Abstract
The extracellular matrix (ECM) presents an evolving set of mechanical cues to resident cells. We developed methacrylated hyaluronic acid (MeHA) hydrogels containing both stable and hydrolytically degradable crosslinks to provide cells with a gradually softening (but not fully degradable) milieu, mimicking physiological events such as fibrosis regression. To demonstrate the utility of this cell culture system, we studied the phenotype of rat hepatic stellate cells, the major liver precursors of fibrogenic myofibroblasts, within this softening environment. Stellate cells that were mechanically primed on tissue culture plastic attained a myofibroblast phenotype, which persisted when seeded onto stiff (~ 20 kPa) hydrogels. However, mechanically primed stellate cells on stiff-to-soft (~ 20 to ~ 3 kPa) hydrogels showed reversion of the myofibroblast phenotype over 14 days, with reductions in cell area, expression of the myofibroblast marker alpha-smooth muscle actin (α-SMA), and Yes-associated protein/Transcriptional coactivator with PDZ-binding motif (YAP/TAZ) nuclear localization when compared to stellate cells on stiff hydrogels. Cells on stiff-to-soft hydrogels did not fully revert, however. They displayed reduced expression of glial fibrillary acidic protein (GFAP), and underwent abnormally rapid re-activation to myofibroblasts in response to re-stiffening of the hydrogels through introduction of additional crosslinks. These features are typical of stellate cells with an intermediate phenotype, reported to occur in vivo with fibrosis regression and re-injury. Together, these data suggest that mechanics play an important role in fibrosis regression and that integrating dynamic mechanical cues into model systems helps capture cell behaviors observed in vivo.
Keywords: hydrogels, dynamic mechanics, liver fibrosis, hepatic stellate cells, YAP/TAZ, myofibroblast
Graphical Abstract
1. Introduction
Tissue fibrosis is prevalent in the developed world and is a contributing factor to over 40% of all deaths.1 Liver fibrosis and primary liver cancer, which in most cases is associated with fibrosis, together account for over 1.7 million deaths annually worldwide.2 Essential to the mechanism of fibrosis is that resident fibroblasts and pericytes undergo myofibroblastic differentiation in response to mechanical and soluble cues, becoming fibrogenic and synthesizing excessive amounts of fibrillar extracellular matrix (ECM) proteins such as type I collagen. The reversibility of this process varies from organ to organ but is particularly notable in the liver, which undergoes significant fibrosis resolution following removal of the injurious stimulus. Interestingly, while recent work using mouse models demonstrated that about half of the myofibroblastic hepatic stellate cells (the major myofibroblast population) in the fibrotic liver undergo apoptosis, the remaining myofibroblasts acquired an “intermediate” phenotype where they appeared similar to healthy stellate cells but were characterized by abnormal gene expression and unusually rapid myofibroblastic re-differentiation after a repeat insult, a finding that may have significant clinical relevance in the care of patients with liver fibrosis.3, 4 Although mechanics play a key role in stellate cell differentiation to myofibroblasts, the role of tissue mechanics in mediating stellate cell phenotypic changes during regression (and repeat differentiation) is unknown.
Given that tissue stiffening is a clinical indicator of fibrosis progression and ECM stiffening may be an important contributing factor to the onset of fibrosis and tumor formation,5-7 it follows that tissue softening may play a role in fibrosis resolution. Hydrogels have proven to be useful substrates for studying cellular mechanotransduction due to their ability to mimic salient ECM characteristics including mechanics, water content, and the facilitation of cell adhesion. However, most cell culture studies use mechanically static materials that do not recapitulate the dynamic nature of processes such as fibrosis progression and resolution.
Several hydrogel systems have been developed that display softening behavior in a temporally-controlled manner, exploiting the use of DNA crosslinking,8 hydrolytically degradable crosslinkers,9, 10 or photocleavable groups.11-13 In particular, the Anseth group has used photocleavable softening hydrogels to investigate stem cell differentiation,11 fibroblast mechanotransduction,12 and cellular mechanical memory in response to varying mechanical doses.13 While these approaches utilized step changes in hydrogel stiffness, tissue mechanics evolve in a gradual manner, so it would be useful to apply a system that mimics this behavior as closely as possible. Zustiak et al. used poly(ethylene glycol)-based hydrogels polymerized with crosslinkers containing hydrolytically-labile ester groups to synthesize a suite of hydrolytically-degradable hydrogels with gradual degradation times on the order of days9 that were suitable for biomedical applications such as protein delivery.10 However, since these materials were fully degradable they were not suitable for studying long-term cell-microenvironment interactions.
In this work, we used a hyaluronic acid (HA) hydrogel system containing both stable and hydrolytically degradable crosslinks to present a gradually softening milieu to cells. HA was chosen for its ease of chemical modification14 and its past utility in mechanobiology studies with several cell types, including hepatic stellate cells.15, 16 We performed mechanical characterization of the hydrogels and investigated the response of activated hepatic stellate cells to the softening environment as a proof-of-concept study of fibrosis regression. Finally, we investigated whether our mechanically dynamic culture platform could capture elements of the clinically-relevant intermediate stellate cell phenotype observed in vivo following fibrosis resolution, both before and after a repeat insult (re-stiffening) event.
