Abstract
Telomeres are nucleoprotein complexes that reside at the ends of linear chromosomes and help maintain genomic integrity. POT1 and TPP1 are telomere-specific proteins that bind as a heterodimer to single-stranded telomere DNA to prevent illicit DNA damage responses and to enhance telomerase-mediated telomere extension. Telomere DNA is guanosine-rich and, as such, can form highly stable secondary structures including G-quadruplexes. G-quadruplex DNA folds into different topologies that are determined by several factors including monovalent ion composition and the precise sequence and length of the DNA. Here, we explore the influence of DNA secondary structure on POT1-TPP1 binding. Equilibrium binding assays reveal that the POT1-TPP1 complex binds G-quadruplex structures formed in buffers containing Na+ with an affinity that is 5-fold higher than for G-quadruplex structures formed in the presence of K+. However, binding of a second heterodimer is insensitive to DNA secondary structure, presumably due to unfolding resulting from binding of the first POT1-TPP1. We further show that the rate constant for POT1-TPP1-induced unfolding of DNA secondary structure is substantially faster for G-quadruplex topologies formed in the presence of Na+ ions. When bound to DNA, POT1-TPP1 forms complexes with similar CD spectra and enhances telomerase activity for all DNA substrates tested, regardless of substrate secondary structure or solution monovalent ion composition. Together, these data indicate that binding of POT1-TPP1 unfolds telomere secondary structure to assist loading of additional heterodimers and to ensure efficient promotion of telomerase-mediated extension.
Graphical abstract
Introduction
Telomeres are nucleoprotein complexes that reside at the ends of linear chromosomes and are comprised of repeating G-rich DNA sequences 1; 2; 3. In mammals, telomere DNA is a repetitive, hexameric sequence of TTAGGG that extends for thousands of bases before ending in shorter single-stranded DNA (ssDNA) overhangs 1; 2; 4. The G-rich telomere DNA is capable of forming stable, secondary structures that include G-quadruplexes. G-quadruplex structures are composed of stacks of G-tetrads, each of which contain four guanines that are organized in a planar arrangement stabilized by a cyclic Hoogsteen hydrogen bonding network 5; 6; 7.
Analysis of G-quadruplexes assembled in vitro has revealed that the central cavities formed within the structures are occupied by monovalent cations, which neutralize the electrostatic repulsion generated by the inwardly pointing keto oxygens of the guanine bases 8. The solvent monovalent ion type is well known to influence G-quadruplex stability and folding topology (reviewed in 9). Similarly, the nucleotide (nt) sequence and length adjacent to G-quadruplex structures formed within telomere DNA sequence can also contribute to structural polymorphism (see 8 and reference therein). In all of these cases, the G-tetrad interaction is maintained, but the orientation of the intercalating nucleotides form strands that can be oriented in parallel, anti-parallel, or hybrid-type (containing both parallel and antiparallel strands) configurations. Importantly, these different G-quadruplex structures can also display functional diversity with respect to ligand binding and specificity 10; 11. In addition to a wealth of data characterizing G-quadruplex structure, stability, and specificity in vitro (see 12), the existence of G-quadruplexes has been confirmed in the telomeres of human cells, within the macronuclei of ciliates, and in Xenopus laevis egg extract 13; 14; 15; 16; 17. However, the mechanisms which proteins and enzymes associate with telomere DNA and the ways they alter its ability to form stable alternative structures are not well understood.
Telomere DNA is synthesized by a unique ribonucleoprotein complex called telomerase, an enzyme that compensates for the inability of replicative polymerases to fully extend the ends of linear chromosomes 18; 19. The ssDNA overhang of telomeres provides the substrate for telomerase, therefore DNA secondary structures that form within this region provide obstacles to telomerase-mediated replication. For example, G-quadruplex structures may function to regulate telomerase activity, primarily by inhibiting telomerase extension 7; 20; 21; 22; 23.
A set of six specialized proteins, collectively termed shelterin, associate with telomere DNA 24; 25; 26; 27. Along with telomerase, the shelterin complex contributes to maintaining the proper structure, function, and overall integrity of telomeres 27; 28; 29. Protection of Telomeres 1 (POT1) protein, along with its binding partner TPP1, binds single-stranded telomere DNA with high affinity. DNA interactions are orchestrated through the N-terminus of POT1, while its C-terminal domain is necessary for TPP1-interaction 30; 31; 32; 33. TPP1 is responsible for recruiting telomerase to the telomere and, together with POT1 enhances telomerase processivity by decreasing the rate constant for dissociation from telomere DNA and increasing the rate constant for telomerase extension activity 34; 35; 36; 37; 38; 39; 40; 41.
The x-ray crystal structure of the POT1 DNA-binding domain in complex with ssDNA reveals that the protein interacts with ten nts of telomere DNA in an extended single-strand conformation 42. The POT1 domain is divided into two oligosaccharide-oligonucleotide binding (OB) folds with the N-terminal domain recognizing a full hexamer of telomere DNA (TTAGGG), while the C-terminal OB-fold binds to the adjacent four nts (TTAG). On longer ssDNA substrates, this recognition sequence is maintained as multiple POT1-TPP1 proteins coat the ssDNA substrate with a protein binding to every two hexamer repeats 43. Of the twelve nts representing two complete hexameric telomere sequences, the last two guanosines are not necessary for POT1-TPP1 binding and recognition 33; 44. These two guanosines do, however, contribute dramatically to the secondary structure of unbound DNA, as they are necessary for G-quadruplex formation.
Although physiological tracts of single-stranded (ss) telomere DNA are ∼50-200 nts in the cell 45, most in vitro studies of DNA-binding and telomerase recruitment by POT1-TPP1 protein to date have been limited to short single-stranded oligonucleotide substrates (<18 nts) for understanding DNA-binding and telomerase recruitment properties of POT1-TPP1 protein 34; 35. As telomere ssDNA needs at least 22-nts to form intramolecular G-quadruplex structures (see 46), less is known regarding the influence of telomere DNA secondary structure on POT1-TPP1 binding events and on telomerase recruitment. For example, recent data demonstrates that POT1-TPP1 induces continuous unfolding of G-quadruplex structures in telomere DNA 47, yet the interaction between protein and DNA is largely dependent on the G-quadruplex topology that is formed 10.
In this report, we interrogate the impact that DNA secondary structure has on multiple POT1-TPP1 binding events. Our data reveal that secondary structure, including G-quadruplexes, impairs binding of POT1-TPP1 to telomere DNA. Furthermore, we demonstrate that POT1-TPP1 binds with variable affinities to polymorphic G-quadruplex structures formed in Na+ versus K+. After the initial loading of a single POT1-TPP1 heterodimer, however, binding of subsequent proteins occurs with similar affinity for all 24-nt substrates investigated, regardless of the secondary structure of the unbound DNA. Consistent with the differential sensitivity to G-quadruplex topology observed for equilibrium binding affinity, time resolved circular dichroism spectroscopy demonstrates that POT1-TPP1 unfolds G-quadruplexes formed in Na+ significantly faster than G-quadruplex structures formed in K+. Finally, we demonstrate that telomerase activity is impaired equally by different G-quadruplex topologies, regardless of monovalent ion composition, and the inclusion of POT1-TPP1 circumvents this obstacle. Together, our results reveal intrinsic biochemical features of molecular recognition that govern the interaction of POT1-TPP1 with ssDNA. This information is important for understanding the in vivo function of POT1-TPP1 as well as providing insights into its central role in telomere maintenance.
Results
An intrinsic feature of single-stranded, G-rich telomere DNA is the ability to form highly stable G-quadruplex structures. The ssDNA-binding heterodimer POT1-TPP1 must necessarily disrupt this stable structure and subsequently interact with telomerase in order to orchestrate proper telomere synthesis. To understand this essential biological function it is important to establish how POT1-TPP1 associates with different intramolecular topologies that telomere DNA can form. To accomplish this goal we quantitatively analyzed the binding of a series of model DNA ligands designed to test the effect of DNA structure on POT1-TPP1 association and its ability to activate telomerase.
