Abstract
Human development and disease are challenging to study because of lack of experimental accessibility to in vivo systems and the complex nature of biological processes. For these reasons researchers turn to the use of model systems, ranging in complexity and scale from single cells to model organisms. While the use of model organisms is valuable for studying physiology and pathophysiology in an in vivo context and for aiding pre-clinical development of therapeutics, animal models are costly, difficult to interrogate, and not always equivalent to human biology. For these reasons, three-dimensional (3D) cell cultures have emerged as an attractive model system that contains key aspects of in vivo tissue and organ complexity while being more experimentally tractable than model organisms. In particular, organ-on-a-chip and organoid models represent orthogonal approaches that have been able to recapitulate characteristics of physiology and disease. Here, we review advances in these two categories of 3D cultures and applications in studying development and disease. Additionally, we discuss development of key technologies that facilitate the generation of 3D cultures, including microfluidics, biomaterials, genome editing, and imaging technologies.
Graphical Abstract
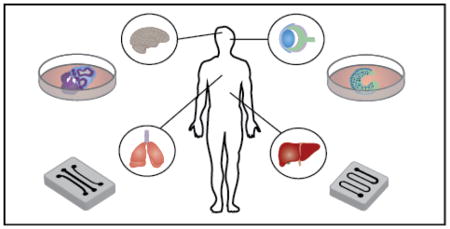
Organ-on-a-chip and organoid culture models present complementary approaches in studying development and disease by balancing experimental tractability and the ability to mimic physiological complexity.
Introduction
Human development and disease are governed by complex mechanisms that are inherently difficult to study because of our inability to often directly observe and perturb the biological processes of interest. As a result, researchers must rely on the use of model systems such as model organisms and in vitro cellular systems. Embryonic development occurs through tightly controlled processes that work in concert within developing organisms to pattern the early embryo and yield formation of specialized tissues. Interest in characterization and understanding of developmental regulation is motivated by the desire to develop a fundamental biological understanding, inform treatment of developmental disorders, and apply knowledge of native processes in engineering stem cell-based regenerative medicine treatments. Likewise, disease pathophysiology has components on the cellular, tissue, and systemic levels, and developing a better understanding will inform development of preventative and therapeutic strategies.
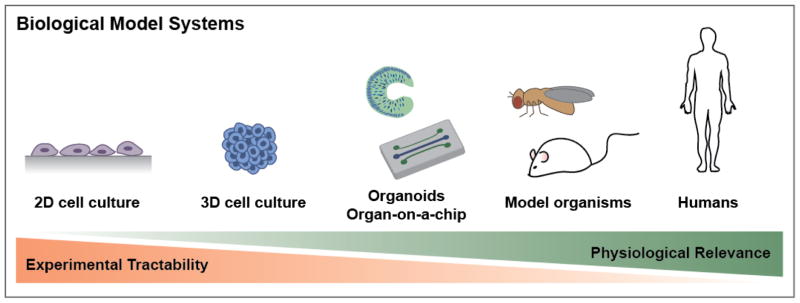
One of the major challenges is finding experimentally tractable, physiologically relevant models as a proxy for studying human biology. Commonly used models include monolayer cell culture, cell aggregates, organoids, organ-on-a-chip systems, tissue explants, and model organisms such as mouse, zebrafish, the fruit fly Drosophila melanogaster, and the nematode Caenorhabditis elegans (Figure 1). These models all have tradeoffs between relevance to human diseases, complexity, experimental accessibility, and cost. Model organisms, for example, provide the advantage that normal physiology and pathology may be studied in a systemic context, using tools such as genetic perturbations, genetic screens, and fluorescent reporters. Many key studies of development have been made in various model organisms,, such as the genetic studies in Drosophila that demonstrated the role of Hox genes in controlling body plan.1 Despite their advantages, however, disease models in animals can be expensive, time-consuming to build, and technically challenging to study. Additionally, there are species-specific differences in the biology of development and disease that can hinder direct translation of findings in animal models to human biology. 2, 3 Further, there remain significant challenges in animal models to recapitulate all aspects of many human diseases, such as microcephaly4, Alzheimer’s5, and autism spectrum disorder.6 On the other hand, models based on two-dimensional (2D) cell culture of primary or established cell lines are easily accessible experimentally in terms of manipulation and analysis. However, 2D cell cultures lack many features of in vivo microenvironments, including extracellular matrix (ECM) and dynamic signaling environments. In this context, three-dimensional (3D) cell culture models, which include organs-on-a-chip, cellular aggregates, tissue explants, and organoids, fall in the middle of the spectrum in terms of experimental accessibility and how well they mimic in vivo physiology. While these types of models are still simplifications of complex in vivo tissue structure and function, they often provide an acceptable tradeoff for studying key aspects of development and disease. However, the biggest challenge that 3D cell culture models still face is accurately recapitulating all of the complexity of native tissues. To this end, improvements in the in vivo relevance of these models will greatly benefit efforts to study development and disease, perform drug testing, and generate cell and tissue types for therapeutic purposes.
Figure 1.
Biological model systems. Model systems for studying human biological range from 2D cell culture to model organisms and lie on a spectrum in terms of their experimental tractability and physiological relevance.
Technologically, development of tools such as microfluidics, biomaterials, genome editing, and imaging methods have contributed to efforts in generating and improving in vitro 3D culture models. In this review, we will highlight contributions in each of these areas towards establishing 3D tissue and organ models. After discussion of these enabling technologies, we highlight recent examples of use of 3D culture models in studying human development and disease.
Enabling Technologies
Approaches in generating tissue and organ models
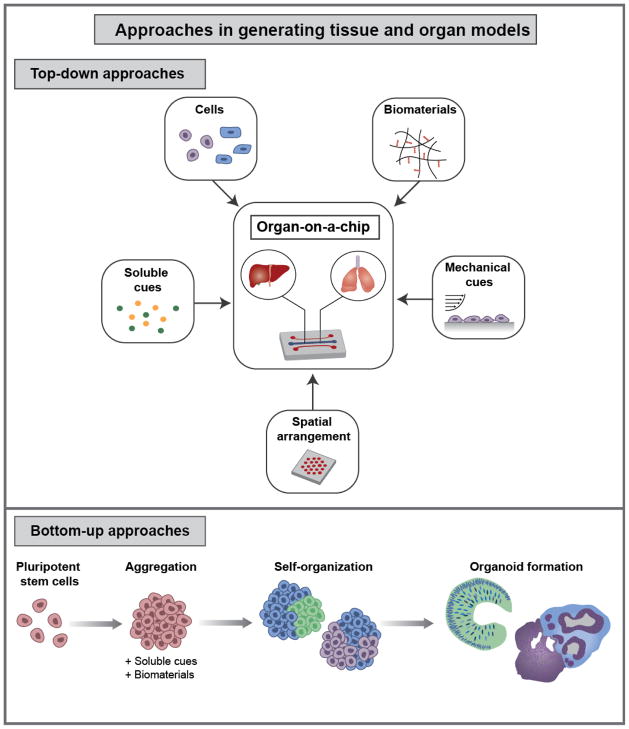
In vivo, cells reside in complex microenvironments where they receive cues from other cells, ECM, local soluble environments, and mechanical environments. These interactions play important roles in maintaining and modulating cellular phenotypes and processes. In engineering model tissue and organ systems, the goal is to mimic many of these interactions in order to generate models with key features of tissue organization and function. There are the two types of general strategies in generating 3D tissue and organ models: bottom-up and top-down approaches (Figure 2).7 In a top-down approach, the strategy employed is to engineer individual components of a tissue environment that, together, mimic and recreate aspects of the system. For example, cellular components can be integrated by co-culturing multiple cell types in defined physical arrangements, 3D organization can be mimicked with biomaterial scaffolds and microfluidic channels, mechanical cues can be presented by biomaterials and fluid flow, and soluble stimuli can be delivered via perfusion. A key example of the use of top-down approaches is in organ-on-a-chip models, which aim to recreate key aspects of organ structure and function in a microfluidic device. In contrast, bottom-up approaches rely on the emergent behavior of biological systems to generate complex tissue- and organ-like constructs. Pluripotent stem cells (PSCs) are primarily used as the cell source due to their capacity for self-organization, through processes of self-assembly, self-patterning, and self-driven morphogenesis.8 A number of organ-like structures, termed “organoids”, have been generated from PSCs for a diverse range of tissue types. Organoids are a rapidly developing area of research with exciting applications in studying development and disease.