2. Materials and methods
2.1 MeHA synthesis
Hyaluronic acid (HA) was modified with methacrylates as previously reported17 (Figure 1A). Sodium hyaluronate (Lifecore, 75 kDa) was dissolved at 2 wt% in deionized water and reacted with methacrylic anhydride (5.6 mL per g NaHA) for 6 h on ice while continuously maintaining pH in the range of 8.0-9.5. The solution was then dialyzed (SpectraPor, 6-8 kDa molecular weight cutoff) at room temperature against deionized water for 7 d and lyophilized. The degree of primary hydroxyl modification with methacrylates was ~ 90% as measured by 1H NMR (Bruker). MeHA was further modified with 1 mM thiolated RGD (GCGYGRGD-SPG, GenScript) via Michael-type addition (45 min, room temperature, pH 9) to permit cell adhesion.
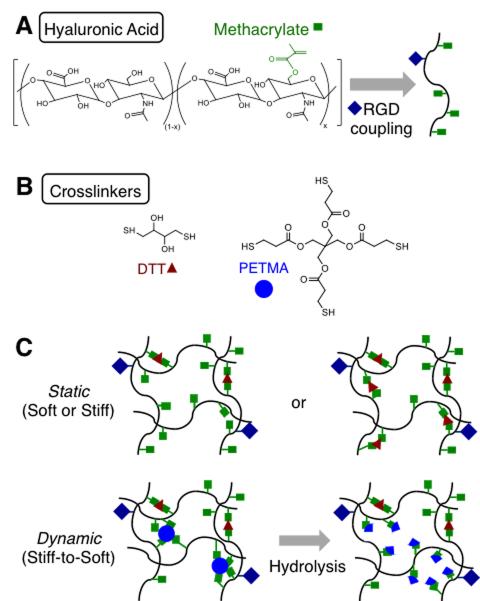
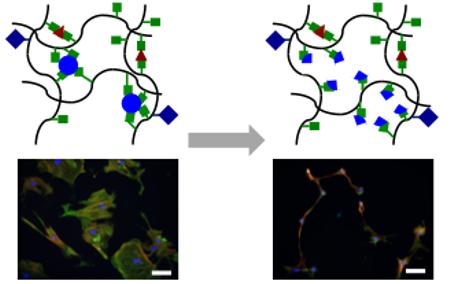
Figure 1. Approach for fabricating gradually softening hydrogel substrates.
(A) Hyaluronic acid (HA) modified with methacrylates (MeHA) and RGD via Michael-type addition between methacrylates on HA and thiols on the RGD peptide. (B) Crosslinkers used for the crosslinking of RGD-modified MeHA to produce stable (DTT) or ester-containing hydrolytically labile (DTT, PETMA) hydrogels. (C) Hydrogels designed as soft or stiff with relatively static mechanical properties through crosslinking with DTT or designed as dynamically softening (stiff-to-soft) by crosslinking with a combination of DTT and PETMA.
2.2 MeHA hydrogel fabrication
RGD-modified MeHA hydrogels were fabricated via Michael type-addition. 3 wt% MeHA was crosslinked using either dithiothreitol (DTT, Sigma) only or a combination of DTT and pentaerythritol tetrakis(mercaptoacetate) (PETMA, TCI Chemical) (Figure 1B). For soft and stiff static groups, DTT was mixed with MeHA to consume 20% and 65% of available network methacrylates respectively. The stiff-to-soft group was fabricated with DTT (0.2 thiol:methacrylate ratio) and PETMA (0.6 thiol:methacrylate ratio). Hydrogel precursor solution (pH 9, 50 μL) was pipetted between a methacrylated coverslip18 (22 × 22 mm) and an untreated coverslip (18 × 18 mm) and then allowed to polymerize at room temperature for 3 h to form hydrogel thin films (~ 100 μm thickness) covalently attached to the methacrylated coverslips.
2.3 Hydrogel mechanics and swelling characterization
Hydrogel elastic moduli were measured using dynamic mechanical analysis (DMA, TA Instruments). Hydrogel samples were stored in PBS at 37°C for 14 days and mechanical analysis was performed at days 0, 1, 3, 5, 7, 10, and 14. Samples were compressed at a strain rate of 10% per min and elastic moduli were calculated from the slope of the stress-strain curve between 10-20% strain. Volumetric swelling ratios (Qv) were calculated from the hydrogel wet weight (Mw) at each time point and the original hydrogel dry weight (Md), using the following equation where the densities of the hydrogel polymer (ρp) and solvent (ρs) were assumed to be 1.23 g mL−1 and 1 g mL−1 respectively19:
2.4 Hydrogel degradation quantification
Hydrogel degradation via hydrolysis was assessed over the course of 14 days. Hydrogels were incubated in PBS at 37°C with buffer changes performed after every mechanical testing time point. Buffer samples were stored at −80°C until analysis. Uronic acid content in the buffer samples was measured to determine HA mass loss.20
2.5 Hepatic stellate cell isolation
Sprague-Dawley rat hepatic stellate cells were isolated as previously described.21 All studies were performed in compliance with the University of Pennsylvania’s Institutional Animal Care and Use Committee in accordance with the National Institutes of Health’s “Guide for the Care and Use of Laboratory Animals” (NIH Publication 85-23, revised 1996). In brief, in situ enzymatic digestion of the liver was performed via sequential perfusion with 0.4% pronase (Roche Diagnostics) and 0.04% collagenase II (Worthington). The resultant slurry was then diluted in minimal essential media (MEM) and filtered through cheesecloth. The total cell suspension was washed twice in 0.002% DNase (Worthington). Stellate cells were isolated from the total cell population through density gradient centrifugation with a 9% Nycodenz (Sigma) solution at 1400 g for 25 min. Stellate cells were then washed in MEM and stored on ice prior to seeding onto hydrogels or tissue culture polystyrene (TCPS)/glass.