The G-quadruplex structures favored by free telomere DNA might enhance POT1-TPP1 recognition allowing it to outcompete other nucleic acid binding proteins (e.g. replication protein A) that are more abundant in the cell. Alternatively, formation of strong non-covalent interactions between POT1-TPP1 and telomere DNA may offset the cost of disrupting stable G-quadruplex structures in the POT1-TPP1-ssDNA complex. Such conformational changes could occur after initial protein binding or by kinetic trapping of transiently unfolded telomere DNA.
Design and characterization of telomere sequences with altered secondary structure
We first designed several substrates that would allow us to determine how secondary structure in telomere DNA might affect POT1-TPP1 recognition and binding (Supplemental Figure 1). The hT24wt DNA is comprised of four telomere repeats and is predicted to form G-quadruplex structures, albeit with different topologies in Na+-versus K+-based solutions 46; 48; 49 (Supplemental Figure 1A). As described above, the guanosine dinucleotides flanking each POT1-TPP1 recognition sequence are not involved in protein binding, but are necessary for G-quadruplex formation 44. As such, these two nts were mutated, which allows the secondary structure of the ssDNA to be manipulated without significantly affecting the stability of the bound DNA-protein complex.
The hT24GG-CC DNA has the two guanosines located outside of the decameric POT1-TPP1 recognition sequence mutated to cytosines. Mutation of the dinucleotide Gs to Cs introduces Watson-Crick base pairing within the ssDNA substrate, thus providing a propensity for it to form intermolecular duplex structures (Supplemental Figure 1B; predicted ΔG of -6.73 kcal/mol 50; 51). Additional substrates were designed in which the two guanosines separating adjacent POT1-recognition sequences were mutated to either inosines or uridines. Inosine lacks the 2-amino group that is a primary contributor to G-quartet, hydrogen-bonding interactions 52. Similarly, substitution of guanosines with uridines will entirely disrupt hydrogen-bonding networks that are essential for G-quadruplex secondary structure (Supplemental Figure 1C).
The structures of all four designed DNA constructs were probed using multiple techniques. Non-denaturing polyacrylamide gel electrophoresis is an effective tool for monitoring the secondary structure of DNA molecules that have similar mass 53; 54. Although some minor bands appear in the non-denaturing gel (e.g. hT24GG-II), the majority of the population (>95%) migrates as a single band that is indicative of the expected shapes for the four DNA substrates investigated (Figure 1A). Under identical conditions, hT24wt and hT24GG-CC migrated faster, due to stable secondary structure formation, than the unfolded hT24GG-II and hT24GG-UU DNAs. Size-exclusion HPLC (SE-HPLC) has also proven to be a powerful tool for assessing the size and shape of oligonucleotides that exhibit structural polymorphism 55. Using this method, we further analyzed the distributions of secondary structure the four 24-nt substrates used in our study. Under identical conditions in either NaCl or KCl buffer, the hT24wt DNA consistently elutes slower that any of the other three substrates (Figure 1B; Supplemental Figure 2). The hT24GG-CC DNA elutes at a lower volume than hT24wt, but higher than the hT24GG-II and hT24GG-UU substrates. The faster mobility of hT24GG-CC indicates a less compact structure relative to hT24wt. The hT24GG-II and hT24GG-UU DNAs elute at the same volume consistent with extended single stranded conformations. Thus, these data support the interpretation that hT24wt forms G-quadruplex structures, hT24GG-CC contains duplex DNA, and hT24GG-II and hT24GGUU have little or no secondary structure.
Figure 1.
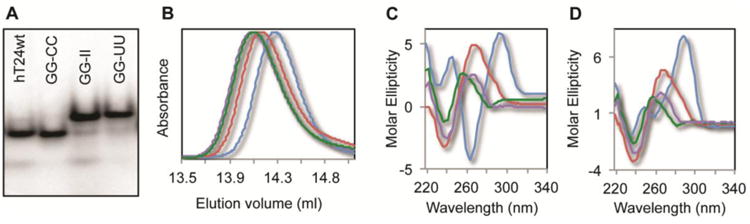
Secondary structure analysis of DNA substrates. (A) Nondenaturing gel electrophoresis reveals different mobility of the various DNA sequences. hT24wt and hT24GG-CC migrate faster, which is indicative of more compact folded structures of these DNA substrates. Conversely, hT24GG-II and hT24GG-UU, which have no internal secondary structure, migrate slower in the gel. (B) Normalized chromatograms of the 24-nt DNA substrates investigated using SE-HPLC in NaCl buffer. hT24wt (blue line) elutes slowest, followed by hT24GG-CC (red line), and hT24GG-II (green line) and hT24GG-UU (purple line) eluting first. Size-exclusion chromatography separates DNA substrates based on shape, with more compact structures eluting later than those that are less compact. CD spectra of hT24wt, hT24GG-CC, hT24GG-II, and hT24GG-UU in (C) Na+ or (D) K+ containing buffer. hT24wt shows a spectrum consistent with antiparallel topology in Na+ ions and hybrid parallel/antiparallel topology in K+ ions. Spectra for hT24GG-CC, hT24GG-II, and hT24GG-UU are similar in Na+ and K+ ions, indicating that their structures are not significantly altered in different monovalent ion solutions. Colors of lines are the same as those in panel (B).
While the native gel and SE-HPLC data supports the notion that the hT24wt DNA forms G-quadruplex structures in both NaCl and KCl, both techniques are limited in assessing structural polymorphism in G-quadruplex topologies. Therefore, the secondary structure of all four DNA substrates was further evaluated using circular dichroism spectroscopy (CD) (Figure 1C and 1D). The identity of the monovalent ion in solution (typically K+ or Na+) is a primary contributing factor to G-quadruplex topology. Each population exhibits a distinct signature in its circular dichroism (CD) spectrum. For example, a G-quadruplex topology that is comprised of antiparallel strands and guanines arranged in alternating syn- and anti- conformations are characterized by a positive band at 295 nm, a smaller positive band at 245 nm, and a negative band at 265 nm in the CD spectra 56. This topology is common for short (22-26 nt) oligonucleotides comprised of telomere DNA sequence in Na+-containing solutions 48. Conversely, G-quadruplex structures formed by short (22-24 nt) oligonucleotides containing telomere DNA sequence tend to fold into multiple stable conformations (i.e. hybrid-type) in K+-containing solutions 48. The hybrid-type, G-quadruplex structures represent a mixture of parallel and antiparallel strands that are characterized by a major band at 290 nm with a shoulder near 268 nm, a weaker band at 250 nm, and a weak negative band near 260 nm in the CD spectrum 48.
In solution containing Na+ the hT24wt DNA sequence revealed a CD spectrum that is consistent with anti-parallel G-quadruplex topology (Figure 1C) and mixed, hybrid-type G-quadruplex structures in the presence of K+ (Figure 1D). In contrast, the CD spectra of the three mutant substrates are indicative of B-form (duplex or single-stranded) DNA, which is characterized by a broad band from 260-280 nm and a negative band near 240 nm 57. CD spectra for hT24GG-CC, hT24GG-II, and hT24GG-UU are all similar in both Na+- and K+-containing solutions indicating that, unlike the hT24wt ssDNA, the secondary structure of these DNA substrates is not sensitive to the identity of solution monovalent ions. Together, our SE-HPLC and CD data are consistent with the hT24wt DNA forming G-quadruplex structures with different topologies in NaCl or KCl buffer, the hT24GG-CC DNA forming a hairpin, and hT24GG-II and hT24GG-UU DNA containing little to no secondary structure as depicted in Supplemental Figure 1.