Figure 2.
Approaches in generating tissue and organ models. In a top-down approach individual components are incorporated into a system to mimic the in vivo tissue environment. Components include multiple cell types, biomaterial scaffolds and ECM, soluble cues, mechanical cues, and microfabricated elements to define spatial arrangement and structure. Organ-on-a-chip models are an example of using a top-down approach. In contrast, a bottom-up approach supplies fewer external cues and instead relies on cellular self-organization to generate tissues with in vivo-like structure and function. Typically PSCs are formed into aggregates and cultured in the presence of soluble and material cues to guide inherent self-organization and yield organoids.
Independent of the approach in developing 3D culture models, a major challenge is in accurately recapitulating key aspects of the in vivo environment. A fundamental understanding of native tissues and organs is first needed before mimicking these in in vitro cultures. Then, the challenge lies in engineering the correct cellular and microenvironment components that will give rise to systems that are structurally and functionally similar to their in vivo counterparts. This is non-trivial, given the complex and dynamic nature of endogenous processes. Continued improvements in culture systems, such as microfluidics based platforms, will facilitate development of more realistic cellular microenvironments. Additional challenges are in the development of tools to assess tissue function and phenotype in various ways. Examples of necessary tools include technologies for genetic manipulation of cells to recreate disease phenotypes, platforms for high-throughput screening, and methods such as tissue clearing for better assessing gene and protein expression within large, intact tissues. Continued development of these areas will improve abilities to apply 3D cultures in studying biological during development and in disease. Here we highlight various tools developed to address these challenges thus far.
Microscale Physiological Systems
Advances in microscale technologies have greatly facilitated the development of 3D cell culture systems. With fabrication techniques borrowed from the microelectronics industry, as well as newer technologies such as 3D printing, microfluidic devices can be rapidly designed and prototyped.9–11 In a common method of fabrication is known as soft lithography, a master is first made in a cleanroom, using photolithography to pattern features on a silicon wafer9. Then, individual devices are casted from the mold in a material such as polydimethylsiloxane (PDMS) and are bonded to a glass slide to create enclosed channels. PDMS is commonly used for biological samples because it is biocompatible and optically transparent, facilitating imaging.12 Inherent features of microfluidics, including the small size scale, on the order of biological samples, and the existence of low Reynolds number, laminar flow together enable enhanced control over soluble and physical aspects of cellular microenvironments. Additionally, microfluidics can increase experimental throughput through assay integration, parallelization, and automation. Together, these capabilities make microfluidics well-suited for engineering 3D culture systems.
Control of the microenvironment
To mimic the in vivo tissue microenvironment within microfluidic-based systems, various strategies can be employed. Specific components of the microenvironment include the structure and arrangement of cell types, the presence of ECM, the physical and fluidic mechanical environment, and the soluble environment. To create defined arrangements of cells in 2D, early work in the field demonstrated that substrates can be micropatterned with ECM and other proteins, to which cells selectively adhere.13–16 This technique can be used to simply facilitate cell adhesion in microfluidic channels or to create arrays of cells in defined positions and shapes. A simplified tissue microenvironment could be constructed by sequential patterning of multiple cell types and co-culturing them. An alternative to patterning cells with adhesive proteins is to physically constrain their position using microfluidic device features such as traps,17–19 chambers,20–22 and channels.23 Traps and chambers are often used to array cell samples for performing assays on the single-cell level. Specifically for constructing on-chip tissue models, channels can be used to simulate different compartments of a tissue: models of the liver sinusoid, commonly use channels to define the sinusoid geometry, with regions of hepatocytes confined by an endothelial cell barrier.23–25 All of these approaches are able to position cells in 2D. In order to create 3D cellular arrangements, cells can be seeded within biomaterials in microfluidic channels.26 Coupling of these various approaches can be used to form multicellular constructs within microfluidics that begin to resemble the composition and arrangement of cells in native tissues. While the resulting systems may be over-simplified models, defining cellular spatial arrangement is an important step in beginning to construct more complex systems, such as in organ-on-a-chip models. Beyond patterning of single cells, microfluidic methods have also been used to manipulate larger samples such as cell aggregates27, 28 and model organisms.29–31 For example, a number of groups have constructed arrays of traps or wells for culturing individual, isolated cellular aggregates.27, 32–34 This provides the ability to screen effects of various factors while gathering single sample level information. Devices for culture and manipulation of cellular aggregates show potential in the area of organoid research.
Upon providing the appropriate cellular components for a tissue model, often the next step is to define cell extrinsic cues, such mechanical cues. The mechanical properties of cell substrates affect a diverse range of processes such as cell differentiation, migration, proliferation, shape, and survival.35 Fluid shear also modulates cellular phenotypes; for example it is implicated in endothelial cell differentiation and vascular growth and remodeling,36, 37 and is implicated in pathophysiology of a number of diseases, such as atherosclerosis.38 Since the mechanical environment plays such an important role in maintaining and modulating cell phenotype, it is important to incorporate these cues within cell culture models.
A final key component of the cellular microenvironment is the presence of soluble cues such as cell metabolites, small molecules, growth factors, and oxygen. Microfluidics can be leveraged to provide spatial and temporal control over the soluble cellular environment because of the highly controllable flow. Perfusion rate can be tuned to optimize optimal media exchange strategies during prolonged cell culture.22, 39 Flow rate can also be changed to modulate autocrine and paracrine signaling.40, 41 For assessing the effects of soluble factors on cell behavior, molecules can be delivered at defined flow rates and concentrations, either uniformly to the entire cell culture or in combinatorial fashion. Specifically, integration of upstream modules can be used to deliver concentration gradients42, 43 or fast temporal delivery of stimuli.44, 45 Beyond delivery of soluble stimuli, devices can also be used to reliably control oxygen concentration in the culture environment. Each of these methods for controlling the soluble cellular environment contributes to the ability to maintain viable on-chip culture, provide soluble stimuli to promote specific cell behaviors, and perform screens of culture conditions. Having these capabilities facilitates the creation of more sophisticated 3D cell culture models.