2.6 Hepatic stellate cell mechanical priming
Following stellate cell isolation, cells were either seeded directly onto soft hydrogels (see next section) or plated onto TCPS (for subsequent experiments on hydrogels) or glass coverslips (for imaging) for 7 days. This extended exposure to a high stiffness culture substrate (E of TCPS/glass > GPa) is widely accepted to result in hepatic stellate cell myofibroblast activation.22 After 7 days of mechanical priming, stellate cells were trypsinized and moved to hydrogels.
2.7 Hepatic stellate cell culture on MeHA hydrogels
In preparation for cell seeding, hydrogels were allowed to swell in PBS overnight at 37°C. Hydrogels were then sterilized using germicidal ultraviolet (UV) irradiation for 2 h and incubated in culture media for at least 30 min prior to stellate cell seeding. Culture media consisted of phenol red-free M199 media (Invitrogen) supplemented with 10 v/v% fetal bovine serum (Sigma), 2 v/v% penicillin streptomycin (Invitrogen), and 1 v/v% fungizone amphotericin B (Invitrogen). Stellate cells were seeded onto sterilized hydrogels placed in 6-well plates at a density of 5 × 103 cells/cm2. Hydrogels were moved to fresh plates and media the following day with subsequent media changes occurring every 3 days.
2.8 Cell imaging, staining, and quantification
Stellate cell spread area was determined using NIH ImageJ analysis of phase contrast images of cell-seeded hydrogels acquired on a Zeiss Axiovert 200 inverted microscope (Hitech Instruments, Inc.). Actin organization (α-SMA) and F-actin), Yes-associated protein/Transcriptional coactivator with PDZ-binding motif (YAP/TAZ) nuclear localization, and glial fibrillary acidic protein (GFAP) expression were determined using fluorescence microscopy. Stellate cell-seeded hydrogels were fixed in 10% buffered formalin for 15 min, permeabilized in 0.1% Triton X-100 for 15 min, and blocked in 3% bovine serum albumin (BSA) in PBS for 1 h at room temperature. Samples were then incubated with primary antibodies (diluted in blocking buffer) overnight at 4°C. Primary antibody targets included α-SMA (mouse monoclonal anti–α-SMA clone 1A4 Ab, Sigma, 1:400), YAP/TAZ (rabbit polyclonal anti-YAP, Santa Cruz Biotechnology, 1:200), or GFAP (rabbit monoclonal anti-GFAP, Abcam, 1:200). Hydrogels were then washed thrice in PBS and incubated either with appropriate secondary antibodies for 2 h at room temperature (AlexaFluor® 488 goat anti-mouse IgG or AlexaFluor® 488 goat anti-rabbit, Invitrogen, 1:200) or with rhodamine phalloidin to visualize F-actin (Invitrogen, 1:200). Finally, hydrogels were washed twice in PBS, treated with DAPI nuclear stain (1:10000) for 1 min, rinsed again in PBS, and stored in fresh PBS at 4°C in the dark until imaging. An Olympus BX51 microscope (B&B Microscopes Limited) was used for all fluorescent imaging. Identical settings were used while imaging (exposure time for each respective channel) as well as during post-image analysis (brightness, contrast levels). NIH ImageJ was used to calculate YAP/TAZ nuclear and cytoplasmic intensities.
2.9 In situ hydrogel re-stiffening
Hydrogels were re-stiffened using an in situ photocrosslinking method we previously described to be cytocompatible in the presence of hepatic stellate cells, since there were remaining methacrylates available after the initial crosslinking.16 Stellate-cell seeded stiff-to-soft hydrogels, following 14 days of softening, were incubated in culture media containing 6.6 mM lithium acylphosphinate (LAP) photoinitiator23, 24 for 30 min at 37°C. LAP-infused hydrogels were then irradiated with blue light for 10 min at 10 mW cm−2 using a dental curing lamp (ESPE Elipar 2500, 3M). Following light irradiation, the re-stiffened hydrogels were rinsed twice in PBS to remove excess LAP and then placed in fresh media.
2.10 Statistical analysis
Data sets were analyzed using student’s t-tests (two groups) or one-way analysis of variance (ANOVA) followed by Tukey-HSD post-hoc tests (more than two groups). At least 3 hydrogels and/or 30 cells were assayed for all analyses. Tukey box plots of single cell data were composed of boxes corresponding to the second and third quartile of the data set with error bars indicating the maximum and minimum values (or 1.5 times the interquartile range, whichever was smaller). Error in bar graphs and scatter plots was standard error of the mean unless otherwise specified. Significance was denoted by *, **, or *** corresponding to P < 0.05, 0.01, or 0.001 respectively.
3. Results and Discussion
3.1 Hydrogels with a combination of stable and degradable crosslinks exhibit gradual softening
We used methacrylate-modified hyaluronic acid (MeHA) to fabricate hydrogels that gradually soften without complete degradation (Figure 1A). MeHA was first modified with RGD to permit cell attachment and was then reacted with a combination of thiolated crosslinkers to form hydrogels (Figure 1B). Dithiothreitol (DTT) and pentaerythritol tetrakis(mercaptoacetate) (PETMA), a tetrafunctional crosslinker containing thiol groups for crosslinking and ester groups for gradual hydrolysis, were used. We anticipated that hydrogels crosslinked with DTT only would remain relatively stable mechanically, while MeHA hydrogels fabricated with a combination of DTT and PETMA would gradually soften due to ester hydrolysis but would not degrade completely because of the presence of the stable DTT crosslinks (Figure 1C).
