Abstract
Malignant ectomesenchymoma (MEM) is an exceedingly rare pediatric sarcoma, with predilection for infants and young children, composed of dual malignant mesenchymal and neuroectodermal components. Microscopically, MEM displays areas of rhabdomyosarcoma (RMS) with intermixed neuronal/neuroblastic foci. The molecular alterations MEM and its relationship with embryonal RMS (ERMS) and malignant peripheral nerve sheath tumor (MPNST) have not yet been elucidated. In this study we use whole transcriptome sequencing in 2 MEM index cases with available frozen tissue, followed by screening the identified genetic abnormalities in 5 additional cases. No candidate fusion genes were detected by FusionSeq analysis, however, the mutation detection algorithms revealed HRAS and PTPRD hot-spot mutations in both index cases, with one case harbouring an additional FBXW7 mutation. As these mutation profiles have been previously described in ERMS we have tested their incidence in a control group of 7 age-matched ERMS. Additionally, the gene signature of MEM was compared to that of RMS, MPNST and neuronal lineage. All 7 MEM patients were male, with a mean age of 7.5 months (range 0.6–17 mo). All except one occurred in the pelvis/urogenital region. Most cases showed ERMS elements, with occasional spindle or undifferentiated/round cell areas. The intermixed neuroectodermal components were mostly scattered ganglion cells, ganglioneuroma, or ganglioneuroblastoma. By Sanger sequencing, 6 of 7 (86%) MEMs had HRAS mutations, with no additional case harboring PTPRD or FBXW7 mutations. The only case lacking HRAS mutation showed neuroblastic micronodules without ganglion cells. The trimethylation at lysine 27 of histone H3 (H3K27me3) expression, typically lost in MPNST, was retained in all cases. In the control ERMS group, 5 of 7 (71%) showed RAS mutations, equally distributed among NRAS, KRAS, and HRAS genes. The expression profiling of MEM showed upregulation of skeletal muscle and neuronal genes, with no significant overlap with MPNST. Our results of common HRAS mutations and composite gene signature with RMS and neuronal/ neuroblastic elements, suggest a closer genetic link of MEM to RMS rather than MPNST.
Keywords: ectomesenchymoma, rhabdomyosarcoma, HRAS, PTPRD, FBXW7
INTRODUCTION
Malignant ectomesenchymoma (MEM) is an exceedingly rare multiphenotypic sarcoma consisting of both mesenchymal and neuroectodermal lines of differentiation 1,2. The mesenchymal component is represented by rhabdomyosarcoma (RMS), frequently of embryonal phenotype and less commonly resembling other variants 3,4. The admixed neuroectodermal component displays elements of (ganglio)neuroblastoma spectrum, varying from primitive neuroblastic cells to mature ganglion cells, with rare cases being reported with malignant peripheral nerve sheath tumor (MPNST), peripheral primitive neuroectodermal tumor, or glioma morphology 5–8. The MEM nomenclature derives from its possible tumorigenesis from the pluripotential neural crest remnants, the so-called ectomesenchyme 1,2.
Although quite limited, most of the existing data on MEM patients, including cytogenetic abnormalities, clinical behavior, and therapeutic response, shows overlapping features with RMS, suggesting that MEM might represent a variant of ERMS 6,9,10. Contradictory to this evidence, a genomic study on the so-called ‘intra-cranial MEM’ revealed an expression profile closer to MPNST 11. Furthermore, the latest edition of World Health Organization classification of soft tissue and bone tumors categorized MEM under nerve sheath tumors 12. As there are no comprehensive studies examining the genetic abnormalities of MEM, our present investigation using whole transcriptome sequencing for novel oncogene discovery and expression signatures aims at addressing the pathogenetic relationship of MEM with ERMS and nerve sheath tumors (i.e. MPNST).
MATERIALS AND METHODS
Case Selection, Pathologic Criteria and Immunohistochemistry
We identified 7 MEM cases from the Surgical Pathology files of MSKCC and Padova University Hospital, Italy. One case had been previously reported 13. The diagnosis of MEM was confirmed by the presence of a biphasic tumor, with histologic evidence of RMS and neuroectodermal elements 6,9. The latter component included variable ganglion cells, ganglioneuroma, ganglioneuroblastoma, or neuroblastoma foci, following the criteria of the International Neuroblastoma Pathology Classification 14. The RMS component was mainly composed of an embryonal RMS (ERMS), although areas that resembled spindle cell RMS or alveolar RMS (ARMS) were also noted and recorded as such, according to the Soft Tissue Sarcoma Committee of the Children’s Oncology Group 15. All cases with round/undifferentiated areas resembling solid or classic ARMS were tested by fluorescence in situ hybridization for FOXO1 gene rearrangements, to exclude a fusion positive ARMS. These compact sheet-like areas of round to undifferentiated cells within a fusion-negative RMS have been recently recognized by Children’s Oncology Group as potential pitfall with ARMS and designated as ‘dense pattern’ of ERMS 15.
The individual components were confirmed by immunohistochemical stains, i.e. desmin and myogenin for RMS; S100 protein for schwannian and synaptophysin for neuronal differentiation. Immunohistochemical staining for neural markers alone without morphologic evidence of neural / neuronal differentiation was considered insufficient for diagnosis 9. Additionally, H3K27me3 (07-449, 1:250, Millipore, Billerica, MA) immunohistochemistry (IHC) was investigated in all MEM cases, as most MPNST show loss of H3K27me3 (trimethylation lysine 27 of histone H3) expression due to frequent loss of function PRC2 complex abnormalities 16. In the control group, 7 patients with ERMS, younger than 5 years of age and with classic morphology, were retrieved from our files. The relevant clinical information was collected from the medical records or communications with the referring pathologists. This study was approved by the individual IRB at each participating institution.
RNA Sequencing and Data Analysis using FusionSeq and Mutation Detection Algorithms
Two index cases (MEM1, MEM2) with frozen tissue were analyzed by whole transcriptome sequencing. Total RNA was processed for RNA sequencing in accordance with the standard Illumina mRNA sample preparation protocol. Briefly, mRNA was isolated with oligo(dT) magnetic beads from total RNA (2μg) and fragmented by incubation at 94°C for 2.5 min in fragmentation buffer. The adaptor-ligated library was then enriched by PCR for 15 cycles and purified. The library was sized and quantified using DNA1000 kit (Agilent) on an Agilent 2100 Bioanalyzer according to the manufacturer’s instructions. Paired-end RNA- sequencing at read lengths of 50 or 51 bp was performed on the HiSeq 2000 platform.