Increasing experimental throughput
Beyond facilitating the generation of defined cellular environments, microfluidics also has the capability to greatly increase experimental throughput through assay integration, parallelization, and automation. Common biological assays, such as immunohistochemistry, in situ hybridization, and live imaging can be performed on cells within microfluidic devices. This enables both cell culture and analysis steps to be performed in the same platform. Additionally, this allows spatial information to be preserved—for example, protein and gene expression can be observed in the intact tissue models. To increase throughput, microfluidic culture chambers and other operations can be parallelized, enabling a large number of samples or conditions to be screened in a single experiment. For example, samples (cells, cell aggregates, or organisms) can be cultured in arrays of traps or chambers and exposed to gradients or different molecules.21, 22 Additionally, automation of device operations can allow high-throughput operations to be performed. This includes things such as performing biochemical reactions, sorting samples, or performing image-based analysis. Reactions such as polymerase chain reaction (PCR) and gene expression analysis, among others, have been implemented in microfluidics with increased efficiency and throughput. A notable example of this is the commercial Fluidigm system. For sorting of biological samples, flow cytometry46 can be implemented on-chip. Additionally, image-based analysis of cells and organisms can also be automated47 and implemented in microfluidics, with applications in performing high-throughput genetic screens.
Biomaterials
Biomaterial approaches offer an orthogonal tool in creating in vitro model cell and tissue models. These laboratory-made and modified naturally occurring materials can be incorporated in culture systems as surface coatings or 3D scaffolds, with the goal of better mimicking the 3D in vivo cellular microenvironment and presenting physical and biochemical cues to instruct cell fate.48, 49 Hydrogels, which are water-swollen polymer networks, are commonly used to mimic ECM in 3D cell culture. Both naturally occurring hydrogels such as collagen, fibrin, hyaluronic acid, alginate, and Matrigel, as well as synthetic hydrogels such as poly(ethylene glycol) (PEG) and poly(vinyl alcohol) (PVA) are widely used. Modification of the mechanical and biochemical properties of these materials50 is leveraged to control the cues that they present to cells. Hydrogel mechanical properties can be tuned by changing parameters such as pore size, crosslinking density, and topology.51 This can be used to mimic the mechanical environments of different in vivo systems or to direct cell differentiation. In order to present biochemical cues to cells, hydrogels can be modified with cell adhesive peptides,52 degradable motifs,53, 54 and covalently linked or sequestered molecules such as growth factors.55, 56 For example, hydrogels can be designed to release growth factors to promote lineage-specific differentiation of stem cells. Microparticles can also be formed from hydrogels and incorporated within cell constructs, in order to present biochemical cues in an “inside-out” approach.57–59
Biomaterials have been used in applications such as tissue scaffolds, cell encapsulation, building organ-on-a-chip models, incorporation in cellular spheroids, and driving stem cell fate, among others. Used alone in culture systems or within microfluidic platforms, they offer additional capabilities in reconstructing aspects of in vivo cellular environments.
Genome Editing
Rapid advances in genome-editing tools in recent years have made it easier to perform genome editing more precisely and with fewer off-target effects. Some of the widely developed systems include TALENs,60 Zinc finger nucleases,61 and CRISPR-Cas9.62, 63 In the context of models of disease and development, these tools have made it easier to probe systems through the creation of fluorescent reporter lines, modulation of gene expression, and introduction of disease-related mutations. This has facilitated the creation of reporter cell lines, cellular and animal disease models, and even in vivo genome editing. In one example, Matano, et al. used CRISPR-Cas9 to introduce mutations associated with colorectal cancer into human intestinal organoids in order to study how mutations in specific pathways affect tumor formation and metastasis.64 Particularly for disease modeling, as we learn more about the specific genetic basis for various diseases, genome editing tools will allow us to create more accurate in vitro cell models. This will enable study of disease pathophysiology and development of pharmacological and other therapeutic strategies.
Imaging Technologies
A major challenge in studying 3D systems is being able to assess gene and protein expression, and spatial organization of these in intact tissues. Performing assays such as quantitative PCR (qPCR) and histological sectioning and staining do not provide spatial phenotypic information, which is useful for providing a more complete picture in characterizing tissues. However, the advent of technologies including tissue clearing65–70 and light sheet microscopy are now enabling characterization of intact 3D tissues. These techniques are still time-consuming and low-throughput, but with continued improvements and incorporation of automation tools, they will be valuable tools for interrogating 3D cultures such as organoid models. Another set of future challenges will be in advancing techniques for live imaging—for example, increasing available reporter cell lines, developing barcoding strategies for imaging many fluorophores at once, and applying high resolution imaging techniques to dense tissues. Improving capabilities for live imaging of tissues will enable visualization of how dynamic processes evolve over time periods of days, e.g. in processes involved in morphogenesis and organogenesis.
3D Culture Models
Organoid cultures
Bottom-up approaches in generating complex tissue models typically rely on biological self-organization, which refers to intrinsic abilities of cellular systems to form tissue structures and signaling environments equivalent to those of in vivo systems.71 Leveraging this ability to generate complex tissue structures from simpler initial cellular structures represents an alternative approach from strategies typically used in areas of engineered tissue or organ systems. Pluripotent stem cells (PSCs) are typically used as a cell source in tissue engineering strategies due to their differentiation potential, and the traditional approach has been to supply cues via activation and inhibition of signaling pathways in order to direct cell fate towards target cell and tissue types. However, a rapidly emerging body of work has shown the capacity of PSCs to self-organize in 3D cultures and form organoid tissues. This has sparked a shift in focus from trying to direct stem cell fate to instead facilitating inherent self-organization.71
Stem cell derived organoids are characterized by the presence of multiple organ-specific cell types, cellular organization similar to that of native tissue, and demonstration of functional characteristics. They share some similarities to embryoid bodies (EBs), which are aggregates of PSCs that undergo differentiation and morphogenesis processes that mimic aspects of early embryonic development.72, 73 Currently, organoid cultures have been developed for a range of systems, including optic cup,74 intestine,75, 76 kidney,77 liver,78 brain,4 anterior pituitary,79 and pancreas.80 Examples of a few of these systems are depicted in Figure 3. Since derivation of organoid cultures is dependent on intrinsic cell behaviors, a major challenge is finding ways to promote and guide self-organization. Approaches shared across most organoid culture examples include a 3D cell aggregation step and culture of cell aggregates in Matrigel. A common strategy is to mimic native developmental processes, such as tissue-specific activation of signaling pathways, to guide organoid development. For example, Spence, et al. generated intestinal organoids from human PSCs in part by sequentially presenting activin A and FGF/Wnt, growth factors associated with endoderm patterning during embryonic development.76 An orthogonal strategy is to culture PSC-derived progenitors or a combination of cells types, which then self-organize. Takebe, et al. used this method in generating liver buds; they co-cultured induced PSC- (iPSC) derived hepatic endoderm cells, human umbilical vein endothelial cells (HUVECs), and mesenchymal stem cells, which self-organized into 3D liver buds (Figure 3C).78
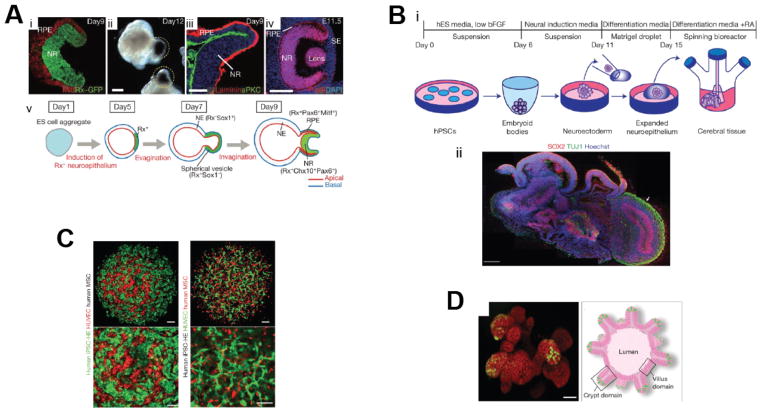
Figure 3.