Previous work showed that hepatic stellate cells, the major precursors of fibrogenic myofibroblasts in the liver, differentiate into fibrogenic myofibroblasts when cultured on stiff hydrogels with stiffness levels similar to fibrotic liver (elastic modulus (E) > 10 kPa), while stellate cells cultured on soft, compliant hydrogels resembling the stiffness of healthy liver (E ~ 1-3 kPa) maintain their normal, quiescent state and do not undergo myofibroblast activation.15, 25, 26 Therefore, for soft and stiff static hydrogels crosslinked with DTT only we targeted starting elastic moduli of ~ 15-20 kPa and ~ 1-3 kPa corresponding to diseased and healthy liver tissue, respectively. For the stiff-to-soft hydrogel we targeted initial mechanics of ~ 15-20 kPa with final mechanics of ~ 1-3 kPa after 14 days. Mechanical characterization of hydrogels over 14 days showed relatively stable mechanics for the soft and stiff groups (Figure 2A). Although the elastic modulus of the stiff group decreased from ~ 19 to ~ 12 kPa, this final stiffness has still been shown to promote hepatic stellate cell myofibroblast differentiation.25
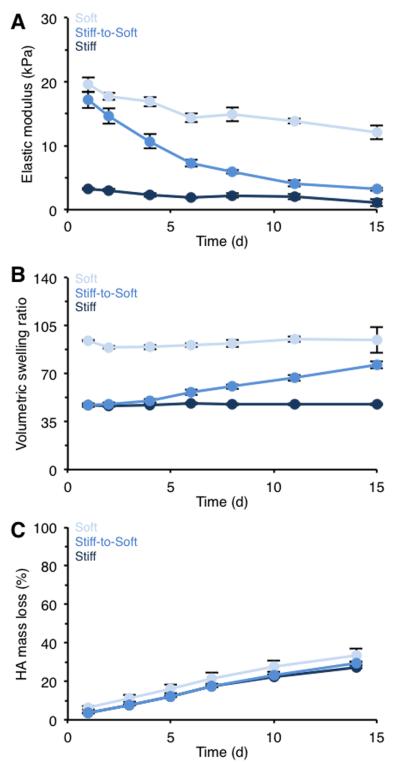
Figure 2. Characterization of static and dynamic hydrogels.
(A) Hydrogel elastic modulus, (B) volumetric swelling ratio, and (C) HA mass loss for hydrogel formulations crosslinked with DTT only (soft or stiff) or with a combination of DTT and PETMA degradable crosslinker (stiff-to-soft). n > 3 hydrogels per group per time point.
Meanwhile, the stiff-to-soft group exhibited a gradual decrease in elastic modulus from an initial stiffness of ~ 17 kPa to ~ 3 kPa with no significant difference in final elastic modulus compared to the soft group at day 14. This gradual decrease in crosslinking density also altered swelling behavior of the hydrogels as expected, with a corresponding increase of swelling observed for the stiff-to-soft group over 14 days (Figure 2B). Importantly, these hydrogels did not degrade completely and maintained their structural integrity over 14 days. All experimental groups showed similar HA mass loss profiles (Figure 2C), confirming that the overall HA network remained similarly stable due to the DTT crosslinks; however, the decreased mechanics indicated reduced crosslink density in the hydrolytically unstable group. Additionally, the elastic moduli (E) data for the stiff-to-soft group displayed pseudo first order exponential decay kinetics corresponding to the gradual decrease in crosslink density due to ester hydrolysis as represented by:
The degradation time constant (k) for the stiff-to-soft group was 0.124 d−1.
3.2 Hepatic stellate cells show reduced spreading when cultured on stiff-to-soft hydrogels
Following mechanical characterization of our hydrogel formulations, we assessed hepatic stellate cell phenotype when cultured on static and dynamic MeHA hydrogels. Since the observed differences in swelling behavior may alter the local density of RGD ligands presented to stellate cells for attachment, we performed control experiments to demonstrate that stellate cell attachment to HA hydrogels was mediated by RGD and that lower RGD concentrations (0.5 mM versus 1 mM) did not result in reduced cell attachment (Figure S1).
In order to evaluate stellate cell response to changing mechanics, we first induced myofibroblast differentiation by seeding freshly isolated stellate cells onto plastic or glass for 7 days (Figure 3A). This mechanical priming step (E of TCPS/glass > GPa) is widely accepted to result in hepatic stellate cell myofibroblast activation.22 Stellate cells were then moved to soft, stiff, or stiff-to-soft hydrogels for an additional 14 days. We hypothesized that the soft and stiff environments would promote a return to the quiescent phenotype or perpetuation of the acquired myofibroblast phenotype, respectively, while the stiff-to-soft group would reflect a gradual return of an intermediate phenotype as observed in vivo where fibrosis resolution takes several weeks.3, 4 As a control, freshly isolated stellate cells were seeded in parallel directly onto soft static hydrogels (no mechanical priming) and remained on these hydrogels for the entire duration of the experiment (21 days total).
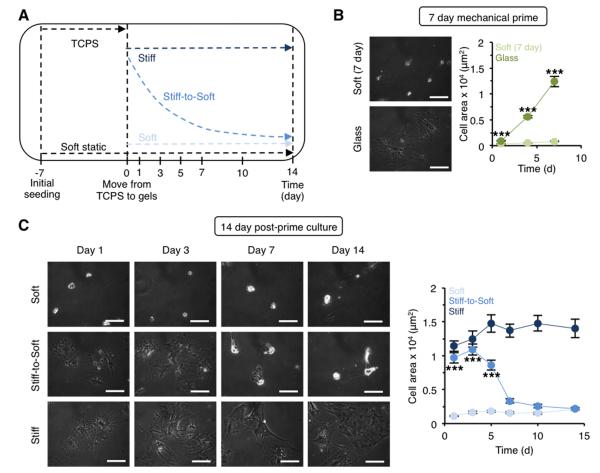
Figure 3. Hepatic stellate cells show gradually decreased spreading when cultured on stiff-to-soft hydrogels.