All reads were independently aligned with the STAR alignment software against the human genome sequence (hg19) and a splice junction library, simultaneously. The mapped reads were converted into Mapped Read Format and analyzed with FusionSeq to identify potential fusion transcripts. RNA sequencing data was also used to gene mutation calls. BAM files were generated by STAR alignment, followed by PicardTools (ver 1.130) standard preprocessing. MuTect (var 1.15) and VarScan (var 2.3.8) variant callers were both applied for mutation detection, followed by vcf2maf for converting VCF into MAF files, with the annotation added by Variant Effect Predictor tool provided by Ensembl. Sanger sequencing validation was performed subsequently.
Gene Expression Signatures using RNAseq data and Affymetrix U133A
We used different datasets to establish gene signatures of RMS and peripheral nerve sheath tumors. The available RNA sequencing data from >100 sarcomas (which includes 9 RMS cases) was analyzed to obtain an RMS gene signature, by using log2FC (fold change) > 1 and p < 0.01 for statistical analysis. We then used our Affymetrix U133A microarray expression data to establish a peripheral nerve sheath tumor enriched gene list, by comparing a group of 14 benign and malignant peripheral nerve sheath tumors (9 schwannomas, 3 neurofibromas, and 2 MPNSTs) with 27 other sarcoma samples 17,18, using a similar log2FC > 1 and p < 0.01 for statistical analysis. From Fredlund et al. study on neuroblastoma, we selected 16 genes implicated in neuronal differentiation, including: MEIS1 (Meis Homeobox 1), PHOX2B (Paired-Like Homeobox 2b), CHGA (Chromogranin A), NTRK1 (Neurotrophic Tyrosine Kinase, Receptor, Type 1), etc 19. Furthermore, the HG-U133A 2-plus microarray expression data of the reported intracranial MEM was compared to our 2 index MEM cases signature with RNAseq data 11.
DNA Extraction and Polymerase Chain Reaction (PCR)
Genomic DNA was extracted from either frozen or formalin-fixed paraffin-embedded tissue, using the phenol/chloroform method or the QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA), respectively. The targeted exon regions of candidate genes were amplified by the PCR using corresponding forward and reverse primers (Supplementary Table 1). PCR was conducted using the Clontech Advantage 2 PCR Enzyme System kit (Clontech, Mountain View, CA). The PCR products were purified using QIAquick PCR Purification Kit (Qiagen) and confirmed by Sanger sequencing. All mutations were verified bidirectionally.
RESULTS
Clinicopathologic Features and Immunohistochemical Findings
The clinicopathologic features of MEM were summarized in Table 1. All seven patients were males, with a median age of 7.5 months (range, 0.6–17); 5 of them being infants. All except one were located in the pelvic/genitourinary area: 2 paratesticular, 1 urinary bladder, 1 prostate, and 2 in the pelvic soft tissue. One case arose in the soft tissues of the hand 13. Two cases were diagnosed as RMS on the biopsy, and subsequently recognized as MEM on the resection specimen. All patients presented with localized disease at diagnosis, except for one who, in addition showed locoregional lymph node metastasis. Microscopically, the lymph node mets revealed both RMS and neuroblastoma areas (MEM6). Six patients received chemotherapy, following the RMS regimen protocols in 5, including COG ARST0531 (3), EpSSG2005 (1), RMS96 (1), and MSKCC P6 protocol, developed for high-risk Ewing sarcoma in the remaining patient. No recurrence or metastasis occurred in 6 cases with available follow-up data during a median period of 5.4 years (range, 0.9–16.7).
Table 1.
Clinicopathological features of malignant ectomesenchymoma.
| Case | Age (month) | Sex | Location | Tumor Status | Size (cm) | Initial Stage | Treatment Protocol | Histological Components | Follow-up (year) | Final Status |
|---|---|---|---|---|---|---|---|---|---|---|
| 1* | 0.6 | M | Paratesticular | Primary | - | - | EpSSG2005 | ERMS-s + GC | 8.0 | NED |
| 2* | 12 | M | Paratesticular | Primary | - | - | None | RRMS-s + GN | 2.0 | NED |
| 3 | 8 | M | Urinary bladder | Post-treatment | - | - | RMS96 | ERMS-c + GN | - | - |
| 4 | 3 | M | Pelvis | Post-treatment | 7.2 | T2bN0M0 | COG ARST0531-A | ERMS-c + GN | 0.9 | DOO |
| 5γ | 17 | M | Hand | Primary | 2.7 | T1aN0M0 | MSKCC P6 | ERMS-d + GNB | 16.7 | NED |
| 6 | 11 | M | Pelvis | Post-treatment | 6.1 | T2bN1M0 | COG ARST 0531-B | ERMS-d + NB | 2.9 | NED |
| 7 | 1.2 | M | Prostate | Post-treatment | 8.0 | T2bN0M0 | COG ARST 0531-B | ERMS-c + GN | 8.1 | NED |
Tested by RNA sequencing; spRMS, spindle cell rhabdomyosarcoma; ERMS-c, embryonal rhabdomyosarcoma, classic pattern; ERMS-d, embryonal rhabdomyosarcoma, dense pattern; GC, ganglion cells; GN, ganglioneuroma; GNB, ganglioneuroblastoma; NB, neuroblastoma; DOO, dead of other causes; NED, no evidence of disease.
previously reported [13]
Microscopically, the RMS and neuroectodermal components were intimately intermixed in all cases. Even when sharp demarcation of the two elements was discerned at low power, the high magnification disclosed both scattered ganglion cells in RMS areas, as well as rhabdomyoblasts within the ganglioneuroma area (Fig. 1). In most cases, both elements were readily identified at low to mid magnification, except in one case (MEM1) where only rare ganglion cells were scattered in a predominant RMS component. The RMS component in all cases was of embryonal type, showing the typical alternating myxoid-cellular (3), spindle cell (2), and dense/round cell (2) pattern (Fig. 2A). The 2 MEMs with a dense/round cell component were negative for FOXO1 gene rearrangements by fluorescence in situ hybridization (data not shown). Indeed, the original classification of these 2 cases favored an ARMS (MEM5 and MEM6); however, upon re-review spindled or epithelioid rhabdomyoblasts with more abundant cytoplasm and irregular nuclear contours were also noted, distinct from the classic monomorphic round cell cytology of ARMS (Fig. 2B). Both cases demonstrated strong and diffuse desmin and myogenin reactivity in the cellular/round cell areas, also reminiscent of ARMS immunostaining pattern (Fig. 2E). The RMS component typically had a brisk mitotic activity (>10/10 HPF) and necrosis. In 4 cases, material from the post-chemotherapy resection was also available, showing therapy-related changes, such as decreased cellularity, more prominent rhabdomyoblastic maturation, and stromal hyalinization (Fig. 3A).