Organoid examples. A: Optic cup organoids. (i–iv) Images show morphology and gene expression of optic cup structures at days 9 and 12. (i,ii) Formation of eye cup structures expressing Rx-GFP, indicating early retinal tissue, on day 9. (i,ii) The outer shell expressed markers resembling retinal epithelium progenitors, including Mitf (i) and accumulated pigment (ii). (iii) Expression of aPKC and laminin demonstrate apical-basal polarity. (iv) E11.5 mouse eye. (v) Schematic of optic cup formation.74 B: Cerebral organoids. (i) Schematic of protocol for cerebral organoid formation. (ii) Sectioning and staining of tissue shows the complex tissue morphology, with regions of neural progenitors (SOX2, red) and neurons (TUJ1, green) (arrow). Scale bars, 200 μm.4 C: Liver buds generated from human iPSCs. Images show presence of human iPSC hepatic endoderm (iPSC-HE) (green) and endothelial networks (red) inside liver buds. Scale bars, 100 μm.78 C: D: Intestinal organoids. On left, confocal image shows intestinal crypts grown for 3 weeks. Lgr-GFP+ stem cells (green) are located at the tips of crypt domains. Scale bar, 50 μm. On right, aschematic of a crypt organoid depicts thestructure.85 All figures reprinted by permission from Macmillan Publishers Ltd: Nature. Respectively: Eiraku et al,74 copyright 2011. Lancaster et al,4 copyright 2013. Takebe et al.,78 copyright 2013. Sato et al,85 copyright 2013.
One of the exciting applications of organoid cultures is indeed in modeling pathology and diseases. Organoids can potentially address a gap in knowledge in understanding diseases that are difficult or impossible to study in animal models—for example because of significant physiological or behavioral differences. Additionally, these culture systems are a cheaper and more tractable alternative for modeling and understanding the pathophysiology of disease for which causes and genetic basis are not well known. An early example of disease modeling in organoids was the use of cerebral organoids generated from patient-derived iPSCs to model microcephaly. Although mutations in several genes have been implicated in this neurodevelopmental disorder, mouse mutants do not exhibit the same severe reduction in brain size that is seen in patients.4 Analysis of patient-derived organoids by Lancaster et al. revealed premature neuronal differentiation, suggesting this as a potential cause for the disease phenotype. Beyond uncovering a previously unknown potential disease mechanism, this study more broadly demonstrated the feasibility of using cerebral organoid cultures for studying human neurodevelopment and disease. In a second example, Mariani, et al. generated telencephalic organoids from autism patient-derived iPSCs and through transcriptome and gene network analysis found neurodevelopmental alterations—specifically overproduction of GABAergic inhibitory neurons mediated by overexpression of FOXG1.6 Autism spectrum disorder (ASD) is very difficult to study in animal models because of the heterogeneity of disease phenotypes and the absence of behavioral phenotypes, highlighting the potential importance of use of patient-derived organoids in studying this disease. Beyond these two brief examples, other general applications of organoid cultures in disease modeling include in infectious diseases of the gut81 and stomach82, in pancreatic80 and prostate83 cancer, and in cystic fibrosis84.
In conjunction with the use of gene editing tools, organoid disease models can aid in the development of therapeutic strategies for diseases with a known genetic basis. For example, cystic fibrosis (CF) is associated with a single-gene mutation in the cystic fibrosis transmembrane conductor receptor (CFTR). Schwank, et al. used intestinal stem cells from CF patients homozygous for the CFTR mutation to generate intestinal organoids that display phenotypes associated with loss of CFTR function.86 They then successfully performed gene editing using CRISPR/Cas9 mediated homologous recombination to correct the CFTR mutation and associated phenotype in the organoids. This study demonstrated the feasibility of using gene editing to correct for single-gene diseases. In a second example, Matano, et al. combined the use of gene editing tools and intestinal organoids to study the roles of specific mutations in colorectal cancer.64 They used CRISPR/Cas9 to introduce mutations commonly associated with human colorectal tumors into human intestinal organoids. By transplanting the organoids into mice and assessing tumorigenicity, the authors found that the mutations supported stem cell maintenance in the tumor niche but were insufficient to cause metastasis. This example illustrates the utility of using organoid cultures for investigating specific disease mechanisms.
Current challenges associated with development of organoid cultures stem from the complexity of the biological processes involved in organogenesis. With current methods, cultures are highly heterogeneous. For self-organization of cells into functional tissues to occur, a complex orchestration of events such as cell differentiation, cell migration, timed activation of signaling pathways, and formation of tissue architecture is required. Developing a better understanding of the cues needed in these processes will aid creation of organoids that even better mimic in vivo biology. To do this, one important step is to better understand endogenous developmental processes and apply these in guiding organogenesis. Another significant challenge is the lack of vascularization of tissues. Once 3D cell cultures grow to certain size they will start to develop a necrotic core due to limited transport of oxygen and nutrients to the center. To address this, a variety of approaches are being developed for creating vascularized tissue constructs. These range from printing vascular networks87 to engineering systems where vasculargenesis and angiogenesis occur.88, 89
Further development is needed to improve upon tools for generating organoid cultures and characterization of their phenotypes. Culture of organoids in Matrigel seems to play some role in their formation, suggesting that use of ECM-mimicking and functionalized biomaterials could perhaps be used more extensively to provide instructive cues. Furthermore, although microfluidic based methods for culturing organoids have been relatively unexplored, microfluidics can potentially be leveraged in defining instructive cues to promote self-organization or in handling of 3D cultures in order to facilitate live tracking of organogenesis. Finally, development of analytical tools will aid improved characterization of intact tissues. This includes things such as live imaging, tissue clearing and quantitative in situ transcriptome and proteomic profiling.
Organ-on-a-chip
In contrast to relying on biological self-organization and assembly, organ-on-a-chip systems employ a top-down approach in constructing tissue and organ models. These microfluidic-based systems utilize a range of tools, including microfabrication and biomaterials to create microphysiological systems that model key aspects of physiology and disease. Engineering approaches are used to integrate aspects of physiology, including cellular organization, ECM, high order tissue structure, mechanical forces, and biochemical signals. Together, these components recapitulate aspects of physiological structure and function.