(A) Schematic of experimental design for stellate cell culture on hydrogels. Stellate cells were initially plated on TCPS/glass or directly onto soft static hydrogels following isolation. After 7 days of mechanical priming, cells were moved to either soft or stiff (static) hydrogels or onto stiff-to-soft hydrogels. As a control, freshly isolated cells that were plated onto soft hydrogels were cultured for an additional 14 days (21 days total) with no mechanical priming. (B) Stellate cell phase contrast images and spread area during 7 days of mechanical priming. ***: P < 0.001, glass group significantly greater than soft (day 7) group. (C) Stellate cell phase contrast images and spread area after plating on hydrogels and culturing for an additional 14 days. ***: P < 0.001, stiff-to-soft group significantly greater than soft group. Scale bars: 100 μm. n > 37 cells per group per time point.
As expected, stellate cells plated onto glass rapidly spread, lost their lipid droplets, and assumed a myofibroblast-like morphology after 7 days, while stellate cells seeded onto soft hydrogels remained rounded and significantly less spread (Figure 3B). Activated cells moved to stiff hydrogels maintained a consistently spread morphology during the 14 day post-prime culture, while activated stellate cells on soft hydrogels quickly adjusted to the new mechanical environment and became rounded (Figure 3C). Meanwhile, stellate cells on stiff-to-soft hydrogels gradually transitioned from a spread, myofibroblast-like phenotype to a more rounded shape similar to the stellate cells on the soft hydrogels. There was no significant difference in spread area between stellate cells on the stiff-to-soft and soft experimental groups after 14 days. While some stellate cells likely died and detached during these experiments, we confirmed through live-dead staining that stellate cells adhered to hydrogels were largely viable (> 95%, Figure S2).
3.3 Mechanically primed hepatic stellate cells lose actin stress fiber organization following culture on stiff-to-soft hydrogels
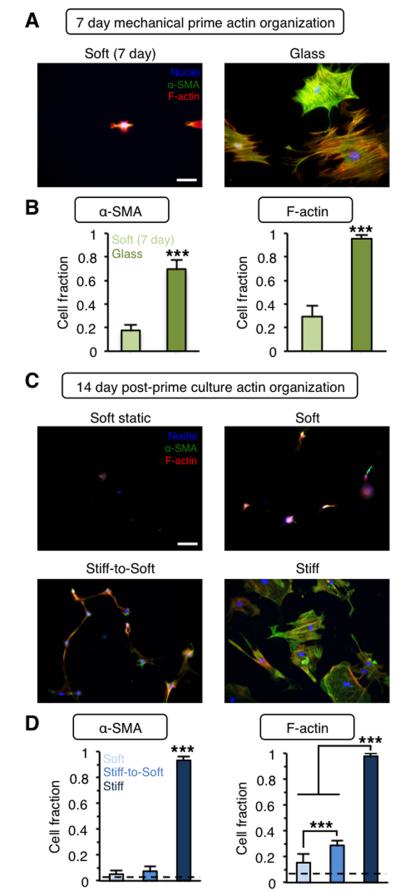
In addition to measuring stellate cell spread area, we also investigated the cytoskeletal features that define fibrogenic myofibroblasts. In particular, α-SMA expression and organization into stress fibers is a hallmark of the myofibroblast phenotype. Here, we examined α-SMA and F-actin organization and stress fiber formation (Figure 4). Following the 7 day mechanical prime, ~ 70% of stellate cells showed organized α-SMA stress fibers with nearly all (~ 95%) displaying organized F-actin stress fibers (Figure 4A,B). In contrast, a significantly lower fraction of stellate cells on soft hydrogels showed α-SMA or F-actin organization in stress fibers. Mechanically primed stellate cells maintained their actin organization on stiff hydrogels with over 90% of cells showing α-SMA organization after an additional 14 days of post-priming culture (Figure 4C,D). In contrast, both the soft and stiff-to-soft groups showed significantly reduced α-SMA and F-actin stress fiber organization after 14 days, reaching levels comparable to the soft static group (dashed lines, Figure 4D). Although a significantly higher fraction of the cells in stiff-to-soft group displayed organized F-actin stress fibers compared to the soft group, it is expected that this gap would close with extended culture time in the soft environment. Additionally, although previous work with lung fibroblasts showed little reduction in α-SMA expression when cells were moved from a stiff (100 kPa) to soft (5 kPa) substrate, this may be due to the extended mechanical priming in the aforementioned work (2 weeks instead of 1 week) as well as differences in the cell types, biomaterial platforms, and stiffnesses investigated.27
Figure 4. Mechanically primed hepatic stellate cells lose actin stress fiber organization following culture on stiff-to-soft hydrogels.
(A) Representative images and (B) quantification of actin organization for freshly isolated stellate cells cultured for 7 days on soft hydrogels or glass. Blue: nuclei, Green: α-SMA, Red: F-actin. (C) Representative images and (D) quantification of actin organization for stellate cells cultured for 21 days total on soft hydrogels (soft static, dashed lines) or mechanically primed stellate cells cultured for an additional 14 days on soft, stiff, or stiff-to-soft hydrogels. Blue: nuclei, Green: α-SMA, Red: F-actin. (Dashed lines: soft static control). ***: P < 0.001. Scale bars: 100 μm. n > 26 cells per group.