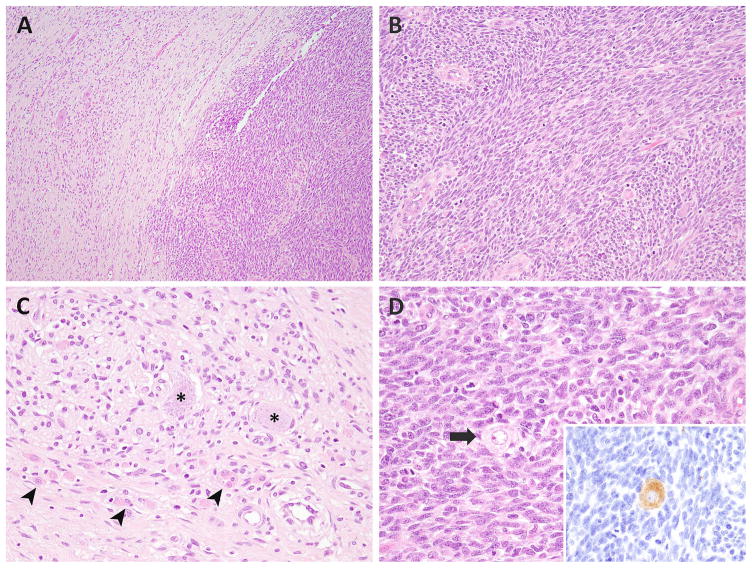
Figure 1. Microscopic features of MEM.
The most common appearance consisted of discrete sharply demarcated RMS (right) and ganglioneuroma (left) components (A, MEM2). The RMS component exhibited interlacing bundles of monomorphic spindle cells (B), while the ganglioneuroma component showed ganglion cells (asterix), nerve bundles, schwannian cells, and intermixed rhabdomyoblasts (arrowheads) (C, MEM2). Few ganglion cells (arrow) were scattered within the mitotically active RMS area (inset: synaptophysin stain) (D).
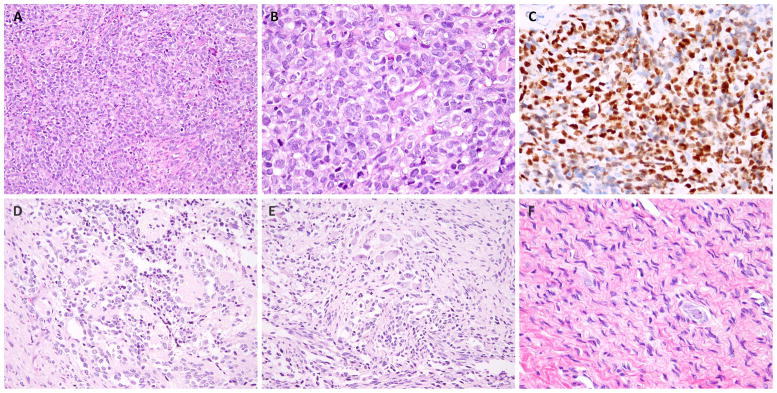
Figure 2. The morphologic spectrum of the RMS and neuroectodermal components in MEM.
One of the unusual patterns of RMS resembled solid variant ARMS and was composed of solid sheets of primitive round cells (so-called dense pattern of ERMS) (A, B; MEM5), which was diffusely positive for myogenin (C). The ganglioneuroblastoma areas in this case were composed of maturing neuroblastic cells with variable amount of cytoplasm, focal rosetting and neuropil stroma (D, MEM5). Some areas showed intermingled mature ganglion cells (upper) and spindled rhabdomyoblastic cells (lower) (E), while others showed pure ganglioneuroma areas (F).
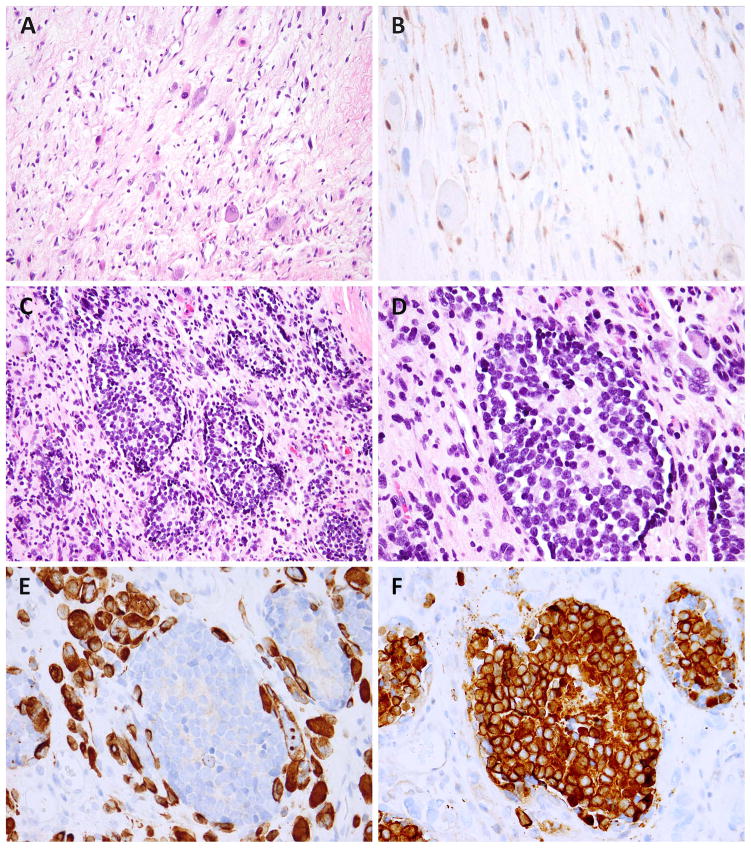
Figure 3.
Post-treatment MEM showed decreased cellularity and scattered rhabdomyoblasts and ganglion cells in a myxoid background (A, MEM4); an S100 stain highlights Schwannian and satellite cells (B). Another post-chemo MEM showed no treatment response, being composed of neuroblastoma islands with a micronodular pattern, within the RMS areas (C, MEM5). The neuroblastic cells have monomorphic round, hyperchromatic nuclei and form Homer-Wright rosettes (D). The desmin stain highlights the rhabdomyoblasts but not the neuroblastoma nodules (E), while synaptophysin stains the neuroblastoma nodules but not the surrounding rhabdomyoblasts (F).