In organ-on-a-chip systems, structure and organization of cellular components can be defined through design of microfluidic channels and compartmentalized chambers. For example, within the liver, blood flows through the sinusoid space which is separated from hepatocytes by an endothelial cell barrier. Lee, et al. modeled the liver sinusoid in a microfluidic device by designing a narrow U-shaped chamber flanked on either side by channels with convective flow (Figure 4B).23 Within the chamber, densely packed hepatocytes could be cultured, and the chamber walls contained narrow, closely spaced parallel channels to mimic the endothelial cell barrier and allow for diffusive transport. This early liver model demonstrates how microfluidic compartmentalization and fluid transport can be leveraged; similar approaches have been used in other on-chip liver models.24, 25 Moving from creating cell and tissue features in 2D to 3D, common approaches include using multilayer microfluidic channels or culturing cells in hydrogels. For example, the Kamm lab has created perfusable microvascular networks within microfluidic channels through the co-culture of endothelial cells (ECs) and stromal cells in 3D fibrin matrices.26 The ECs and stromal cells were cultured separately in respective channels with a fluidic channel in between. This configuration allowed diffusion dependent, contact independent communication between the two cell types. Secretion of pro-angiogenic growth factors and ECM proteins by the stromal cells supported angiogenesis and vasculargenesis of the ECs, producing perfusable networks
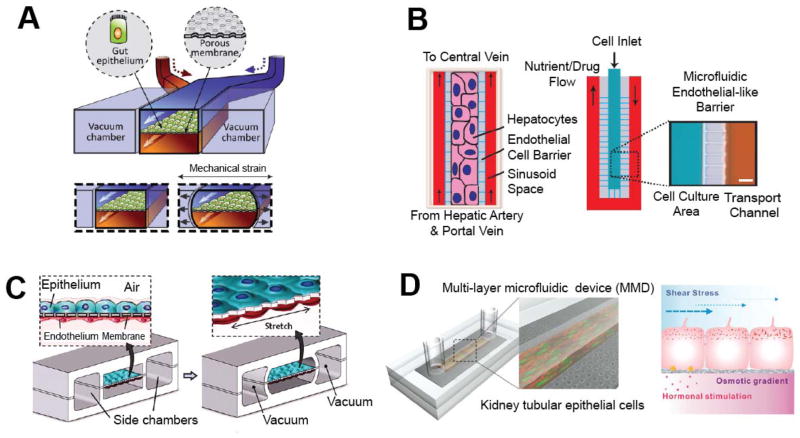
Figure 4.
Organ-on-a-chip examples. A: Schematic shows the design of a human gut-on-a-chip. Human intestinal epithelial cells are cultured on an ECM-coated porous membrane and exposed to low levels of fluid shear stress. Side vacuum chambers are used to apply cyclic strain that mimics physiological peristaltic motions. Reproduced from Kim et al.90 with permission from The Royal Society of Chemistry. B: Schematics show the design of the liver-on-a-chip. Hepatocytes are cultured in a central channel, surrounded by small, closely spaced parallel channels that mimic the endothelial cell barrier. Two side channels deliver nutrients and drugs by diffusive to the central cell culture region. Reproduced from Lee et al.23 with permission from Wiley Periodicals. Copyright 2007, Wiley Periodicals, Inc. C: A human lung-on-a-chip models the alveolar-capillary interface. Alveolar epithelial cells and pulmonary microvascular endothelial cells are cultured on opposite sides of a PDMS membrane. Vacuum is applied to lateral side chambers to stretch the PDMS membrane and mimic physiological breathing. From Hung et al.91 Reprinted with permission from AAAS.D: Schematics show the design of a kidney duct model. Renal cells are cultured within the channel and exposed to fluid shear stress, osmotic gradients, and hormonal stimulation. Reproduced from Jang et al.92 with permission from The Royal Society of Chemistry.
Incorporating controllable mechanical forces such as fluid shear and mechanical strain into organ-on-a-chip systems enables studies of how these play a role in tissue physiology and disease. For example, Jang, et al. looked at the effects of fluid shear stress on renal tubular epithelial cells.92, 93 These cells have osmoregulatory functions in the kidney and are exposed to fluidic environments, but the role of shear stress is not well understood. Using a microfluidic system in which the cells were cultured in a channel and exposed to fluid shear (Figure 4D), the authors found that fluid shear stress plays a role in cytoskeletal reorganization and trafficking of aquaporin-2, a water transport protein. Huh, et al. used a different type of approach to examine the role of mechanical strain in a lung model.94 The authors recreated the alveolar-capillary interface of the lung within microfluidic channels by culturing alveolar epithelial cells and pulmonary microvascular endothelial cells on opposing sides of a PDMS membrane (Figure 4C). Air was introduced into the channel on the epithelial side to mimic the alveolar airspace. By applying vacuum to two additional side chambers, the PDMS membrane could be stretched, allowing cyclic mechanical strain typical of normal respiration to be applied to the tissue interface. This platform was applied in studying how mechanics influence pulmonary inflammation and immune response.
To date, a wide range of organ systems have been modeled thus far, including examples such as liver,23–25 kidney,92 intestine,90, 95 lung,91, 94, 96 heart,97 muscle,98 angiogenesis,99 and bone marrow.100 A few examples of these are shown in Figure 4. As organ-on-a-chip systems become more sophisticated and able to recreate key features of physiology, they have increasing utility in studying physiology, modeling disease, and screening therapeutics. Kim, et al have developed a human gut-on-a-chip model90 and applied it to studying how intestinal inflammation and bacterial overgrowth contribute to pathophysiology of intestinal diseases.95 Using a device similar to that of the group’s lung-on-a-chip model (Figure 4A),94 human intestinal epithelial cells were cultured on an ECM-coated membrane and exposed to luminal flow and peristalsis-like mechanical deformations. Under these conditions, the cells form intestinal villi structures. Using this model, they co-cultured the intestinal cells with commensal microbes and immune cells and assessed how the gut microbiome, inflammatory cells, and cyclic mechanical deformations contribute to intestinal bacterial overgrowth and inflammation. These studies have great relevance in understanding mechanisms of intestinal disease such as inflammatory bowel disease (IBS), Crohn’s disease, and ulcerative colitis. Another example of disease-relevant studies is the use of perfusable microvascular networks for studying tumor cell extravasation. Jeon, et al. looked at extravasation of human metastatic breast cancer cells in a microvascularized bone-mimicking microenvironment.88 Human bone marrow derived mesenchymal stem cells (hBM-MSCs), osteo-differentiated hBM-MSCs, and human umbilical vein endothelial cells (HUVECs) were cultured within a fibrin gel and went on to form microvascular networks. Metastatic breast cancer cells were perfused through the networks and exhibited rolling, adhesion, and extravasation into the surrounding ECM. The authors found that blocking breast cancer cell A3 adenosine receptors resulted in higher extravasation rates, implicating adenosine in reducing extravasation. This example highlights the utility of this type of model for studying organ-specific metastasis, identifying molecular pathways involved, and screening for potential targeted therapeutics.