DNA secondary structure impairs the initial POT1-TPP1 binding event to 24-nt substrates
Multiple protein binding events on a single substrate can be independently separated and quantified using EMSA 43; 44; 58. When performed under equilibrium binding conditions, Kd values can be determined for each bound state. As such, EMSA was employed to examine the sequential binding reactions of multiple POT1-TPP1 proteins associating with ssDNA substrates exhibiting different secondary structures. In NaCl, POT1-TPP1 binds to G-quadruplex hT24wt DNA with two distinct populations (Figure 2A). The relative abundance of each protein-DNA complex population was quantified and plotted as a function of increasing POT1-TPP1 concentrations. The data were fit to a simple sequential, two-site equilibrium binding model and the equilibrium dissociation constants (K1 and K2) were determined by globally fitting the concentration dependent changes in the populations of the free DNA and the complexes containing either one or two POT1-TPP1 proteins (Figure 2B). EMSA assays and data fitting were also performed for hT24GG-CC, hT24GG-II, and hT24GG-UU substrates (Supplemental Figures 3-6).
Figure 2.
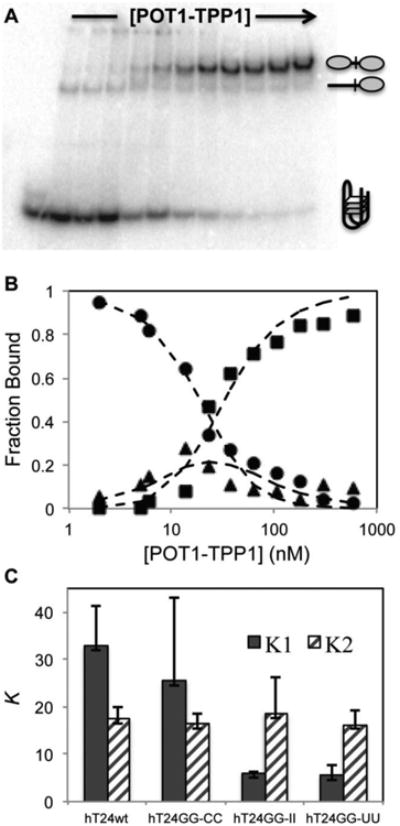
The binding of POT1-TPP1 to adjacent sites on DNA substrates in NaCl containing buffer. (A) EMSAs were performed under equilibrium binding conditions to determine the effects of DNA secondary structure on initial and subsequent binding of POT1-TPP1 proteins. The gel shown here reveals the binding profile for POT1-TPP1 interactions with hT24wt DNA. (B) The bands in the EMSA were quantified by autoradiography and plotted. The fraction of total protein was plotted for zero (●), one (▲), or two (■POT1-TPP1 binding stoichiometry as a function of increasing protein concentration. Dashed lines are the least-square global fit to a sequential binding mechanism (Scheme 1) used to estimate the equilibrium dissociation constants K1 and K2. Three independent binding experiments were analyzed to calculate average values and standard deviations for each DNA substrate (Table 1). (C) Summary of the EMSA fitting data. The average K1 (filled columns) and K2 (hashed colums) values (in nM) and their standard deviations are shown for each DNA substrate.
Comparison of values of the equilibrium dissociation constants K1 and K2 for each substrate revealed an overall trend in which stable secondary structure in the unbound DNA correlates with lower affinity (higher K1) for initial POT1-TPP1 binding, but does not affect the affinity of the second binding event (i.e. the magnitude of K2; Figure 2C). In NaCl buffer, the hT24wt and hT24GG-CC DNAs display an average K1 value that is about five-fold higher (meaning 5-fold lower affinity) than that of the unstructured hT24GG-II and hT24GG-UU DNAs (Figure 2C and Table 1). In contrast, the K2 value was similar for POT1-TPP1 binding to all four DNA substrates.
Table 1.
Equilibrium dissociation constants for POT1-TPP1 binding to DNAs exhibiting different secondary structure.
| Substrate | K1 (nM); NaCl | K2 (nM); NaCl | K1 (nM); KCl | K2 (nM); KCl |
|---|---|---|---|---|
| hT24wt | 33.0 ± 8.4 | 17.5 ± 2.6 | 129.8 ± 30.5 | 25.2 ± 5.0 |
| hT24GG-CC | 25.5 ± 17.5 | 16.3 ± 2.3 | 42.1 ± 9.6 | 22.4 ± 3.3 |
| hT24GG-II | 6.0 ± 0.4 | 18.6 ± 7.7 | 4.3 ± 1.0 | 9.5 ± 3.0 |
| hT24GG-UU | 5.5 ± 2.2 | 16.3 ± 3.1 | 7.1 ± 2.5 | 9.2 ± 2.2 |
To test whether POT1-TPP1 binding is sensitive to the type and/or stability of G-quadruplex structure formed in the free DNA, we repeated the gel shift experiments using protein and DNA complexes assembled in KCl buffer as opposed to NaCl buffer. Gel shifts were quantified and plotted and K1 and K2 values calculated as described above (Supplemental Figures 3-6). The results demonstrate that POT1-TPP1 binds hT24GGCC, hT24GG-II, and ht24GG-UU DNA with similar affinities independent of the identity of the solution monovalent ions. In contrast, G-quadruplex structures formed by hT24wt DNA in K+ impair the initial POT1-TPP1 binding significantly when compared to those G-quadruplex structures formed in Na+ (Figure 3). There was no difference in the apparent affinity of subsequent protein binding events, as the second POT1-TPP1 binds to all hT24wt, hT24GG-CC, hT24GG-II, and hT24GG-UU in NaCl and KCl with similar K2 values (Figure 3; Table 1).
Figure 3.
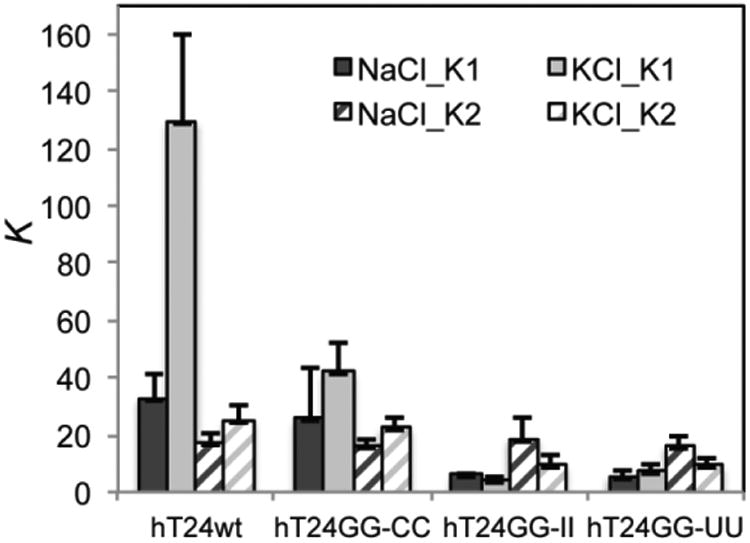
Different G-quadruplex structures alter the equilibrium binding affinity of POT1-TPP1. Sequential POT1-TPP1 protein binding reactions were evaluated by EMSA (Supplemental Figures 3-6) as described in Figure 2. Binding of POT1-TPP1 protein was evaluated with four DNA substrates in either 150 mM NaCl or 150 mM KCl. The affinity of the first protein binding event to G-quadruplex structures formed by hT24wt DNA in KCl is significantly hindered compared to all other DNA substrates analyzed, including G-quadruplex structures formed by hT24wt in NaCl. In contrast, binding affinities of subsequent protein binding events was similar for all DNA substrates in NaCl or KCl. All data represent results obtained from three independent experiments.
POT1-TPP1 binding alters the secondary structure of the bound DNA
The high resolution x-ray crystal model of human POT1 bound to a 10-nt strand of telomere DNA indicates that the nucleic acid is bound in an extended conformation that is incompatible with G-quadruplex topology 42. Other studies using longer DNA revealed that POT1 protein disrupts G-quadruplex structure, making the DNA template more accessible for telomerase-mediated extension 20; 59 or, as a POT1-TPP1 complex, to prevent binding of replication protein A 10. To better understand how POT1-TPP1 influences the structure of the bound telomere DNA, we compared the CD spectra of the free telomere model DNAs and protein to that of the bound DNA-protein complex.