Ganglioneuroma was the most prevalent neuroectodermal component, being present in 4 cases, while scattered ganglion cells, ganglioneuroblastoma, or neuroblastoma occurred in one case each. In one case the ganglioneuroma component was quite focal and located at the periphery of RMS areas (MEM2). In another case, the neuroblastic component was present as small islands embedded in a RMS background, imparting a micronodular pattern (Fig. 3C, MEM6). The most frequent morphologic appearance was that of typical ERMS admixed with ganglioneuroma or ganglion cells (Table 1). Interestingly, the dense pattern ERMS was accompanied by either ganglioneuroblastoma or neuroblastoma, suggesting a synchronous level of differentiation among the two components. One of these 2 cases, showed a heterogeneous appearance, including scattered ganglion cells in dense ERMS, ganglioneuroblastoma within differentiating ERMS, and mature ganglioneuroma (Fig. 2, MEM5). The neuroblastic elements showed corresponding immunoprofiles based on the different components, such as S100 protein positivity in schwannian cells, GFAP for neuropil, and synaptophysin for ganglion cells or neuroblastoma. No heterologous differentiation was found. All cases retained H3K27me3 expression in both RMS and neuroectodermal regions.
Novel HRAS, PTPRD, and FBWX7 Mutations Identified by RNA Sequencing
No fusion candidate was identified in the 2 index cases by the FusionSeq algorithm. Instead, the bioinformatic mutation detection tools (MuTect and VarScan) revealed recurrent HRAS (exon 2, p.G13R) and PTPRD (exon 20: p.V892A in MEM1; p.V847L in MEM2) mutations in both index cases and FBWX7 (exon 10, p.R505H) mutations in MEM2 (Fig. 4). These mutations were first validated by direct sequencing and then screened in the remaining 5 MEM cases. Based on these RNAseq findings and the previously reported hotspot mutations in RMS we have investigated all 7 MEM and the control RMS group for the following hot spots: HRAS exon 3, NRAS exons 2 and 3, KRAS exon 2, PIK3CA exons 10 and 21, PTPRD exon 25, FBWX7 exon 10 and CTNNB1 exon 3 20–22. In total, 6 of 7 (86%) MEMs harbored HRAS mutations, 4 with p.G13R mutation and 2 with p.Q61L. Three of the p.G13R mutations demonstrated nearly homozygous pattern; but no normal DNA was available to confirm their germline genotype. The cases containing p.G13R mutation (MEM1–3, 7) exhibited the more common combination of classic ERMS and ganglion cells or ganglioneuroma, while the cases with p.Q61L mutation (MEM4, 5) showed either classic or dense ERMS and ganglioneuroma or ganglioneuroblastoma. The only case lacking RAS mutations (MEM6) demonstrated dense ERMS and neuroblastoma without ganglion cells. In the control RMS group, we detected 5 RAS mutations in 7 cases (71%), including 1 HRAS (p.Q61K), 2 NRAS (p.Q61K; p.Q61H), and 2 KRAS (p.G12D) mutations (Fig. 4B).
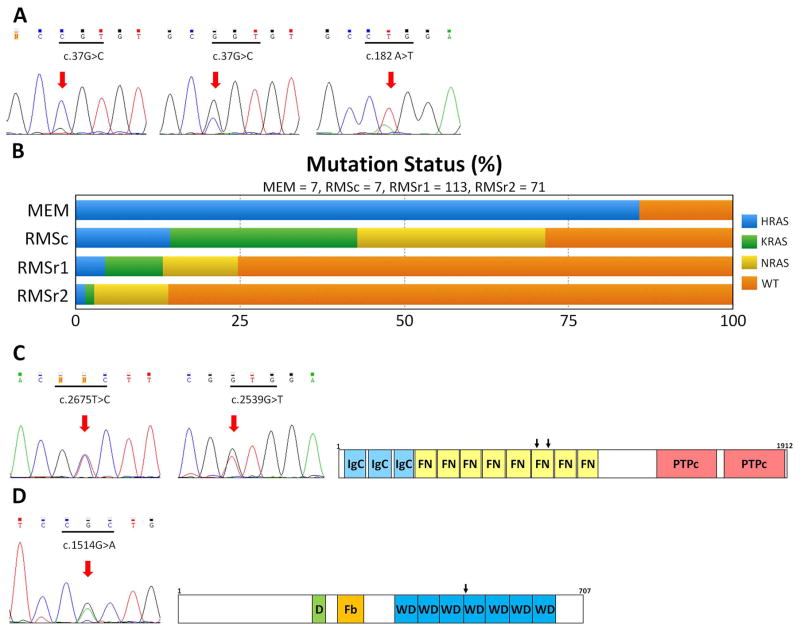
Figure 4. Novel oncogenic mutations identified in MEM.
(A) HRAS mutation: homozygous p.G13R mutation (left), heterozygous p.G13R mutation (middle), heterozygous p.Q61L mutation (right). (B) Distribution and frequency (%) of RAS family members mutations in MEM and RMS [RMSc, control RMS group; RMSr1, RMS meta-analysis 20,21,31 (limited to patients 5 years of age); RMSr2, RMS from same meta-analysis (> 5 years of age)]. (C) PTPRD mutations (p.V892A and p.V847L) and protein domains (IgC, immunoglobulin-like C2 type domains; FN, fibronectin type III domain; PTPc, phosphatase catalytic domain). (D) FBXW7 mutation (p.R505H) and protein domains (D, dimerization domain; Fb, F-box domain; WD, tryptophan–aspartic acid repeat domain).
No additional PTPRD or FBWX7 mutations were identified in the remaining MEM or the control RMSs. Both PTPRD exon 20 mutations were located in the 6th fibronectin type III domain. The FBXW7 missense mutation occurred at one of the critical 3 arginine residues in the WD (tryptophan-aspartic acid) repeat, which mediate substrates binding to the Skp1-Cullin-F-box (SCF) ubiquitin-ligase complex 23. No PIK3CA or CTNNB1 hot spot mutations were identified in the MEM cohort. No PRC2 component mutations (i.e. in EED and SUZ12 genes) nor decreased expression was identified in the 2 index MEM cases with available RNAseq data.