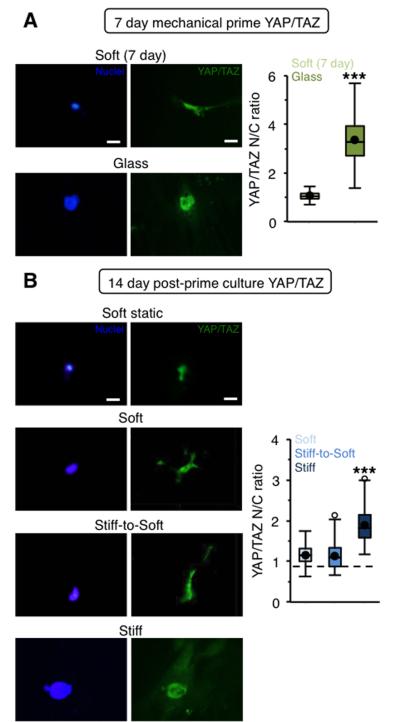
3.4 Mechanically primed hepatic stellate cells show reduced nuclear YAP/TAZ on stiff-to-soft hydrogels
We next investigated signaling pathways involved in hepatic stellate cell mechanotransduction. The YAP/TAZ mechanosensing complex has been implicated as a critical regulator of mechanotransduction through its role in relaying extracellular signals to the nucleus to initiate downstream transcription events.28 In recent years YAP/TAZ has been shown to play a critical role in diverse cellular functions including mesenchymal stem cell (MSC) differentiation28 and the induction of fibrosis.29, 30 Interestingly, YAP/TAZ has also been identified as an intracellular rheostat associated with MSC mechanical memory; MSCs that have prolonged exposure to stiff microenvironments display persistent activation of YAP/TAZ signaling as measured by nuclear localization and are biased towards osteogenic differentiation.13 As expected, stellate cells mechanically primed for 7 days showed significantly higher nuclear YAP/TAZ compared to stellate cells on soft hydrogels (Figure 5A). While activated stellate cells cultured on stiff hydrogels for an additional 14 days maintained elevated nuclear YAP/TAZ localization, the soft and stiff-to-soft groups both showed an approximately equal distribution of YAP/TAZ between the nucleus and cytoplasm, which was similar to that of cells cultured on soft hydrogels without mechanical priming (dashed line) (Figure 5B). This indicates that YAP/TAZ may not be a marker of mechanical memory of myofibroblasts for the mechanical priming time investigated.
Figure 5. Mechanically primed hepatic stellate cells show reduced nuclear YAP/TAZ on stiff-to-soft hydrogels.
(A) Representative images and quantification of YAP/TAZ staining for freshly isolated stellate cells cultured for 7 days on soft hydrogels or glass. Blue: nuclei, Green: YAP/TAZ. (B) Representative images and quantification of YAP/TAZ staining for stellate cells cultured for 21 days total on soft hydrogels (soft static, dashed line) or mechanically primed stellate cells cultured for an additional 14 days on soft, stiff, or stiff-to-soft hydrogels. Blue: nuclei, Green: YAP/TAZ. (Dashed line: soft static control). ***: P < 0.001, significantly greater than other experimental groups. Scale bars: 20 μm. n > 26 cells per group
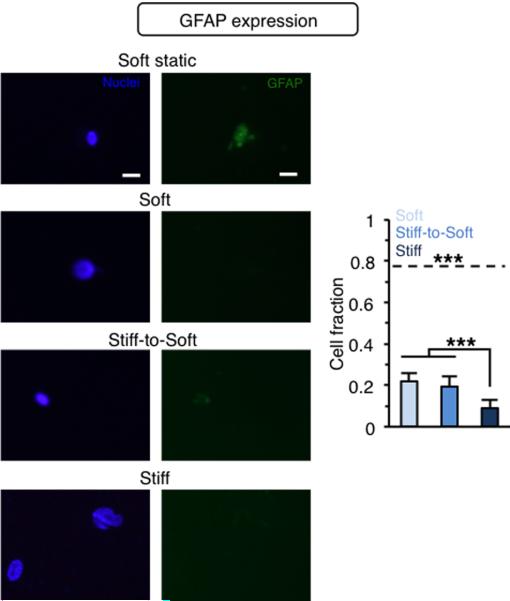
3.5 Stellate cells on stiff-to-soft hydrogels display intermediate phenotype as measured by GFAP expression
Although stellate cells did not maintain elevated levels of nuclear YAP/TAZ on stiff-to-soft hydrogels, we also assessed the expression of other markers that might indicate an intermediate phenotype between activated myofibroblasts and quiescent stellate cells. A previous study describing the adoption of an intermediate phenotype by stellate cells in vivo during fibrosis regression showed that, while cells appeared quiescent and most markers (such as α-SMA) returned to healthy levels during resolution, others, such as GFAP, did not.3 GFAP is a marker of the stellate cell population within the liver that is down-regulated during myofibroblast differentiation. However, GFAP expression remained down-regulated even after one month of resolution in the aforementioned study.3 Nearly 80% of hepatic stellate cells cultured on the soft static hydrogels for the duration of our experiments stained positive for GFAP (dashed line, Figure 6). However, a significantly lower fraction of GFAP-positive cells (< 25%) was observed for all three experimental groups that underwent mechanical priming for 7 days on TCPS before being plated on hydrogels (soft, stiff, stiff-to-soft). Critically, these results suggest that changes in substrate mechanics may play an important role in the intermediate stellate cell phenotype observed in vivo.
