulem P, Thorlacius S, Gylfason A, Steinberg S, Helgadottir A, Ingason A, Steinthorsdottir V, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Pedersen O, Aben KK, Witjes JA, Swinkels DW, den Heijer M, Franke B, Verbeek AL, Becker DM, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Gulcher J, Kiemeney LA, Kong A, Thorsteinsdottir U, Stefansson K. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 18.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, Illig T, Hackett R, Heid IM, Jacobs KB, Lyssenko V, Uda M, Boehnke M, Chanock SJ, Groop LC, Hu FB, Isomaa B, Kraft P, Peltonen L, Salomaa V, Schlessinger D, Hunter DJ, Hayes RB, Abecasis GR, Wichmann HE, Mohlke KL, Hirschhorn JN. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, Mangino M, Freathy RM, Perry JR, Stevens S, Hall AS, Samani NJ, Shields B, Prokopenko I, Farrall M, Dominiczak A, Johnson T, Bergmann S, Beckmann JS, Vollenweider P, Waterworth DM, Mooser V, Palmer CN, Morris AD, Ouwehand WH, Zhao JH, Li S, Loos RJ, Barroso I, Deloukas P, Sandhu MS, Wheeler E, Soranzo N, Inouye M, Wareham NJ, Caulfield M, Munroe PB, Hattersley AT, McCarthy MI, Frayling TM. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minami S, Kamegai J, Sugihara H, Suzuki N, Wakabayashi I. Growth hormone inhibits its own secretion by acting on the hypothalamus through its receptors on neuropeptide Y neurons in the arcuate nucleus and somatostatin neurons in the periventricular nucleus. Endocr J. 1998;45(Suppl):S19–S26. doi: 10.1507/endocrj.45.suppl_s19. [DOI] [PubMed] [Google Scholar]
- 21.Campbell CD, Lyon HN, Nemesh J, Drake JA, Tuomi T, Gaudet D, Zhu X, Cooper RS, Ardlie KG, Groop LC, Hirschhorn JN. Association studies of BMI and type 2 diabetes in the neuropeptide Y pathway: a possible role for NPY2R as a candidate gene for type 2 diabetes in men. Diabetes. 2007;56:1460–1467. doi: 10.2337/db06-1051. [DOI] [PubMed] [Google Scholar]
- 22.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genom Hum Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Yoon D, Lee MH, Kim DJ, Park M, Cha SH, Kim JW, Han BG, Min H, Ahn Y, Park MS, Han HR, Jang HY, Cho EY, Lee JE, Cho NH, Shin C, Park T, Park JW, Lee JK, Cardon L, Clarke G, McCarthy MI, Lee JY, Oh B, Kim HL. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 24.Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N, Wilson SG, Andrew T, Falchi M, Gwilliam R, Ahmadi KR, Valdes AM, Arp P, Whittaker P, Verlaan DJ, Jhamai M, Kumanduri V, Moorhouse M, van Meurs JB, Hofman A, Pols HA, Hart D, Zhai G, Kato BS, Mullin BH, Zhang F, Deloukas P, Uitterlinden AG, Spector TD. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Center JR, Nguyen TV, Bagger Y, Gulcher JR, Eisman JA, Christiansen C, Sigurdsson G, Kong A, Thorsteinsdottir U, Stefansson K. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 26.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Snorradottir S, Center JR, Nguyen TV, Alexandersen P, Gulcher JR, Eisman JA, Christiansen C, Sigurdsson G, Kong A, Thorsteinsdottir U, Stefansson K. New sequence variants associated with bone mineral density. Nat Genet. 2009;41:15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 27.Xiong DH, Liu XG, Guo YF, Tan LJ, Wang L, Sha BY, Tang ZH, Pan F, Yang TL, Chen XD, Lei SF, Yerges LM, Zhu XZ, Wheeler VW, Patrick AL, Bunker CH, Guo Y, Yan H, Pei YF, Zhang YP, Levy S, Papasian CJ, Xiao P, Lundberg YW, Recker RR, Liu YZ, Liu YJ, Zmuda JM, Deng HW. Genome-wide association and follow-up replication studies identified ADAMTS18 and TGFBR3 as bone mass candidate genes in different ethnic groups. Am J Hum Genet. 2009;84:388–398. doi: 10.1016/j.ajhg.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]