MEM Expression Profiling in Relationship to RMS and MPNST
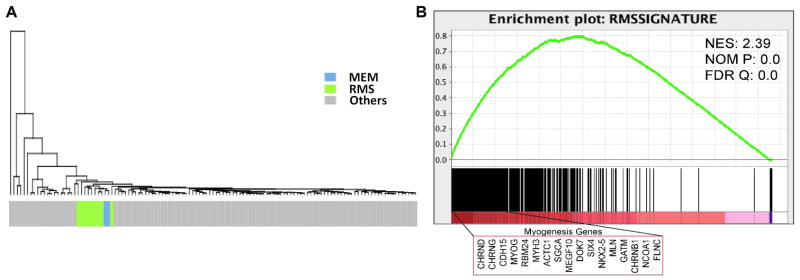
The unsupervised hierarchical clustering using the RNAseq dataset of a large panel of >100 different soft tissue tumors showed MEM and RMS tightly grouped together, separate from all other tumor types (Fig. 5A). Using the same dataset, we obtained a RMS signature of 279 genes (log2FC > 1 and p < 0.01) by comparing the 9 RMS with all other sarcoma types (excluding the 2 MEM cases). Gene set enrichment analysis confirmed that both RMS and MEM were enriched in the RMS signatures (Fig. 5B). We then established a peripheral nerve sheath tumor signature, comparing a group of 3 neurofibromas, 9 schwannomas, and 2 MPNSTs with a group of different sarcoma types available on the Affymetrix Human Genome U133A array (437 genes, log2FC > 1 and p < 0.01)17,18 and subsequently validated by gene set enrichment analysis (Supplemental Fig. 1A). This 473 peripheral nerve sheath tumor gene signature was then applied for supervised clustering of the RNAseq samples, which showed no specific cluster correlation, indicating a distinct expression profile from MEM (Supplemental Fig. 1B). We additionally investigated a 20 highly expressed gene signature reported in an intra-cranial MEM by HG-U133A 2-plus microarray analysis 11. However, only 2 genes: SCN7A (Sodium Channel, Voltage Gated, Type VII Alpha Subunit) and C7 (Complement Component 7) were differentially overexpressed in our 2 MEM cases compared to RMS, but statistically non-significant (log2FC = 2.08 and 3.52; p = 0.333 and 0.511, respectively). Of the 4 highly overexpressed genes in their intra-cranial MEM: MAGP2 (MFAP5, Microfibrillar Associated Protein 5), SULT1E1(Sulfotransferase Family 1E, Estrogen-Preferring, Member 1), PDGFRL (Platelet-Derived Growth Factor Receptor-Like), and CXCL13 (Chemokine, C-X-C Motif, Ligand 13), all had low level of expression in our 2 index MEM and 9 RMS cases with RNAseq data (Supplemental Fig. 1C).
Figure 5. Overlapping gene signature of MEM and RMS.
By hierarchical clustering of a large panel of sarcomas available on the RNAseq using the 279 RMS-enriched gene list, MEM grouped closely with RMS (A). Gene set enrichment analysis of combined RMS and MEM cases showed uniform enrichment of RMS signature genes, including many myogenesis genes (B).
As MEM and RMS cases grouped together upon unsupervised hierarchical clustering, we obtained a common MEM-RMS gene signature of 307 differentially expressed genes by comparing with all other sarcoma types on RNAseq data set (log2FC >3.5 and p < 0.001). Among the top upregulated genes in both MEM and RMS, there were many involved in skeletal muscle-function, such as MYOG (Myogenin), CHRND (Cholinergic Receptor, Nicotinic, Delta), MRLN (Myoregulin), and MEGF10 (Multiple EGF-Like-Domains 10). When comparing MEM to RMS, there were several neuronal developmental genes among the top-ranked gene list, such as EPHA3 (EPH Receptor A3), ADM (Adrenomedullin), NEFM (Neurofilament, Medium Polypeptide), SPOCK2 (Sparc/Osteonectin, Cwcv And Kazal-Like Domains Proteoglycan)(Supplemental Fig. 1D). However, the main neuronal developmental genes described as up-regulated in neuroblastomas, such as MEIS1, PHOX2B, CHGA, and NTRK1, were not overexpressed in MEM 19.
DISCUSSION
The first description of a tumor with dual RMS and ganglioneuroma components was in the cerebellopontine angle of a 15 month-old boy under the designation of ‘gangliorhabdomyosarcoma’24. Naka et al. subsequently proposed the term malignant ectomesenchymoma (MEM) for a retroperitoneal tumor arising in a 2 year-old girl showing ganglioneuroblastoma and a variety of mesenchymal components, including RMS, liposarcoma, and chondroid tissue 1. The MEM terminology was used thereafter to describe tumors with mixed malignant mesenchymal and neuroectodermal elements 2,4, including primary central nervous system tumors containing heterologous differentiation 25. Approximately 64 cases of MEM have been so far documented in the literature, mostly as single case reports 3,4. The majority (82%) of cases occurred in the first decade of life, with a slight male preponderance (M:F ratio of 1.38). The most common anatomic site is pelvic/perineal or intra-abdominal/retroperitoneal region (50%), followed by intra-cranial site (17%), head and neck (17%), extremities (14%), and mediastinum (2%). Since the broader definition (i.e. including heterologous elements) seemingly encompasses various entities, we restrict our study selection criteria for cases exhibiting RMS and neuroblastic differentiation according to the two larger published studies 6,9.
The present study investigating 7 MEM patients demonstrates a remarkable demographic overlap with ERMS, including male predominance, young age (<2 years), and pelvic/urogenital distribution. Five of them received RMS treatment regimens and all 3 cases with long-term follow-up pursued uneventful courses. Histologically, these cases have various ERMS patterns and neuroblastic elements and none contained additional heterologous differentiation. Most cases showed obvious rhabdomyoblastic differentiation reminiscent of ERMS, with a variety of growth patterns, including alternating myxoid-cellular areas, fascicular spindle cell growth, or compact round cell sheets. The latter growth pattern, negative for FOXO1 fusion, has been designated as ‘dense pattern’ of ERMS by the Children’s Oncology Group 15, to be distinguished from the solid variant of ARMS by their angulated nuclei and variably prominent nucleoli. In a large retrospective Children’s Oncology Group analysis, among the 225 ARMS reviewed, 84 (33%) cases were reclassified as ERMS; half of these cases displaying a uniformly dense cellularity, resembling solid ARMS-like pattern 15. Moreover, the dense ERMS areas often show strong myogenin expression, a further overlapping feature with ARMS. Of note, ARMS has been previously described in MEM by two cooperative studies, either alone or mixed with ERMS 6,9; however, no FOXO1 gene rearrangements has been identified to date 4. Our current results strongly suggest that the ARMS-like pattern described in MEM represents the dense ERMS. A botryoid ERMS subtype has also been described in a vaginal MEM 26.