Using a split-cell cuvette for CD analysis we tested how POT1-TPP1 interactions altered the secondary structure of both the hT24wt and the hT24GG-II DNA substrates and in Na+ or K+ containing solutions. First, a CD spectrum with POT1-TPP1 protein in one compartment and DNA in the other allowed the measurement of the sum of the independent and unmixed spectral contributions of free protein and DNA (Figure 4A). The combined spectrum from the split cell revealed a dominant, negative band near 240 nm that could be attributed to the POT1-TPP1 protein. At higher wavelengths in the split cell configuration, the combined spectrum of free POT1-TPP1 and hT24wt DNA maintain signatures that are characteristic of DNA alone. Specifically, in Na+ based solutions, the spectrum retains a negative band at 265 nm and a positive band at 295 nm, whereas the K+-containing solutions remain associated with a positive peak at 290 nm connected to a shoulder and another positive value near 255 nm (Figure 4A). In the split cell, the hT24GG-II substrate retains a maximum positive peak at 260 nm in either Na+ or K+ solutions that is indicative of its unstructured, B-form topology (Figure 4A).
Figure 4.
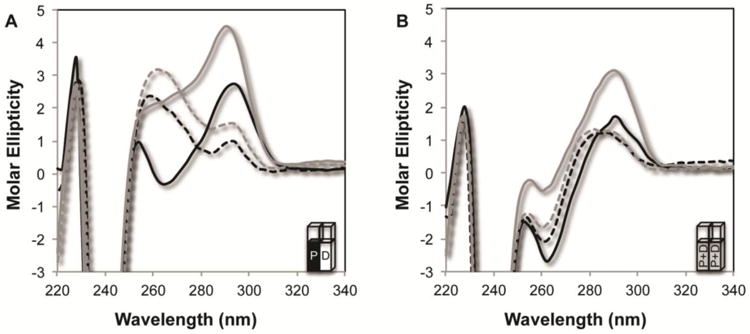
Combined CD spectra of free POT1-TPP1 with free DNA and POT1-TPP1-DNA complexes. (A) CD spectra obtained using a split cell containing POT1-TPP1 in one chamber and either hT24wt or hT24GG-II DNA in the second chamber. The combined spectra contain the CD signatures of both free protein and free DNA. Spectra were taken with 10 μM POT1-TPP1 in one chamber and the second chamber containing 2 μM of either hT24wt in 150 mM NaCl (solid black line), hT24wt in 150 mM KCl (solid grey line), hT24GG-II DNA in 150 mM NaCl (dashed black line), or hT24GG-II in 150 mM KCl (dashed grey line). (B) To measure the changes in CD spectra due to protein-DNA complex formation, solutions from the two chambers in the split cell were mixed together and incubated for 20 minutes before the CD spectra of the resulting DNA-protein complexes were measured. Sample labels are the same as in panel (A).
Having established the independent additive contributions of free DNA and protein to the observed CD spectrum, the two solutions from both sides of the split cell were mixed so that both cells contained identical mixtures of POT1-TPP1-DNA complexes. The CD spectra of the resulting protein-DNA complexes are similar for both hT24wt- and hT24GG-II-DNA complexes with positive peaks near 255 and 290 nm and a negative peak near 260 nm (Figure 4B). While the intensity of the peaks varied among the substrates and solutions analyzed, this signature was observed for all DNA-protein complexes in Na+ and K+ containing solutions. Furthermore, the CD spectra of the protein-DNA complexes is unique compared to the additive contributions of free DNA and protein observed spectrum when they are in separate cells. The similar CD profile for all protein-DNA complexes suggests that all the nucleoprotein complexes analyzed adopt similar structures. Higher resolution structural information on the apparently similar nucleoprotein complexes will require alternative methods that are more powerful than CD spectroscopy. Nonetheless, our data reveal a clear alteration in both hT24wt and hT24GG-II DNA secondary structure, upon POT1-TPP1 binding, in both Na+ and K+ solutions.
POT1-TPP1 unfolds different G-quadruplex topologies with different rate constants
Knowing that POT1-TPP1 alters the secondary structure of G-quadruplex and disordered telomere DNA at equilibrium, we next analyzed the kinetics of the changes that occur upon mixing DNA and protein. To do so, we measured the CD spectra of the POT1-TPP1-DNA solution over a period of twenty minutes, with measurements taken in two-minute intervals. The concentration of protein was in excess of DNA to insure complete loading of two POT1-TPP1 heterodimers, and both the protein and DNA concentrations were in excess of K1 and K2 to drive the binding reaction to completion. The results for the disordered, hT24GG-II telomere DNA were the same in either NaCl or KCl (Figure 5A and 5B). All structural changes were observed within the first two-minutes with no significant changes in the CD spectra occurring after that, demonstrating that the reaction had approached equilibrium. Similarly, the hT24wt DNA forming G-quadruplex structures in NaCl demonstrated a significant change in the CD spectrum within the first two minutes of mixing POT1-TPP1 protein with DNA, followed by only very minor changes in the CD spectra taken between two and twenty minutes (Figure 5C). In contrast, the hT24wt that forms G-quadruplex structures in KCl displayed a large change in the spectrum recorded in the first two minutes of mixing, followed by additional and continual changes in the spectra recorded throughout the duration of the experiment (Figure 6D). These data suggest that POT1-TPP1 binds to disordered telomere DNA and to G-quadruplex structures formed in NaCl relatively quickly to induce conformational changes completely within the first two minutes. Conversely, POT1-TPP1 binding to G-quadruplex structures formed in KCl induces conformational changes throughout the course of the twenty-minute experiment.
Figure 5.
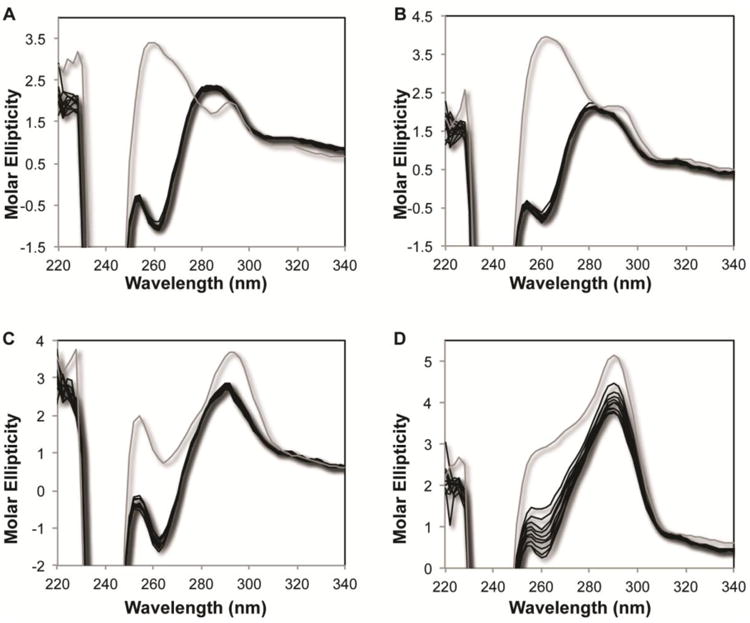
Time-dependent changes in the CD spectra of POT1-TPP1-DNA complexes. Time points of CD spectra of hT24GG-II DNA mixed with POT1-TPP1 in (A) NaCl or (B) KCl containing buffer and for hT24wt DNA mixed with POT1-TPP1 in (C) NaCl or (D) KCl containing buffer. In all panels, the grey line represents time=0 with CD spectra recorded for POT1-TPP1 protein and DNA in separate cells of the cuvette. After mixing the protein with the DNA in both chambers of the split-cell cuvette, subsequent CD spectra (shown in black lines) were recorded every two minutes for twenty minutes total.
Figure 6.