While organ-on-a-chip models are valuable tools for investigating detailed mechanisms of physiology and disease and for performing cheaper, faster drug screening, a continuing challenge is to make these systems more reflective of physiological function. Existing models are successful in recreating specific individual aspects of organ function, but they still are oversimplifications of whole-organ physiology. One reason why it is challenging to recreate organ physiology in vitro is its inherent biological complexity. Additionally, while some physiological systems are well-characterized, many are not, making it difficult to design models in a top-down approach. These issues motivate the strategy of using self-organizing biological systems such as organoid cultures to create tissues in a bottom-up type approach. Instead of having to define the various components of a tissue environment, cells can be guided to undergo self-organization and morphogenesis to form tissues structures. Incorporating aspects of emergent systems in creating organ-on-a-chip could perhaps help in creating more complex and physiologically relevant tissues.
An additional challenge is in designing strategies to model interactions between different tissue and organ systems, which can be important in studying systemic aspects of disease pathophysiology and in screening for drug toxicity. These so-called “body-on-a-chip” systems typically consist of organ-specific cell types cultured in individual microfluidic modules and connected by circulating flow.101–103 Some of the design considerations important for these systems include how to incorporate dynamic organ-organ interactions, the exchange of metabolites between different tissues, and how to mimic the circulatory system. A critical part of improving upon body-on-a-chip models will be to increase the complexity of the individual organ modules so that they better recreate essential organ functions. While many of these challenges have yet to be completely addressed, all of these are active areas of research.
Conclusions and Perspectives
3D culture models are important tools for studying human development and disease, as both supplements and substitutes to animal models. In particular, organ-on-a-chip and organoid cultures demonstrate the ability to recapitulate many key aspects of normal and diseased tissue physiology. These systems show exciting potential but still face a number of challenges, which if addressed will further improve their complexity, in vivo relevance, and utility in various applications.
The inadequacy of in vitro tissue models to recapitulate various aspects of native tissues can be addressed by two general approaches. First, by continuing fundamental studies to learn how morphogenesis and organogenesis are executed during embryonic development, we can apply these principles in better guiding emergent cell behavior. By supplying the proper cues to cell cultures, more mature and native-like tissues can be formed. Second, generation of better tissue models will be furthered by developments in technologies that can be used to engineer cell microenvironments and guide cell fate, including but not limited to, microfluidic techniques and biomaterials. An example of use of these two approaches is in addressing the challenges of low yield efficiency and high heterogeneity in organoid cultures. The culture heterogeneity likely stems from a combination of insufficient external cues, environmental variability, and intrinsic cellular heterogeneity. To improve upon this, better knowledge of the required external cues and better control of the cellular environment with technological approaches such as microfluidics can potentially help.
Developing improved methods for characterizing 3D culture models will also be important. Morphological and biochemical characterization is important for understanding the composition of generated tissues, assessing responses to perturbations (such as in drug screening), and in investigating mechanisms of disease. Observing phenotypes, such as gene and protein expression, in intact tissues is still challenging, but continued improvements in areas such as live imaging, gene editing, tissue clearing, and high resolution microscopy are beginning to address these challenges.
Thus far, efforts in the areas of organs-on-a-chip and organoids have been largely independent. However, work in improving in vitro tissue and organ models will benefit from incorporating aspects of both approaches. Organoid systems are able to generate tissues with arguably more complexity and similarities to native tissues; however, a major challenge is the lack of control over these self-emergent systems. Conversely, while one of the advantages of organ-on-a-chip approaches is having control over each aspect of the tissue environment, these models are still relatively simplified. Thus, there is potential in a combined approach: relying in part on cellular self-emergence to generate complex tissues, but also using tools to better define the cellular microenvironment in order to reproducibly guide and direct cell behavior.
3D cell cultures have exciting potential for use in studying physiology, modeling disease, and screening therapeutics. Particularly exciting applications of these systems include studying complex and non-genetic diseases, which often cannot be recreated in animal models. Having the ability to generate patient-cell derived tissue and organ models will surely enable large-scale screens to identify disease mechanisms, understand systemic aspects of disease, and develop therapeutic approaches.
Supplementary Material
Insight Statement.
Challenges in studying human development and disease stem from our inability to experimentally access human biology in vivo. Many researchers rely on model systems, each of which has significant tradeoffs. Three-dimensional cell culture models have emerged as a system with sufficient complexity for studying key aspects of tissue- and organ-level physiology and pathophysiology while still remaining experimentally tractable. In vitro models such as organs-on-chips and organoids use a range of strategies to create organ models indicative of endogenous structure and function, which has been enabled by key technologies, including microfluidics, biomaterials, genome editing, and imaging technologies. By integrating these technologies and further understanding fundamental developmental biology, the research community will be able to address many important human diseases.
Acknowledgments
The authors would like to acknowledge the following funding sources: NSF Graduate Research Fellowship (DGE-1148903) to ELJ; NSF EBICS Science and Technology Center (CBET 0939511) to HL and funding from NIH (NS096581, GM088333, EB021676, EB020424, and AG050304) to HL.
Notes and references
- 1.Mallo M, Wellik DM, Deschamps J. Developmental biology. 2010;344:7–15. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Z, Huangfu D. Development. 2013;140:705–717. doi: 10.1242/dev.086165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elsea SH, Lucas RE. ILAR journal/National Research Council, Institute of Laboratory Animal Resources. 2002;43:66–79. [Google Scholar]