Figure 6. GFAP staining of stellate cells on stiff-to-soft hydrogels display intermediate phenotype.
Representative images and quantification of GFAP staining for stellate cells cultured for 21 days total on soft hydrogels (soft static, dashed line) or mechanically primed stellate cells cultured for an additional 14 days on soft, stiff, or stiff-to-soft hydrogels. (Dashed line: soft static control). Blue: nuclei, Green: GFAP. ***: P < 0.001. Scale bars: 20 μm. n > 23 cells per group.
3.6 Re-stiffening hydrogel results in rapid myofibroblast activation
Another important (and clinically relevant) characteristic of the intermediate stellate cell phenotype is the ability to rapidly undergo myofibroblast re-differentiation in response to a repeat injury.3 In order to model a repeat injury, we leveraged our hydrogel design to re-stiffen the cellular environment by photopolymerization of the unreacted methacrylates in the network16 (Figure 7A). In this scenario, stellate cells were mechanically primed for 7 days, cultured on stiff-to-soft hydrogels for an additional 14 days, and then the hydrogels were re-stiffened and the culture was extended for another 2 days. Mechanical characterization indicated that the stiff-to-soft hydrogels were re-stiffened to their original stiffness corresponding to diseased liver (Figure 7B). Stellate cells on re-stiffened hydrogels rapidly spread and re-assumed a myofibroblast phenotype within 2 days, showing significantly higher spread area, α-SMA and F-actin stress fiber organization, and nuclear YAP/TAZ compared to stellate cells on stiff-to-soft hydrogels (Figure 7C,D,E). While the stellate cell phenotype observed only 2 days after re-stiffening was similar to the phenotype observed after 7 days of mechanical priming, this behavior could be explained by the extended culture time.16 Nevertheless, these results mirrored observed in vivo trends and indicated that stellate cells on stiff-to-soft hydrogels may have the ability to more rapidly re-differentiate into myofibroblasts.
Figure 7. Re-stiffening hydrogel results in rapid myofibroblast activation.
(A) Scheme for re-stiffening of hydrogels by in situ radical polymerization of unreacted hydrogel methacrylates. (B) Elastic moduli of stiff-to-soft hydrogels and re-stiffened hydrogels before and after light exposure. (C) Stellate cell spread area before and after light exposure (day 14 stiff-to-soft cell area data reproduced from Fig. 3 for comparison). (D) Representative image and quantification of actin organization for stellate cells on re-stiffened hydrogel. Blue: nuclei, Green: α-SMA, Red: F-actin (stiff-to-soft actin organization data reproduced from Fig. 4 for comparison). (E) Representative image and quantification of YAP/TAZ nuclear localization for stellate cells on re-stiffened hydrogel. Blue: nuclei, Green: YAP/TAZ (stiff-to-soft YAP/TAZ data reproduced from Fig. 5 for comparison). ***: P < 0.001, significantly greater than stiff-to-soft group. Scale bars: 100 μm (C, D), 20 μm (E). n > 51 cells per group.
4. Conclusions
Hydrogel platforms that capture the dynamic mechanical properties of the ECM are needed to facilitate better understanding of how cells interpret microenvironmental signals in the context of development, wound repair, and disease development. In this work we developed a hydrogel platform combining stable and hydrolytically degradable crosslinks to present a gradually softening cellular microenvironment. We showed that the hydrogel degradation kinetics followed pseudo first order exponential decay kinetics due to ester hydrolysis. We then applied this system in proof-of-concept studies to investigate hepatic stellate cell phenotype in the context of fibrosis resolution. Activated stellate cells cultured on stiff-to-soft hydrogels showed reductions in spreading, nuclear YAP/TAZ, and actin organization after 14 days, displaying a phenotype that appeared similar to stellate cells on soft hydrogels. However, these cells also displayed characteristics of an intermediate phenotype observed in vivo following fibrosis resolution with reduced expression of GFAP and rapid re-activation in response to re-stiffening (re-injuring) the microenvironment. Together, these data suggest that mechanical properties may be important in mediating phenotypic changes observed during fibrosis progression and resolution in vivo. Additionally, these results underscore the importance of presenting a dynamic mechanical environment to cells during in vitro studies to more faithfully capture observed in vivo phenomena. We envision that dynamic hydrogel platforms such as the one described here will also be useful for studies of stem cell differentiation, mechanical memory and epigenetics, and fibrosis in other organs.
Supplementary Material
Insight, innovation, integration.
There is a tremendous need to engineer hydrogel systems that recapitulate the dynamic mechanical nature of physiological and pathological events. This work presents a gradually softening and re-stiffening hydrogel system to capture changes in microenvironmental mechanics during fibrosis resolution and repeat injury. We showed that hepatic stellate cells, the major fibrogenic cells in liver fibrosis, responded to gradual softening by re-organizing their actin cytoskeleton to reduce spreading and YAP/TAZ nuclear localization. Stellate cells cultured on softening hydrogels also display a pathologically-relevant intermediate phenotype as seen in vivo. Our results underscore the importance of presenting a dynamic mechanical environment to cells during in vitro studies to more faithfully capture in vivo phenomena.
Acknowledgements
The authors would like to acknowledge Shannon Tsai for isolation of rat stellate cells and Dr. Chris Highley, Dr. Adrianne Rosales, and Chris Rodell for assistance with NMR and for helpful discussions. The David and Lucile Packard Foundation (JAB) and the National Institutes of Health (F32 DK103463, SRC; DK058123, RGW) supported this work.