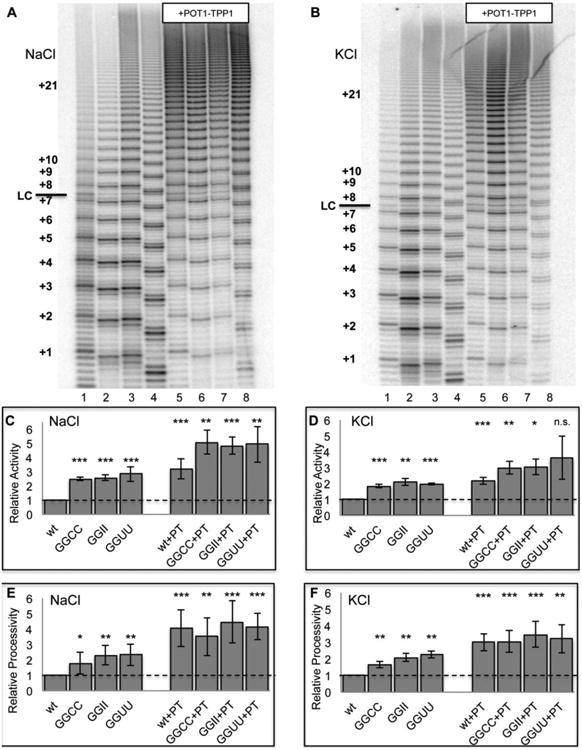
POT1-TPP1 binding overcomes telomerase inhibition caused by G-quadruplex structures. (A) Direct telomerase assay on 24-nt substrates in 150 mM NaCl. Lane 1 = hT24wt; lane 2 = hT24GG-CC; lane 3 = hT24GG-II; lane 4 = hT24GG-UU; lane 5 = hT24wt+POT1-TPP1; lane 6 = hT24GG-CC+POT1-TPP1; lane 7 = hT24GG-II+POT1-TPP1; lane 8 = hT24GG-UU+POT1-TPP1. Hexameric repeats of telomere DNA substrates are labeled on the left side. LC = loading control. (B) Quantification of total telomerase activity for samples analyzed in panel (A). (C) Quantification of telomerase repeat addition processivity for samples analyzed in panel (A). (D) Direct telomerase assay on 24-nt substrates in 150 mM KCl. Sample description and lane numbers are the same as described in panel (A). (E) Quantification of total telomerase activity for samples analyzed in panel (D). (F) Quantification of telomerase repeat addition processivity for samples analyzed in panel (D).
Fitting the data shows that the reaction in KCl is biphasic, which is consistent with several alternative models such as a sequential mechanism or parallel pathways. The rate constant for the first phase is too fast to measure and the rate constant for the second is approximately 0.07 min-1 (Supplemental Figure 7). In contrast, unfolding of the other substrates occurs with a rate constant at least ten-fold faster since the reaction is complete before the first time point. In relation to the determined equilibrium binding constants of POT1-TPP1 for the various ssDNA substrates, these data reveal that a fifteen-minute incubation was sufficient for full equilibrium for all substrates and conditions with the exception of hT24wt ssDNA in K+. Our kinetic data demonstrate that full binding of POT1-TPP1 to hT24wt in K+ approaches equilibrium after 15 minutes, and has reached >90% completion after a 20-minute incubation period. Therefore, the equilibrium binding constants for hT24wt in K+ are accurate, but likely to be slightly higher than those reported in Table 1.
POT1-TPP1 enhances telomerase activity and processivity equally for all G-quadruplex topologies
G-quadruplex structures impair telomerase activity and they are not extended effectively by telomerase 20; 41; 47. The inclusion of POT1-TPP1 rescues telomerase activity, presumably due to disruption of the G-quadruplex secondary structure. The POT1-TPP1 heterodimer also enhances telomerase processivity and extension kinetics independent of overcoming G-quadruplex inhibition, 34; 35; 36; 41; 47. We performed direct telomerase extension assays in the presence and absence of POT1-TPP1 on the 24-nt substrates with different secondary structures to differentiate between the effects of POT1-TPP1 enhancement of telomerase activity by increasing the rate of nucleotide addition versus increasing processivity (Figure 6). Experiments were performed in either NaCl or KCl buffer and results were quantified as total activity and as repeat addition processivity for each condition (Methods and Materials).
In the absence of POT1-TPP1 protein, and in reactions containing either NaCl or KCl buffer, telomerase extended a 24-nt telomere DNA substrate forming G-quadruplex structures two to three fold less efficiently than substrates that do not form G-quadruplex structures (Figures 6A and 6B). The inhibitory effect of G-quadruplex structures was observed for both relative activity (Figure 6C and 6D) and relative repeat addition processivity (Figure 6E and 6F) in reactions containing either Na+ or K+. Telomerase relative repeat addition processivity was three-fold higher for the unstructured hT24GGII and hT24GG-UU substrates when compared to the hT24wt DNA. In comparison, telomerase repeat addition processivity on the hT24GG-CC substrate was only twice as high as that of hT24wt (Figure 6E and 6F). We suspect that the inherent ability of hT24GG-CC to form a hairpin structure may slightly impair telomerase processivity when compared to the unstructured hT24GG-UU and hT24GG-II substrates.
The inclusion of POT1-TPP1 enhanced telomerase relative activity by about 3-fold for G-quadruplex-forming hT24 substrates (Figures 6C and 6D). POT1-TPP1 also enhanced overall activity for all of the substrates that do not form G-quadruplex structures, but to a lesser degree (∼1.6-fold). When POT1-TPP1 protein was included in the reactions, telomerase repeat addition processivity was nearly identical for all four 24-nt substrates investigated, regardless of buffer composition or starting secondary structure (Figures 6E and 6F). Together, these data indicate that secondary structure of the telomere DNA substrate adversely affects telomerase processivity in the absence of POT1-TPP1 protein. Conversely, the inclusion of POT1-TPP1 makes telomerase processivity equivalent for all substrates, regardless of secondary structure in the unbound DNA. Our results further demonstrate that telomerase activity and processivity are nearly identical in K+- and Na+-containing solutions, even though the G-quadruplex morphology differs in the two solutions. These data reveal that diverse G-quadruplex structures decrease telomerase activity and processivity equally, and that the inclusion of POT1-TPP1 overcomes this barrier independent of monovalent ion identity.
Discussion
Our data reveal the effects of DNA secondary structure on initial POT1-TPP1 protein binding, multimerization, and telomerase activation. In each of the 24-nt ssDNA substrates examined under equilibrium binding conditions, only the first protein binding event was sensitive to DNA secondary structure, with POT1-TPP1 demonstrating the highest affinity for unstructured ssDNA, followed by DNA forming a hairpin loop, and finally G-quadruplex structures are bound with the lowest affinity. Conversely, the binding affinity for the second POT1-TPP1 binding event was similar for all 24-nt substrates examined. CD spectroscopy supports unfolding of structured DNA during POT1-TPP1 binding. For the G-quadruplex structures formed in KCl, however, our CD experiments reveal that the G-quadruplex structures continue to slowly undergo conformational changes at POT1-TPP1 protein concentrations that result in relatively rapid unfolding in the presence of Na+. Together, these results indicate that there are significant general effects as well as differential mechanistic features for POT1-TPP1 binding to telomere DNA exhibiting different secondary structures.