- 4.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster SJ, Bachstetter AD, Nelson PT, Schmitt FA, Van Eldik LJ. Frontiers in genetics. 2014;5:88. doi: 10.3389/fgene.2014.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, Gerstein M, Grigorenko Elena L, Chawarska K, Pelphrey Kevin A, Howe James R, Vaccarino Flora M. Cell. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutmacher DW, Holzapfel BM, De-Juan-Pardo EM, Pereira BA, Ellem SJ, Loessner D, Risbridger GP. Curr Opin Biotechnol. 2015;35:127–132. doi: 10.1016/j.copbio.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Sasai Y. Cell Stem Cell. 2013;12:520–530. doi: 10.1016/j.stem.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Annual Review of Biomedical Engineering. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 10.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 11.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 12.Sia SK, Whitesides GM. Electrophoresis. 2003;24:3563–3576. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- 13.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Biomaterials. 1999;20:2363–2376. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 14.Singhvi R, Kumar A, Lopez G, Stephanopoulos G, Wang D, Whitesides G, Ingber D. Science. 1994;264:696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 15.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 16.Mrksich M, Dike LE, Tien J, Ingber DE, Whitesides GM. Experimental Cell Research. 1997;235:305–313. doi: 10.1006/excr.1997.3668. [DOI] [PubMed] [Google Scholar]
- 17.Chung K, Rivet CA, Kemp ML, Lu H. Analytical Chemistry. 2011;83:7044–7052. doi: 10.1021/ac2011153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinh N-D, Chiang Y-Y, Hardelauf H, Baumann J, Jackson E, Waide S, Sisnaiske J, Frimat J-P, Thriel Cv, Janasek D, Peyrin J-M, West J. Lab on a Chip. 2013;13:1402–1412. doi: 10.1039/c3lc41224e. [DOI] [PubMed] [Google Scholar]
- 19.Torisawa YS, Mosadegh B, Luker GD, Morell M, O’Shea KS, Takayama S. Integr Biol (Camb) 2009;1:649–654. doi: 10.1039/b915965g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung PJ, Lee PJ, Sabounchi P, Aghdam N, Lin R, Lee LP. Lab Chip. 2005;5:44–48. doi: 10.1039/b410743h. [DOI] [PubMed] [Google Scholar]
- 21.Titmarsh DM, Hudson JE, Hidalgo A, Elefanty AG, Stanley EG, Wolvetang EJ, Cooper-White JJ. PloS one. 2012;7:e52405. doi: 10.1371/journal.pone.0052405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lecault V, Vaninsberghe M, Sekulovic S, Knapp DJ, Wohrer S, Bowden W, Viel F, McLaughlin T, Jarandehei A, Miller M, Falconnet D, White AK, Kent DG, Copley MR, Taghipour F, Eaves CJ, Humphries RK, Piret JM, Hansen CL. Nature methods. 2011;8:581–586. doi: 10.1038/nmeth.1614. [DOI] [PubMed] [Google Scholar]
- 23.Lee PJ, Hung PJ, Lee LP. Biotechnology and Bioengineering. 2007;97:1340–1346. doi: 10.1002/bit.21360. [DOI] [PubMed] [Google Scholar]
- 24.Toh YC, Lim TC, Tai D, Xiao G, van Noort D, Yu H. Lab Chip. 2009;9:2026–2035. doi: 10.1039/b900912d. [DOI] [PubMed] [Google Scholar]
- 25.Nakao Y, Kimura H, Sakai Y, Fujii T. Biomicrofluidics. 2011;5:022212. doi: 10.1063/1.3580753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, Lee H, Chung M, Jeon NL. Lab on a Chip. 2013;13:1489–1500. doi: 10.1039/c3lc41320a. [DOI] [PubMed] [Google Scholar]
- 27.Cimetta E, Figallo E, Cannizzaro C, Elvassore N, Vunjak-Novakovic G. Methods. 2009;47:81–89. doi: 10.1016/j.ymeth.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frey O, Misun PM, Fluri DA, Hengstler JG, Hierlemann A. Nature communications. 2014;5:4250. doi: 10.1038/ncomms5250. [DOI] [PubMed] [Google Scholar]
- 29.Chung K, Kim Y, Kanodia JS, Gong E, Shvartsman SY, Lu H. Nature methods. 2011;8:171–176. doi: 10.1038/nmeth.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levario TJ, Zhao C, Rouse T, Shvartsman SY, Lu H. Scientific Reports. 2016;6:21366. doi: 10.1038/srep21366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung K, Zhan M, Srinivasan J, Sternberg PW, Gong E, Schroeder FC, Lu H. Lab on a Chip. 2011;11:3689–3697. doi: 10.1039/c1lc20400a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vrij EJ, Espinoza S, Heilig M, Kolew A, Schneider M, van Blitterswijk CA, Truckenmuller RK, Rivron NC. Lab Chip. 2016;16:734–742. doi: 10.1039/c5lc01499a. [DOI] [PubMed] [Google Scholar]
- 33.Fung WT, Beyzavi A, Abgrall P, Nguyen NT, Li HY. Lab Chip. 2009;9:2591–2595. doi: 10.1039/b903753e. [DOI] [PubMed] [Google Scholar]
- 34.Khoury M, Bransky A, Korin N, Konak LC, Enikolopov G, Tzchori I, Levenberg S. Biomedical microdevices. 2010;12:1001–1008. doi: 10.1007/s10544-010-9454-x. [DOI] [PubMed] [Google Scholar]
- 35.Daley WP, Peters SB, Larsen M. Journal of Cell Science. 2008;121:255–264. doi: 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 36.Ahsan T, Nerem RM. Tissue engineering Part A. 2010;16:3547–3553. doi: 10.1089/ten.tea.2010.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitsumata M, Fishel RS, Nerem RM, Alexander RW, Berk BC. American Journal of Physiology - Heart and Circulatory Physiology. 1993;265:H3–H8. doi: 10.1152/ajpheart.1993.265.1.H3. [DOI] [PubMed] [Google Scholar]
- 38.Nerem RM. Journal of biomechanical engineering. 1992;114:274–282. doi: 10.1115/1.2891384. [DOI] [PubMed] [Google Scholar]
- 39.Kim L, Vahey MD, Lee HY, Voldman J. Lab on a Chip. 2006;6:394–406. doi: 10.1039/b511718f. [DOI] [PubMed] [Google Scholar]
- 40.Przybyla LM, Voldman J. Proc Natl Acad Sci U S A. 2012;109:835–840. doi: 10.1073/pnas.1103100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellison D, Munden A, Levchenko A. Molecular bioSystems. 2009;5:1004–1012. doi: 10.1039/b905602e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keenan TM, Folch A. Lab on a Chip. 2008;8:34–57. doi: 10.1039/b711887b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeon NL, Dertinger SKW, Chiu DT, Choi IS, Stroock AD, Whitesides GM. Langmuir. 2000;16:8311–8316. [Google Scholar]
- 44.He L, Kniss A, San-Miguel A, Rouse T, Kemp ML, Lu H. Lab on a Chip. 2015;15:1497–1507. doi: 10.1039/c4lc01070a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chingozha L, Zhan M, Zhu C, Lu H. Analytical Chemistry. 2014;86:10138–10147. doi: 10.1021/ac5019843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hur SC, Tse HT, Di Carlo D. Lab Chip. 2010;10:274–280. doi: 10.1039/b919495a. [DOI] [PubMed] [Google Scholar]
- 47.Crane MM, Stirman JN, Ou CY, Kurshan PT, Rehg JM, Shen K, Lu H. Nature methods. 2012;9:977–980. doi: 10.1038/nmeth.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guvendiren M, Burdick JA. Current Opinion in Biotechnology. 2013;24:841–846. doi: 10.1016/j.copbio.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lutolf MP, Hubbell JA. Nat Biotech. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Rodrigues J, Tomas H. Chemical Society reviews. 2012;41:2193–2221. doi: 10.1039/c1cs15203c. [DOI] [PubMed] [Google Scholar]
- 51.Cha C, Kim SY, Cao L, Kong H. Biomaterials. 2010;31:4864–4871. doi: 10.1016/j.biomaterials.2010.02.059. [DOI] [PubMed] [Google Scholar]