The neuroectodermal elements represented in MEM cover the entire spectrum of neuroblastic phenotype, ranging from scattered ganglion cells, mature ganglioneuroma, intermediate ganglioneuroblastoma, to primitive neuroblastoma. The mature ganglioneuroma pattern is the predominant morphology. The neuroblastic components are typically intimately associated with RMS. One of our cases (MEM5) exhibited a unique micronodular neuroblastoma pattern within the RMS background, which was previously reported in the Intergroup Rhabdomyosarcoma Study Groups study 9. However, the genetic alterations often present in neuroblastoma, such as N-MYC amplification or ALK exon 23 or 25 mutation, were not identified in this MEM case (data not shown). Rare case reports describing peripheral primitive neuroectodermal tumor in MEM without molecular support raise skepticism regarding the diagnosis 6,27,28. In one case, the peripheral primitive neuroectodermal tumor component was positive for CD56 and S100, however, no CD99 status was investigated 27. These reports illustrate the inconsistent diagnostic criteria for MEM diagnosis 6,29. Indeed, the Intergroup Rhabdomyosarcoma Study Groups and Cooperative Weichteilsarkom Studiengruppe suggest that MEM could be either under or over-diagnosed, respectively 6,9.
Aside from the similar clinicopathologic features, our MEM cohort also shows a high frequency (86%) of HRAS mutations, in keeping with a homogeneous pathologic and molecular entity. An activated RAS signaling pathway through oncogenic mutations is also observed in the control pediatric ERMS cases (71%); however, the spectrum of mutations seen spans all NRAS, KRAS, and HRAS members. Notably, the HRAS mutation (p.Q61K) detected in one RMS results in a different amino acid substitution than the one seen in MEM (p.Q61L). The lysine (K) amino acid shows a positive-charged side chain rather than the hydrophobic side chains of leucine (L). The prevalence of RAS mutations in ERMS ranges from 11.7% to 22.6% 20,21,30,31. Among the 44 RAS-mutant ERMS from 4 series 20,21,30,31, NRAS mutations are the most common (27, 61%), followed by KRAS (11, 25%) and HRAS mutations (6, 14%). The hot spots for both NRAS and HRAS mutations in ERMS occur in the codon 61 11,20,21,30. There are 3 ERMS cases reported with identical HRAS p.G13R and p.Q61L mutations as detected in our MEM cases 20,21,32. Interestingly, the HRAS mutations seen in MEM are different than the ones described in Costello syndrome, a rare multisystem disorder caused by heterozygous germline HRAS mutation and showing a phenotype characterized by craniofacial dysmorphology, intellectual disabilities, cardiac malformations, and short stature 33,34. Costello syndrome confers tumor predisposition, including RMS, neuroblastoma, and urothelial cell carcinoma of urinary bladder 35. The HRAS mutation genotype appears to correlate with the phenotype and even with the risk of malignancy 33.
Ras proteins are well-known proto-oncogenes which are somatically mutated in a wide variety of human cancers 36. The Ras family, including HRAS, KRAS and NRAS, belongs to the small G protein superfamily and involves the Ras/MAPK (mitogen-activated protein kinase) pathway, which transduces the extracellular input to the intracellular compartment through the GDP/GTP-regulated switches. The cancer-related somatic mutations in Ras members often occur at amino acids 12, 13, or 61, which are the conserved sites for modulating GDP/GTP binding and hydrolysis. There is increasing evidence that Ras proteins have isoform-specific biologic functions and tumorigenic effects, possibly attributed to the C-terminal hypervariable region, and that different mutation codons might induce different transformation potential. Furthermore, different cancers show diverse RAS mutation predominance: KRAS in colorectal and pancreatic cancers; NRAS in hematopoietic neoplasm and malignant melanoma; HRAS in salivary gland and urinary tract carcinoma 22,36. In addition, HRAS mutations were detected in both naive and treated MEMs, suggesting an intrinsic event rather than a chemotherapy-induced process. As previously reported37 and seen in 2 of our cases, a diagnosis of MEM was rendered only after the surgical specimen was examined, the biopsy material showing only ERMS areas.
Other notable mutations detected in rare MEM involve the PTPRD and FBXW7 genes. PTPRD is one of the 107-member family of protein tyrosine phosphatases, which induce a rapid turnover of the phosphate moiety on the phosphorylated tyrosine residues caused by protein tyrosine kinases 38. Therefore, several protein tyrosine phosphatase members are candidate tumor suppressors. Inactivation of PTPRD by deletion, mutation, or epigenetic methylation has been described in glioblastoma, melanoma, head and neck squamous cell carcinoma, and lung cancer 39,40. Interestingly, recurrent microdeletions of PTPRD are also reported in neuroblastoma 41. Most PTPRD mutations are located at conserved domains, such as immunoglobulin-like C2 type, fibronectin type III, or phosphatase catalytic domains. The two PTPRD mutations detected in MEMs occur in the fibronectin type III domain. In contrast, RMS show frequent PTEN methylation (70%) or rare PTPN11 mutation (3.3%), the latter belongs to the non-receptor protein tyrosine phosphatases21. FBXW7 gene encodes a member of the F-box protein family 23. The FBXW7 proteins form dimers in the Skp1-Cullin-F-box (SCF) ubiquitin ligase complex, which function in ubiquitin-mediated proteolysis. FBXW7 mutations are widely found in human cancers and most mutations occur in the three arginine residues (R465, R479, R505) that are critical for substrate interaction. FBXW7 mutations were occasionally described in fusion-negative RMS (2.4–6.4%) 20,21.
The unsupervised hierarchical clustering showed that the 2 index MEM cases grouped tightly with RMS, findings which contradict the prior report of an intracranial MEM clustering with MPNST 11. This phenomenon is further consolidated by the overlapping RMS signature and correlated by gene set enrichment analysis. Nearly all of the reported top overexpressed genes in the intracranial MEM were not detected as upregulated in our MEM and RMS group. Moreover, the retained H3K27me3 expression in MEM is also distinct from most MPNSTs, which show frequent loss-of-function somatic alterations of the PRC2 components (EED or SUZ12) 16. The MEM gene signature does not include only the upregulation of myogenesis genes (at comparable expression levels with the RMS group), but also genes implicated in neuronal development.