Mechanistically, POT1-TPP1 interacts with telomere DNA using two individual OB-fold domains that form the POT1 DNA-binding motif 42. Although changes in telomere DNA sequence can alter the order of events, protein binding to telomere DNA is orchestrated by the second OB-fold of POT1 interacting with DNA first, followed by interactions between DNA and the first OB-fold of POT1 protein 47. In the context of unstructured ssDNA, our data indicate that binding of two POT1-TPP1 proteins bind the two recognition sites independently and non-cooperatively (similar K1 and K2). For polymorphic G-quadruplex structures, however, it is likely that the second OB-fold of POT1 binds first to one of the TTA loops and this interaction weakens the G-quadruplex hydrogen bonding network, allowing for partial unfolding that is weakened further by binding of the second POT1 OB-fold. Binding of both OB-folds would eventually result in the telomere DNA occupying a fully extended state as is seen in the crystal structure with a 10-nt DNA substrate that is too short to form G-quadruplex structures 42 (Figure 7). Alternatively, POT1-TPP1 binding may simply trap the unfolded DNA conformation. In this model unfolding of the G-quadruplex occurs in a pre-equilibrium step that is followed by POT1-TPP1 binding which prevents refolding. These slower kinetics of POT1-TPP1 unfolding of G-quadruplex structures formed in K+ correlates well with the greater stability and slower unfolding of these structures in the absence of protein binding 60. Importantly, the data presented here provide an essential framework for determining the identity of intermediates in this process and quantifying the rate constants for their interconversion using rapid kinetic and biophysical methods.
Figure 7.
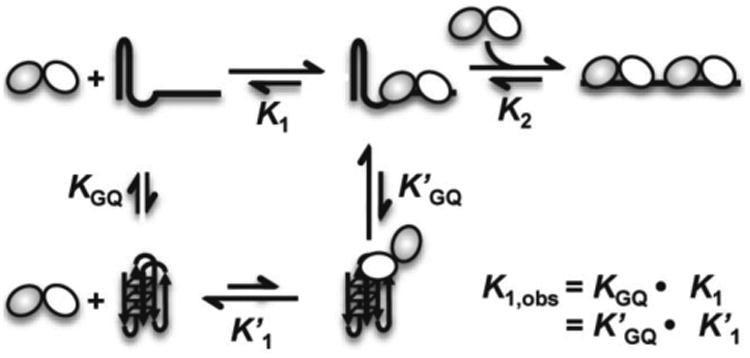
Proposed model for two POT1-TPP1 proteins binding with and unfolding 24-nt telomere DNA. The effect of G-quadruplex formation results in a K1,obs > K1. Note that each protein heterodimer consists of two OB-fold domains (shown as white and grey circles) that recognize telomere DNA and can bind independently.
Consistent with the kinetic results, our findings show that POT1-TPP1 binding to G-quadruplex structures formed in NaCl occurs with an affinity that is nearly 5-fold higher than for those topologies formed in KCl. The discrepancy in G-quadruplex unfolding in KCl over NaCl can also be attributed to a well-known increased stability of G-quadruplex structures formed in K+ 61. An alternative, or additional, explanation might be due to the difference in G-quadruplex topology formed by hT24wt in NaCl or KCl buffers. In this case, perhaps the TTA loop necessary for POT1-TPP1 recognition is more accessible in the G-quadruplex topologies formed in NaCl buffer. As the CD unfolding experiments were performed with protein concentrations significantly higher than the calculated K2 value, then the final bound state of the reaction consists of two POT1-TPP1 proteins bound to each 24-nt substrate, including G-quadruplex structures in NaCl and KCl. In this case, and particularly for the hT24wt DNA forming G-quadruplex structures in KCl buffer, it is possible that each POT1-TPP1 protein binds partially via a single OB-fold that unfolds the G-quadruplex or traps it in a ssDNA conformation so that the second OB-fold within each protein can eventually bind.
Despite significant differences in affinity of POT1-TPP1 binding to telomere DNA with G-quadruplex topologies, telomerase activity and processivity show equivalent sensitivities to both G-quadruplex structures. The addition of POT1-TPP1 unfolds all G-quadruplex topologies to enhance telomerase activity, which is consistent with previous studies showing that POT1, with or without TPP1, disrupts G-quadruplex structures for telomerase accessibility 20; 41; 47. In addition to disrupting G-quadruplex structures, the POT1-TPP1 heterodimer enhances telomerase function by aiding in translocation and slowing primer dissociation 36 and by increasing its rate of extension 41; 47. Our data reveal additional insight for the diverse contributions of POT1-TPP1 in enhancing telomerase activity and processivity. Specifically, POT1-TPP1 binding relieves secondary structure of telomere substrates to enhance telomerase activity by two- to three-fold and, once bound to the DNA substrate, the heterodimer enhances telomerase processivity an additional ∼1.5-fold.
In addition to G-quadruplex structures, telomere DNA can also form T-loops, structures whose formation is governed by interactions with shelterin proteins 62. Since not all telomeres end in T-loops, it is plausible to speculate that the secondary structure of the DNA, along with unfolding of those structures by shelterin proteins, governs T-loop formation. This feat is especially true when considering that T-loop formation is believed to require separation of the duplex telomere DNA and subsequent invasion of the single-stranded overhang, both processes that would be altered by secondary structure of the DNA. Similarly, POT1-TPP1 binding at the telomere prevents the induction of ATR-mediated DNA damage signaling pathways 63; 64. In light of our data, perhaps the secondary structure of telomere DNA is an overlooked yet important regulator that contributes to the ability of POT1-TPP1 binding to outcompete replication protein A to prevent induction of the ATR pathway.
Materials and Methods
POT1-TPP1 expression and purification
Human full-length POT1 and an N-terminal 6× His-tagged TPP1 were co-expressed in Spodoptera frugiperda 9 (Sf9) insect cells using a recombinant baculovirus expression system as previously described 44. The TPP1 protein is a well-characterized construct that comprises both its POT1- and telomerase interaction domains (residues 89-334). Sf9 cells were grown in suspension to a density of approximately 1 million cells/ml and then infected at a multiplicity of infection of one virus per insect cell. After 72 hours, infected cells were pelleted and stored at -80°C. Frozen cell pellets were resuspended in lysis buffer containing 25 mM HEPES pH 8.0, 150 mM NaCl, 5 mM DTT, 5 mM benzamidine, 1 mM PMSF, and 1 cOmplete ULTRA protease inhibitor cocktail tablet (Roche Diagnostics, Indianapolis, IN). Cells were then lysed using sonication and incubated with two units/ml of RQ1 DNase (Promega, Madison, WI) for 30 minutes before pelleting cellular debris through ultracentrifugation at 36K rpm for 45 minutes at 4°C. Following ultracentrifugation, supernatant was applied to a gravity filtration column with buffer washed high-density nickel cross-linked beads (Gold Biotechnology, St. Louis, MO). Bead binding was performed at 4°C, then rinsed with 25 mM HEPES pH 8.0 and 150 mM NaCl or 150 mM KCl (pH was adjusted using NaOH or KOH, respectively) buffer and subsequently washed with the same buffer containing 20 mM imidazole. Protein was eluted with buffer containing 200 mM imidazole and then further purified with an ACTA HPLC system and size-exclusion chromatography using a Superdex 200 HiLoad 16/60 chromatography column (GE Healthcare). Protein fractions were pooled, concentrated with a Millipore Amicon Ultra 10K centrifugal column, and flash-frozen in 10-100 μl aliquots to be stored at -80°C.
DNA Oligonucleotides and 5′ end-labeling
The 24-nt oligonucleotides investigated in this study were all synthesized by Invitrogen Life Technologies. Each DNA sequence contains two separate POT1-TPP1 recognition sequences (GGTTAGTTAG) separated by dinucleotides that were designed to disrupt or alter the DNA secondary structure of native telomere DNA without altering either of the POT1-TPP1 binding motifs. The primary sequences of the different substrates are as: hT24wt=(5′-GGTTAGGGTTAGGGTTAGGGTTAG-3′); hT24GG-CC=(5′-CCTTAGGGTTAGCCTTAGGGTTAG-3′); hT24GG-II=(5′- IITTAGGGTTAGIITTAGGGTTAG -3′); and hT24GG-UU=(5′- UUTTAGGGTTAGUUTTAGGGTTAG -3′). 5′ end-labeling was performed using 25 pmols of each oligonucleotide reacted with 5′-end labeled using ATP[γ-32P] (Perkin Elmer) and T4 Polynucleotide Kinase (New England BioLabs) as previously described 43. Labeled products were purified from unreacted nucleotide using illustra MicroSpin G-25 columns (GE Healthcare).