References
- 1.Wynn TA. J Pathol. 2008;214:199–210. doi: 10.1002/path.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.W. H. Organization Cause specific mortality: regional estimates 2000-2011. 2014.
- 3.Kisseleva T, Cong M, Paik Y, Scholten D, Jiang C, Benner C, Iwaisako K, Moore-Morris T, Scott B, Tsukamoto H, Evans SM, Dillmann W, Glass CK, Brenner DA. Proc Natl Acad Sci U S A. 2012;109:9448–9453. doi: 10.1073/pnas.1201840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Troeger JS, Mederacke I, Gwak GY, Dapito DH, Mu X, Hsu CC, Pradere JP, Friedman RA, Schwabe RF. Gastroenterology. 2012;143:1073–1083. e1022. doi: 10.1053/j.gastro.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, Yamauchi M, Gasser DL, Weaver VM. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georges PC, Hui JJ, Gombos Z, McCormick ME, Wang AY, Uemura M, Mick R, Janmey PA, Furth EE, Wells RG. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1147–1154. doi: 10.1152/ajpgi.00032.2007. [DOI] [PubMed] [Google Scholar]
- 7.Liu F, Mih JD, Shea BS, Kho AT, Sharif AS, Tager AM, Tschumperlin DJ. J Cell Biol. 2010;190:693–706. doi: 10.1083/jcb.201004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Previtera M, Trout K, Verma D, Chippada U, Schloss R, Langrana N. Ann Biomed Eng. 2012;40:1061–1072. doi: 10.1007/s10439-011-0483-2. [DOI] [PubMed] [Google Scholar]
- 9.Zustiak SP, Leach JB. Biomacromolecules. 2010;11:1348–1357. doi: 10.1021/bm100137q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zustiak SP, Leach JB. Biotechnol Bioeng. 2011;108:197–206. doi: 10.1002/bit.22911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Tibbitt MW, Langer SJ, Leinwand LA, Anseth KS. Proceedings of the National Academy of Sciences. 2013;110:19336–19341. doi: 10.1073/pnas.1306369110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang C, Tibbitt MW, Basta L, Anseth KS. Nat Mater. 2014;13:645–652. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Highley CB, Prestwich GD, Burdick JA. Current Opinion in Biotechnology. 2016;40:35–40. doi: 10.1016/j.copbio.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Guvendiren M, Perepelyuk M, Wells RG, Burdick JA. Journal of the Mechanical Behavior of Biomedical Materials. 2014;38:198–208. doi: 10.1016/j.jmbbm.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caliari SR, Perepelyuk M, Cosgrove BD, Tsai SJ, Lee GY, Mauck RL, Wells RG, Burdick JA. Scientific Reports. 2016;6:21387. doi: 10.1038/srep21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burdick JA, Chung C, Jia X, Randolph MA, Langer R. Biomacromolecules. 2005;6:386–391. doi: 10.1021/bm049508a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guvendiren M, Yang S, Burdick JA. Adv Funct Mater. 2009;19:3038–3045. [Google Scholar]
- 19.Burdick JA, Chung C, Jia X, Randolph MA, Langer R. Biomacromolecules. 2005;6:386–391. doi: 10.1021/bm049508a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bitter T, Muir HM. Analytical Biochemistry. 1962;4:330–334. doi: 10.1016/0003-2697(62)90095-7. [DOI] [PubMed] [Google Scholar]
- 21.Uemura M, Swenson ES, Gaca MD, Giordano FJ, Reiss M, Wells RG. Mol Biol Cell. 2005;16:4214–4224. doi: 10.1091/mbc.E05-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geerts A, Vrijsen R, Rauterberg J, Burt A, Schellinck P, Wisse E. Journal of Hepatology. 1989;9:59–68. doi: 10.1016/0168-8278(89)90076-7. [DOI] [PubMed] [Google Scholar]
- 23.Majima T, Schnabel W, Weber W. Die Makromolekulare Chemie. 1991;192:2307–2315. [Google Scholar]
- 24.Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Biomaterials. 2009;30:6702–6707. doi: 10.1016/j.biomaterials.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen AL, Bloomer SA, Chan EP, Gaca MD, Georges PC, Sackey B, Uemura M, Janmey PA, Wells RG. Am J Physiol Gastrointest Liver Physiol. 2011;301:G110–118. doi: 10.1152/ajpgi.00412.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saums MK, Wang W, Han B, Madhavan L, Han L, Lee D, Wells RG. Langmuir. 2014;30:5481–5487. doi: 10.1021/la4047758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balestrini JL, Chaudhry S, Sarrazy V, Koehler A, Hinz B. Integr Biol (Camb) 2012;4:410–421. doi: 10.1039/c2ib00149g. [DOI] [PubMed] [Google Scholar]
- 28.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 29.Mannaerts I, Leite SB, Verhulst S, Claerhout S, Eysackers N, Thoen LFR, Hoorens A, Reynaert H, Halder G, van Grunsven LA. J Hepatol. 63:679–688. doi: 10.1016/j.jhep.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 30.Liu F, Lagares D, Choi KM, Stopfer L, Marinković A, Vrbanac V, Probst CK, Hiemer SE, Sisson TH, Horowitz JC, Rosas IO, Fredenburgh LE, Feghali-Bostwick C, Varelas X, Tager AM, Tschumperlin DJ. Am J Physiol Lung Cell Mol Physiol. 2015;308:L344–L357. doi: 10.1152/ajplung.00300.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.