Among the 9 intracranial cases reported, including cerebrum (6), cerebellum (1), falx cerebri (1), and cerebellopontine angle (1) 8,25, only the cerebellopontine tumor showed a combined RMS and ganglioneuroma 24. Although the appearance of ganglion cells or neuroblastoma and rhabdomyoblasts was noted in 5 cases, the predominant mesenchymal component was described as non-specific spindle-shaped cells with focal rhabdomyoblast-like cells, supported by focal actin or desmin positivity, but not myogenin 3,11,29,42. In one of the cases the diagnosis was revised in a follow-up study 6,29. None of the remaining 3 cases showed RMS differentiation and one case had an oligoastrocytoma instead of neuroblastic elements 8,25,43. The same skepticism resides in the adult MEM cases reported, most of them may represent “malignant mesenchymoma” 9.
In conclusion, we report 7 cases of MEM, defined by composite RMS and neuroblastic differentiation. All the RMS components resembled ERMS with classic, spindle cell, or dense pattern; while the neuroblastic elements ranged from scattered ganglion cells, ganglioneuroma, ganglioneuroblastoma, or neuroblastoma. The MEM patients showed a homogeneous clinical presentation overlapping that of ERMS, of very young male children, mostly of infants, occurring in the pelvis/urogenital region. The high frequency of HRAS mutations identified in MEM, corroborated with a skeletal muscle-related gene signature and retained H3K27me3 expression, suggest a closer relationship to RMS than MPNST.
Supplementary Material
Gene set enrichment analysis confirmed the enrichment of 437 gene signature in peripheral nerve sheath tumors (A). MEM and RMS exhibited randomly distributed in clustering analysis using the 437-gene panel of peripheral nerve sheath tumors (B). The expression levels of 4 up-regulated genes described in an intracranial MEM 11 were not found to be statistically different in MEM and RMS (C). The heatmap of the neuronal gene signature shows up-regulation in MEM (blue) but not in RMS (green) (D).
Acknowledgments
Supported in part by: P50 CA140146-01 (CRA); P30-CA008748 (CRA); Kristen Ann Carr Foundation (CRA); Cycle for Survival (CRA).
References
- 1.Naka A, Matsumoto S, Shirai T, et al. Ganglioneuroblastoma associated with malignant mesenchymoma. Cancer. 1975;36:1050–1056. doi: 10.1002/1097-0142(197509)36:3<1050::aid-cncr2820360331>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 2.Karcioglu Z, Someren A, Mathes SJ. Ectomesenchymoma. A malignant tumor of migratory neural crest (ectomesenchyme) remnants showing ganglionic, schwannian, melanocytic and rhabdomyoblastic differentiation. Cancer. 1977;39:2486–2496. doi: 10.1002/1097-0142(197706)39:6<2486::aid-cncr2820390627>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 3.Freitas AB, Aguiar PH, Miura FK, et al. Malignant ectomesenchymoma. Case report and review of the literature. Pediatr Neurosurg. 1999;30:320–330. doi: 10.1159/000028818. [DOI] [PubMed] [Google Scholar]
- 4.Nael A, Siaghani P, Wu WW, et al. Metastatic malignant ectomesenchymoma initially presenting as a pelvic mass: report of a case and review of literature. Case Rep Pediatr. 2014;2014:792925. doi: 10.1155/2014/792925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawamoto EH, Weidner N, Agostini RM, Jr, et al. Malignant ectomesenchymoma of soft tissue. Report of two cases and review of the literature. Cancer. 1987;59:1791–1802. doi: 10.1002/1097-0142(19870515)59:10<1791::aid-cncr2820591018>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 6.Dantonello TM, Leuschner I, Vokuhl C, et al. Malignant ectomesenchymoma in children and adolescents: report from the Cooperative Weichteilsarkom Studiengruppe (CWS) Pediatr Blood Cancer. 2013;60:224–229. doi: 10.1002/pbc.24174. [DOI] [PubMed] [Google Scholar]
- 7.Yohea MEGE, Balarezoc FS, Timothy BR, Wua AC, Fincka CM, Orseya AD. A novel case of pediatric thoracic malignant ectomesenchymoma in an infant. J Pediatr Surg Case Rep. 2013;1:3. [Google Scholar]
- 8.Kun Y, Duan Z, Mei X, et al. A rare case of malignant pediatric ectomesenchymoma arising from the cerebrum. Int J Clin Exp Pathol. 2015;8:8545–8550. [PMC free article] [PubMed] [Google Scholar]
- 9.Boue DR, Parham DM, Webber B, et al. Clinicopathologic study of ectomesenchymomas from Intergroup Rhabdomyosarcoma Study Groups III and IV. Pediatr Dev Pathol. 2000;3:290–300. doi: 10.1007/s100249910039. [DOI] [PubMed] [Google Scholar]
- 10.Floris G, Debiec-Rychter M, Wozniak A, et al. Malignant ectomesenchymoma: genetic profile reflects rhabdomyosarcomatous differentiation. Diagn Mol Pathol. 2007;16:243–248. doi: 10.1097/PDM.0b013e3180645105. [DOI] [PubMed] [Google Scholar]
- 11.Kleinschmidt-DeMasters BK, Lovell MA, Donson AM, et al. Molecular array analyses of 51 pediatric tumors shows overlap between malignant intracranial ectomesenchymoma and MPNST but not medulloblastoma or atypical teratoid rhabdoid tumor. Acta Neuropathol. 2007;113:695–703. doi: 10.1007/s00401-007-0210-0. [DOI] [PubMed] [Google Scholar]
- 12.Coffin CM. Ectomesenchymoma. In: Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F, editors. WHO Classification of Tumours of Soft Tissue and Bone. Lyon: IARC; 2013. p. 191. [Google Scholar]
- 13.Oppenheimer O, Athanasian E, Meyers P, et al. Malignant ectomesenchymoma in the wrist of a child: case report and review of the literature. Int J Surg Pathol. 2005;13:113–116. doi: 10.1177/106689690501300117. [DOI] [PubMed] [Google Scholar]
- 14.Shimada H, Ambros IM, Dehner LP, et al. Terminology and morphologic criteria of neuroblastic tumors: recommendations by the International Neuroblastoma Pathology Committee. Cancer. 1999;86:349–363. [PubMed] [Google Scholar]