Size-exclusion HPLC
Oligonucleotides were prepared in the elution buffer (25 mM Hepes, pH8, 150 mM NaCl or KCl) by heating 20 μM solutions to 95°C for 5 min, followed by gradual cooling (1°C/min) to 4°C and then stored at -20°C. The DNA concentrations were estimated on the basis of absorbance readings obtained using a NanoDrop spectrometer (Thermo Fisher Scientific, Inc., Waltham, MA) and were confirmed on the AP π*-180 Instrument. Frozen samples were thawed on ice and then brought to room temperature before HPLC injection. For HPLC runs, 20 μL of DNA was injected into a Shimadzu SPD-20A HPLC and separated using a SEC-300 size-exclusion column (Thermo Scientific™ Acclaim™). All SE-HPLC runs were conducted at room temperature using a flow-rate of 0.2 ml/min and constant UV absorbance monitoring at a wavelength of 260 nm.
Electrophoretic Mobility Shift Assays
Electrophoretic Mobility Shift Assays (EMSA) were performed under equilibrium binding conditions with an excess of protein and limiting DNA concentrations. 300 pM 32P-labeled DNA oligonucleotides were incubated with increasing concentrations of purified POT1-TPP1 protein in the range of 0-600 nM. All reactions were prepared in a solution containing 1.2 μg/ml tRNA, 4 μg/ml BSA, 60 mM Tris (pH 8.0), 5 mM DTT (dithiothreitol) and 90 mM NaCl or KCl, as indicated. Protein-DNA complexes were incubated at room temperature for fifteen minutes before being separated by gel electrophoresis. Free DNA and POT1-TPP1-bound DNA were resolved on a 4-20% Tris/Borate/EDTA (TBE) nondenaturing, gradient gel (BioRad) run at 4°C in 1× TBE buffer. Gels were dried and exposed to phosphorimager plates, which were imaged on a Typhoon Phosphorimager (GE Healthcare). Data were quantitated using ImageQuant TL (GE Healthcare) and processed with Microsoft Excel software. All experiments were performed in triplicate.
Determination of equilibrium dissociation constants for POT1-TPP1 binding to DNA
The data from EMSA experiments were analyzed using a sequential, two-site binding model (Scheme 1) in order to determine the equilibrium dissociation constants for initial and subsequence POT1 binding events.
Scheme 1.

In Scheme 1 a single ssDNA (D) containing two binding sites for POT1-TPP1 protein (P) binds two proteins sequentially governed by two equilibrium dissociation constants K1 and K2 that describe the affinity of the first and second binding events, respectively. Under equilibrium binding conditions in which the concentration of free protein is in excess of the DNA in the binding reaction the fractions of total DNA in the D (Fd), DP (Fdp) and DP2 (Fdp2) species are given by,
| Eq 1 |
| Eq 2 |
| Eq 3 |
The data for the fraction of total DNA in the D, DP and DP2 species was globally fit using Microsoft Excel to Eq. 1-3 in order to calculate the K1 and K2 values for each experiment. Averages and standard deviations from at least three measurements were used to determine the values reported in the text and Table 1.
Circular Dichroism Spectroscopy
CD spectra were obtained using an Applied Photophysics π*-180 (Applied Photophysics Ltd, United Kingdom) spectrometer. A quartz split-cell cuvette (Hellma USA Inc., Plainview, NY) with 2×4.375 mm path length was used to acquire CD spectra from 220–400 nm with a 2-nm bandwidth, 2-nm step size, and 2-s collection time per data point. The telomere DNA samples (hT24wt and hT24GG-II) were diluted to 10 μM stock in protein buffer as described above for SE-HPLC. DNA samples were thawed on ice and diluted to 2 μM final concentrations in appropriate buffers and placed in one compartment of the split-cell. POT1-TPP1 protein was thawed and diluted in appropriate buffers (NaCl or KCl) to 10 μM concentrations for a 1:5 molar ratio of DNA:protein. However, each 24-nt substrate contained two separate POT1-TPP1 recognition sites, so the stoichiometry of protein:DNA binding sites was 2.5:1. The POT1-TPP1 protein solution, diluted in either NaCl or KCl protein buffer, was placed into the second chamber of the split cell. The CD spectra for DNA and protein in adjacent cells were scanned three times, and the average spectrum represents the depicted data. Spectra were additionally obtained for DNA or POT1-TPP1 protein alone, under similar conditions.
For the POT1-TPP1 protein and DNA binding reactions, samples were taken from each chamber in the split cell and mixed thoroughly in a microfuge tube before being equally distributed in each chamber of the split cell. All spectra are presented after applying proper buffer corrections. For DNA unfolding experiments, CD spectra were recorded at two-minute intervals (as described above) over a course of twenty minutes.
Preparation of cell lysate for telomerase assay
Plasmids expressing hTR and hTERT, were transiently transfected into HEK 293T cells with Lipofectamine® 2000 Reagent (Invitrogen) as previously described 65. Cells were grown in nine-well plates and transient transfections were conducted at ∼1 million cells per well. 48 hours post-transfection, cells were harvested by trypsinization. Cells were then pelleted and lysed with ice-cold CHAPS lysis buffer (10 mM Tris-HCl pH 7.5, 1 mM MgCl2, 1 mM EGTA, 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 10% glycerol, 400 mM NaCl) at 4°C for 30 min. Each sample also contained 3 μl of RNasin (Promega) and protease inhibitor cocktail EASYpack (Roche). Cell lysates were then passed three times through a 27-gauge needle, flash frozen, and stored at -80°C.
Direct Telomerase Incorporation Assay
Telomerase activity assays were performed by mixing 2 μl of hTR and hTERT transfected HEK 293T cell lysate into a 16 μl reaction containing: 35 mM Tris-HCl pH 8.0, 0.7 mM MgCl2, 1.8 mM β-mercaptoethanol, 0.7 mM spermidine, 35 mM KCl or NaCl, 500 μM dTTP, 500 μM dATP, 2.9 μM dGTP, 2 μl [α-32P]-dGTP (10 μCi/μl, 3000 Ci/mmol, Perkin-Elmer) and 1 μM desired primer. Purified POT1-TPP1 protein was added to a final concentration of 1 μM in 150 mM NaCl or 150 mM KCl, plus 25 mM HEPES, pH 8.0, to those reactions containing POT1-TPP1 protein. For those reactions without POT1-TPP1 protein, an equivalent volume of the appropriate (KCl or NaCl) protein buffer was used instead. The telomerase reaction was carried out for 30 min at 30°C and then quenched by adding 100 μl 3.6 M NH4OAc, 20 μg of glycogen, 4 μl of EDTA 10 mM and a 5′-32P-labeled hT69 primer ((TTAGGG)11TTA) as a loading control. The radioactivity of the loading control was determined by liquid scintillation counting and 400 cpm were loaded into each reaction. All ssDNA products synthesized in the assay were ethanol-precipitated and then analyzed on a 12% polyacrylamide/7 M urea/1× TBE denaturing gel. The gel was dried and subjected to densitometry. Product intensities were detected on a Typhoon Trio PhosphorImager (GE Healthcare) and quantified with ImageQuant TL 1D v8.1 software (GE Healthcare).
Quantification of telomerase assay products was performed as described previously 36; 38. Briefly, relative intensities for each hexamer repeat was determined and normalized against loading control for each lane. Total activity was reported as total lane counts by summing relative intensity of all normalized bands within a lane. Repeat addition processivity was calculated by first correcting for the number of radiolabeled Gs incorporated within each hexamer repeat and calculating the fraction left behind (FLB) by dividing the sum of intensities for each round of extension (1-n) by the entire sum of intensities for the total lane count. The ln(1-FLB) was plotted against the repeat number of telomerase extension and the slope was fitted to the linear portion of those data, which was represented as repeat round numbers 5-20 in the telomerase assay. Repeat addition processivity was defined as -0.693/slope.
Supplementary Material
Research Highlights.