- 52.Hern DL, Hubbell JA. Journal of biomedical materials research. 1998;39:266–276. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 53.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA. Proceedings of the National Academy of Sciences. 2003;100:5413–5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hudalla GA, Murphy WL. Advanced Functional Materials. 2011;21:1754–1768. doi: 10.1002/adfm.201002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee K, Silva EA, Mooney DJ. Journal of the Royal Society, Interface/the Royal Society. 2011;8:153–170. doi: 10.1098/rsif.2010.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bratt-Leal AM, Nguyen AH, Hammersmith KA, Singh A, McDevitt TC. Biomaterials. 2013;34:7227–7235. doi: 10.1016/j.biomaterials.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hettiaratchi MH, Miller T, Temenoff JS, Guldberg RE, McDevitt TC. Biomaterials. 2014;35:7228–7238. doi: 10.1016/j.biomaterials.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen AH, McKinney J, Miller T, Bongiorno T, McDevitt TC. Acta Biomaterialia. 2015;13:101–110. doi: 10.1016/j.actbio.2014.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joung JK, Sander JD. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 62.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. Nature Medicine. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 65.Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Nature neuroscience. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 66.Erturk A, Becker K, Jahrling N, Mauch CP, Hojer CD, Egen JG, Hellal F, Bradke F, Sheng M, Dodt HU. Nature protocols. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- 67.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, Pak S, Bernstein H, Ramakrishnan C, Grosenick L, Gradinaru V, Deisseroth K. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, Abe T, Kiyonari H, Shimizu Y, Miyawaki A, Yokota H, Ueda HR. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 69.Tomer R, Ye L, Hsueh B, Deisseroth K. Nature protocols. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, Shah S, Cai L, Gradinaru V. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woodford C, Zandstra PW. Current Opinion in Biotechnology. 2012;23:810–819. doi: 10.1016/j.copbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lancaster MA, Knoblich JA. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 73.Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, Soreq H, Benvenisty N. Molecular medicine (Cambridge, Mass) 2000;6:88–95. [PMC free article] [PubMed] [Google Scholar]
- 74.Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, Sasai Y. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 75.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 76.Spence JR, Mayhew CN, Rankin SA, Kuhar M, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, Shroyer NF, Wells JM. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takasato M, Er PX, Becroft M, Vanslambrouck JM, Stanley EG, Elefanty AG, Little MH. Nature Cell Biology. 2014;16:118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- 78.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, Aoyama S, Adachi Y, Taniguchi H. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 79.Suga H, Kadoshima T, Minaguchi M, Ohgushi M, Soen M, Nakano T, Takata N, Wataya T, Muguruma K, Miyoshi H, Yonemura S, Oiso Y, Sasai Y. Nature. 2011;480:57–62. doi: 10.1038/nature10637. [DOI] [PubMed] [Google Scholar]
- 80.Boj Sylvia F, Hwang C-I, Baker Lindsey A, Chio Iok In C, Engle Dannielle D, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector Mona S, Gracanin A, Oni T, Yu Kenneth H, van Boxtel R, Huch M, Rivera Keith D, Wilson John P, Feigin Michael E, Öhlund D, Handly-Santana A, Ardito-Abraham Christine M, Ludwig M, Elyada E, Alagesan B, Biffi G, Yordanov Georgi N, Delcuze B, Creighton B, Wright K, Park Y, Morsink Folkert HM, Molenaar IQ, Borel Rinkes Inne H, Cuppen E, Hao Y, Jin Y, Nijman Isaac J, Iacobuzio-Donahue C, Leach Steven D, Pappin Darryl J, Hammell M, Klimstra David S, Basturk O, Hruban Ralph H, Offerhaus George J, Vries Robert GJ, Clevers H, Tuveson David A. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Finkbeiner SR, Zeng XL, Utama B, Atmar RL, Shroyer NF, Estes MK. mBio. 2012;3:e00159–00112. doi: 10.1128/mBio.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McCracken KW, Cata EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, Wells JM. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gao D, Vela I, Sboner A, Iaquinta Phillip J, Karthaus Wouter R, Gopalan A, Dowling C, Wanjala Jackline N, Undvall Eva A, Arora Vivek K, Wongvipat J, Kossai M, Ramazanoglu S, Barboza Luendreo P, Di W, Cao Z, Zhang Qi F, Sirota I, Ran L, MacDonald Theresa Y, Beltran H, Mosquera J-M, Touijer Karim A, Scardino Peter T, Laudone Vincent P, Curtis Kristen R, Rathkopf Dana E, Morris Michael J, Danila Daniel C, Slovin Susan F, Solomon Stephen B, Eastham James A, Chi P, Carver B, Rubin Mark A, Scher Howard I, Clevers H, Sawyers Charles L, Chen Y. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I, Janssens HM, de Winter-de Groot KM, Brandsma AM, de Jong NWM, Bijvelds MJC, Scholte BJ, Nieuwenhuis EES, van den Brink S, Clevers H, van der Ent CK, Middendorp S, Beekman JM. Nat Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 85.Sato T, Clevers H. Science. 2013;340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 86.Schwank G, Koo B-K, Sasselli V, Dekkers Johanna F, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent Cornelis K, Nieuwenhuis Edward ES, Beekman Jeffrey M, Clevers H. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 87.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DHT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Nat Mater. 2012;11:768–774. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jeon JS, Bersini S, Gilardi M, Dubini G, Charest JL, Moretti M, Kamm RD. Proceedings of the National Academy of Sciences. 2015;112:214–219. doi: 10.1073/pnas.1417115112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vickerman V, Blundo J, Chung S, Kamm RD. Lab on a chip. 2008;8:1468–1477. doi: 10.1039/b802395f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim HJ, Huh D, Hamilton G, Ingber DE. Lab on a Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 91.Huh D, Fujioka H, Tung YC, Futai N, Paine R, 3rd, Grotberg JB, Takayama S. Proc Natl Acad Sci U S A. 2007;104:18886–18891. doi: 10.1073/pnas.0610868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jang KJ, Suh KY. Lab on a Chip. 2010;10:36–42. doi: 10.1039/b907515a. [DOI] [PubMed] [Google Scholar]
- 93.Jang KJ, Cho HS, Kang DH, Bae WG, Kwon TH, Suh KY. Integrative Biology. 2011;3:134–141. doi: 10.1039/c0ib00018c. [DOI] [PubMed] [Google Scholar]
- 94.Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim HJ, Li H, Collins JJ, Ingber DE. Proceedings of the National Academy of Sciences. 2016;113:E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Douville NJ, Zamankhan P, Tung YC, Li R, Vaughan BL, Tai CF, White J, Christensen PJ, Grotberg JB, Takayama S. Lab Chip. 2011;11:609–619. doi: 10.1039/c0lc00251h. [DOI] [PubMed] [Google Scholar]
- 97.Grosberg A, Alford PW, McCain ML, Parker KK. Lab Chip. 2011;11:4165–4173. doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Grosberg A, Nesmith AP, Goss JA, Brigham MD, McCain ML, Parker KK. Journal of pharmacological and toxicological methods. 2012;65:126–135. doi: 10.1016/j.vascn.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Song JW, Munn LL. Proceedings of the National Academy of Sciences. 2011;108:15342–15347. doi: 10.1073/pnas.1105316108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Torisawa Y-s, Spina CS, Mammoto T, Mammoto A, Weaver JC, Tat T, Collins JJ, Ingber DE. Nat Meth. 2014;11:663–669. doi: 10.1038/nmeth.2938. [DOI] [PubMed] [Google Scholar]
- 101.Esch MB, King TL, Shuler ML. Annu Rev Biomed Eng. 2011;13:55–72. doi: 10.1146/annurev-bioeng-071910-124629. [DOI] [PubMed] [Google Scholar]
- 102.Viravaidya K, Sin A, Shuler ML. Biotechnol Prog. 2004;20:316–323. doi: 10.1021/bp0341996. [DOI] [PubMed] [Google Scholar]
- 103.Imura Y, Sato K, Yoshimura E. Anal Chem. 2010;82:9983–9988. doi: 10.1021/ac100806x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.