- 15.Rudzinski ER, Teot LA, Anderson JR, et al. Dense pattern of embryonal rhabdomyosarcoma, a lesion easily confused with alveolar rhabdomyosarcoma: a report from the Soft Tissue Sarcoma Committee of the Children’s Oncology Group. Am J Clin Pathol. 2013;140:82–90. doi: 10.1309/AJCPA1WN7ARPCMKQ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee W, Teckie S, Wiesner T, et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46:1227–1232. doi: 10.1038/ng.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antonescu CR, Yoshida A, Guo T, et al. KDR activating mutations in human angiosarcomas are sensitive to specific kinase inhibitors. Cancer Res. 2009;69:7175–7179. doi: 10.1158/0008-5472.CAN-09-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajdu M, Singer S, Maki RG, et al. IGF2 over-expression in solitary fibrous tumours is independent of anatomical location and is related to loss of imprinting. J Pathol. 2010;221:300–307. doi: 10.1002/path.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fredlund E, Ringner M, Maris JM, et al. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. Proc Natl Acad Sci U S A. 2008;105:14094–14099. doi: 10.1073/pnas.0804455105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shern JF, Chen L, Chmielecki J, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4:216–231. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seki M, Nishimura R, Yoshida K, et al. Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat Commun. 2015;6:7557. doi: 10.1038/ncomms8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26:455–464. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holimon JL, Rosenblum WI. “Gangliorhabdomyosarcoma”: a tumor of ectomesenchyme. Case report. J Neurosurg. 1971;34:417–422. doi: 10.3171/jns.1971.34.3.0417. [DOI] [PubMed] [Google Scholar]
- 25.Ito A, Kumabe T, Saito R, et al. Malignant pediatric brain tumor of primitive small round cell proliferation with bland-looking mesenchymal spindle cell elements. Brain Tumor Pathol. 2013;30:109–116. doi: 10.1007/s10014-012-0106-0. [DOI] [PubMed] [Google Scholar]
- 26.Howley S, Stack D, Morris T, et al. Ectomesenchymoma with t(1;12)(p32;p13) evolving from embryonal rhabdomyosarcoma shows no rearrangement of ETV6. Hum Pathol. 2012;43:299–302. doi: 10.1016/j.humpath.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Yohe ME, Girard ED, Balarezo FS, et al. A novel case of pediatric thoracic malignant ectomesenchymoma in an infant. Journal of Pediatric Surgery Case Reports. 2013;1:20–22. [Google Scholar]
- 28.Stolnicu S, Goyenaga P, Hincu M, et al. Embryonal (botryoides) rhabdomyosarcoma of the uterus harboring a primitive neuroectodermal tumor component. Int J Gynecol Pathol. 2012;31:387–389. doi: 10.1097/PGP.0b013e31823ff3e6. [DOI] [PubMed] [Google Scholar]
- 29.Weiss E, Albrecht CF, Herms J, et al. Malignant ectomesenchymoma of the cerebrum. Case report and discussion of therapeutic options. Eur J Pediatr. 2005;164:345–349. doi: 10.1007/s00431-005-1646-7. [DOI] [PubMed] [Google Scholar]
- 30.Martinelli S, McDowell HP, Vigne SD, et al. RAS signaling dysregulation in human embryonal Rhabdomyosarcoma. Genes Chromosomes Cancer. 2009;48:975–982. doi: 10.1002/gcc.20702. [DOI] [PubMed] [Google Scholar]
- 31.Shukla N, Ameur N, Yilmaz I, et al. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. 2012;18:748–757. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kratz CP, Steinemann D, Niemeyer CM, et al. Uniparental disomy at chromosome 11p15.5 followed by HRAS mutations in embryonal rhabdomyosarcoma: lessons from Costello syndrome. Hum Mol Genet. 2007;16:374–379. doi: 10.1093/hmg/ddl458. [DOI] [PubMed] [Google Scholar]
- 33.Gripp KW, Lin AE. Costello syndrome: a Ras/mitogen activated protein kinase pathway syndrome (rasopathy) resulting from HRAS germline mutations. Genet Med. 2012;14:285–292. doi: 10.1038/gim.0b013e31822dd91f. [DOI] [PubMed] [Google Scholar]
- 34.Aoki Y, Niihori T, Kawame H, et al. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 35.Gripp KW. Tumor predisposition in Costello syndrome. Am J Med Genet C Semin Med Genet. 2005;137C:72–77. doi: 10.1002/ajmg.c.30065. [DOI] [PubMed] [Google Scholar]
- 36.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sebire NJ, Ramsay AD, Malone M, et al. Extensive posttreatment ganglioneuromatous differentiation of rhabdomyosarcoma: malignant ectomesenchymoma in an infant. Pediatr Dev Pathol. 2003;6:94–96. doi: 10.1007/s10024-002-0047-4. [DOI] [PubMed] [Google Scholar]
- 38.Julien SG, Dube N, Hardy S, et al. Inside the human cancer tyrosine phosphatome. Nat Rev Cancer. 2011;11:35–49. doi: 10.1038/nrc2980. [DOI] [PubMed] [Google Scholar]
- 39.Veeriah S, Brennan C, Meng S, et al. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc Natl Acad Sci U S A. 2009;106:9435–9440. doi: 10.1073/pnas.0900571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solomon DA, Kim JS, Cronin JC, et al. Mutational inactivation of PTPRD in glioblastoma multiforme and malignant melanoma. Cancer Res. 2008;68:10300–10306. doi: 10.1158/0008-5472.CAN-08-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stallings RL, Nair P, Maris JM, et al. High-resolution analysis of chromosomal breakpoints and genomic instability identifies PTPRD as a candidate tumor suppressor gene in neuroblastoma. Cancer Res. 2006;66:3673–3680. doi: 10.1158/0008-5472.CAN-05-4154. [DOI] [PubMed] [Google Scholar]
- 42.Papos M, Pekrun A, Herms JW, et al. Somatostatin receptor scintigraphy in the management of cerebral malignant ectomesenchymoma: a case report. Pediatr Radiol. 2001;31:169–172. doi: 10.1007/s002470000404. [DOI] [PubMed] [Google Scholar]
- 43.Altenburger DL, Wagner AS, Eslin DE, et al. A rare case of malignant pediatric ectomesenchymoma arising from the falx cerebri. J Neurosurg Pediatr. 2011;7:94–97. doi: 10.3171/2010.10.PEDS10261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene set enrichment analysis confirmed the enrichment of 437 gene signature in peripheral nerve sheath tumors (A). MEM and RMS exhibited randomly distributed in clustering analysis using the 437-gene panel of peripheral nerve sheath tumors (B). The expression levels of 4 up-regulated genes described in an intracranial MEM 11 were not found to be statistically different in MEM and RMS (C). The heatmap of the neuronal gene signature shows up-regulation in MEM (blue) but not in RMS (green) (D).