POT1 and TPP1 bind exclusively to the single-stranded overhang of telomeres to protect telomere DNA and to regulate telomerase activity. The G-rich telomere DNA is capable of forming diverse structures, which likely control POT1-TPP1 recognition and telomere function.
POT1-TPP1 affinity for G-quadruplex topologies formed in NaCl is 5-times higher than for G-quadruplex structures formed in KCl.
Kinetics of POT1-TPP1 unfolding of different G-quadruplex topologies varies greatly.
POT1-TPP1 enhances telomerase processivity, regardless of telomere DNA secondary structure.
This work provides important biochemical features of molecular recognition that govern the interactions of POT1-TPP1 with structured telomere DNA to mediate telomerase activity.
Acknowledgments
This manuscript is dedicated to the memory of Michael R. Mullins. We thank Drs. Andrea Putnam and Eckhard Jankowski for assistance with global fitting of multiple POT1-TPP1 binding events. This work was supported by grants from the NIH (CA186571 to DJT and GM056740 to MEH) and the American Cancer Society (RSG-13-211-01-DMC) to DJT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988;85:6622–6. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cross SH, Allshire RC, McKay SJ, McGill NI, Cooke HJ. Cloning of human telomeres by complementation in yeast. Nature. 1989;338:771–4. doi: 10.1038/338771a0. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–73. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 4.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–9. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderson E, Hardin CC, Walk SK, Tinoco I, Jr, Blackburn EH. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 6.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–9. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 7.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–80. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 8.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–15. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryan TM, Baumann P. G-quadruplexes: from guanine gels to chemotherapeutics. Mol Biotechnol. 2011;49:198–208. doi: 10.1007/s12033-011-9395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ray S, Bandaria JN, Qureshi MH, Yildiz A, Balci H. G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding. Proc Natl Acad Sci U S A. 2014;111:2990–5. doi: 10.1073/pnas.1321436111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S, Wu Y, Zhang W. G-quadruplex structures and their interaction diversity with ligands. Chem Med Chem. 2014;9:899–911. doi: 10.1002/cmdc.201300566. [DOI] [PubMed] [Google Scholar]
- 12.Murat P, Balasubramanian S. Existence and consequences of G-quadruplex structures in DNA. Curr Opin Genet Dev. 2014;25:22–9. doi: 10.1016/j.gde.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 13.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem. 2013;5:182–6. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci U S A. 2001;98:8572–7. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–54. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 16.Hansel R, Lohr F, Foldynova-Trantirkova S, Bamberg E, Trantirek L, Dotsch V. The parallel G-quadruplex structure of vertebrate telomeric repeat sequences is not the preferred folding topology under physiological conditions. Nucleic Acids Res. 2011;39:5768–75. doi: 10.1093/nar/gkr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moye AL, Porter KC, Cohen SB, Phan T, Zyner KG, Sasaki N, Lovrecz GO, Beck JL, Bryan TM. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat Commun. 2015;6:7643. doi: 10.1038/ncomms8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–13. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 19.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–31. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 20.Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci U S A. 2005;102:10864–9. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Liu JQ, Chen Z, Zheng KW, Chen CY, Hao YH, Tan Z. G-quadruplex formation at the 3′ end of telomere DNA inhibits its extension by telomerase, polymerase and unwinding by helicase. Nucleic Acids Res. 2011;39:6229–37. doi: 10.1093/nar/gkr164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–20. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 23.Oganesian L, Bryan TM. Physiological relevance of telomeric G-quadruplex formation: a potential drug target. Bioessays. 2007;29:155–65. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- 24.O'Connor MS, Safari A, Xin H, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci U S A. 2006;103:11874–9. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279:51338–42. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 26.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–10. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 27.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–34. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 28.Blackburn EH. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579:859–62. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 29.Diotti R, Loayza D. Shelterin complex and associated factors at human telomeres. Nucleus. 2011;2:119–35. doi: 10.4161/nucl.2.2.15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–80. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 31.Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–54. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol. 2004;14:1621–31. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 33.Loayza D, Parsons H, Donigian J, Hoke K, de Lange T. DNA binding features of human POT1: a nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J Biol Chem. 2004;279:13241–8. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- 34.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–10. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 35.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–62. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 36.Latrick CM, Cech TR. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. Embo J. 2010;29:924–33. doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol. 2010;30:2971–82. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012;492:285–9. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sexton AN, Youmans DT, Collins K. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J Biol Chem. 2012;287:34455–64. doi: 10.1074/jbc.M112.394767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012;150:481–94. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hwang H, Opresko P, Myong S. Single-molecule real-time detection of telomerase extension activity. Sci Rep. 2014;4:6391. doi: 10.1038/srep06391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–9. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 43.Taylor DJ, Podell ER, Taatjes DJ, Cech TR. Multiple POT1-TPP1 Proteins Coat and Compact Long Telomeric Single-Stranded DNA. J Mol Biol. 2011;410:10–7. doi: 10.1016/j.jmb.2011.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corriveau M, Mullins MR, Baus D, Harris ME, Taylor DJ. Coordinated interactions of multiple POT1-TPP1 proteins with telomere DNA. J Biol Chem. 2013;288:16361–70. doi: 10.1074/jbc.M113.471896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–66. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 46.Phan AT. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010;277:1107–17. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 47.Hwang H, Buncher N, Opresko PL, Myong S. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure. 2012;20:1872–80. doi: 10.1016/j.str.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–35. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg Med Chem. 2006;14:5584–91. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 50.Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–62. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuker M, Mathews DH, Turner DH. Algorithms and Thermodynamics for RNA Secondary Structure Prediction: A Practical Guide. In: Barciszewski J, Clark BFC, editors. RNA Biochemistry and Biotechnology NATO Science Series. Vol. 70. Kluwer Academic Publishers; Dordrecht, the Netherlands: 1999. pp. 11–43. [Google Scholar]
- 52.Risitano A, Fox KR. Inosine substitutions demonstrate that intramolecular DNA quadruplexes adopt different conformations in the presence of sodium and potassium. Bioorg Med Chem Lett. 2005;15:2047–50. doi: 10.1016/j.bmcl.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 53.Tran PL, Mergny JL, Alberti P. Stability of telomeric G-quadruplexes. Nucleic Acids Res. 2011;39:3282–94. doi: 10.1093/nar/gkq1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24:613–22. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Largy E, Mergny JL. Shape matters: size-exclusion HPLC for the study of nucleic acid structural polymorphism. Nucleic Acids Res. 2014;42:e149. doi: 10.1093/nar/gku751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dapic V, Abdomerovic V, Marrington R, Peberdy J, Rodger A, Trent JO, Bates PJ. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kypr J, Kejnovska I, Renciuk D, Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–25. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Senear DF, Brenowitz M. Determination of binding constants for cooperative site-specific protein-DNA interactions using the gel mobility-shift assay. J Biol Chem. 1991;266:13661–71. [PubMed] [Google Scholar]
- 59.Torigoe H, Furukawa A. Tetraplex structure of fission yeast telomeric DNA and unfolding of the tetraplex on the interaction with telomeric DNA binding protein Pot1. J Biochem. 2007;141:57–68. doi: 10.1093/jb/mvm011. [DOI] [PubMed] [Google Scholar]
- 60.Zhao Y, Kan ZY, Zeng ZX, Hao YH, Chen H, Tan Z. Determining the folding and unfolding rate constants of nucleic acids by biosensor. Application to telomere G-quadruplex. J Am Chem Soc. 2004;126:13255–64. doi: 10.1021/ja048398c. [DOI] [PubMed] [Google Scholar]
- 61.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–14. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 63.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–71. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 64.Guo X, Deng Y, Lin Y, Cosme-Blanco W, Chan S, He H, Yuan G, Brown EJ, Chang S. Dysfunctional telomeres activate an ATMATR-dependent DNA damage response to suppress tumorigenesis. Embo J. 2007;26:4709–19. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. Embo J. 2006;25:565–74. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


