Abstract
Nectar is a primary nutrient reward for a variety of pollinators. Recent studies have demonstrated that nectar also has defensive functions against microbial invasion. In this study, the Liriodendron tulipifera nectary was first examined by scanning electron microscopy, and then the nectar was analyzed by two-dimensional gel electrophoresis and liquid chromatography–tandem mass spectrometry, which led to identification of 42 nectar proteins involved in various biological functions. Bioinformatic analysis was then performed on an identified novel rubber elongation factor (REF) protein in L. tulipifera nectar. The protein was particularly abundant, representing ∼60% of the major bands of 31 to 43 kDa, and showed high, stage-specific expression in nectary tissue. The REF family proteins are the major allergens in latex. We propose that REF in L. tulipifera nectar has defensive characteristics against microorganisms.
Keywords: Liriodendron tulipifera, morphology, nectar protein, proteomics, defense
Introduction
Nectar secretion is an evolutional adaptation of many angiosperms to attract pollinators especially flying insects—for outcrossing, and nectar serves as a nutrient reward and energy source for pollinators (Nicolson and Thornburg, 2007a). Nectar is a very complex solution that is secreted by a specialized organ called the nectary. The secretion of nectar begins when flowers blossom and is usually under developmental control (Carter et al., 1999). After pollination, the nectar is frequently resorbed (Búrquez and Corbet, 1991). Floral nectar is rich in carbohydrates and amino acids (Zha et al., 2012). Other substances such as organic acids (Baker and Baker, 1975), glycosides (Roshchina and Roshchina, 1993), vitamins (Griebel and Hess, 1940), alkaloids (Deinzer et al., 1977) and flavonoids (Rodriguez-Arce and Diaz, 1992) are also present at low concentrations.
Evidence from >1500 species of flowering plants has demonstrated that the occurrence of amino acids in nectar is universal among nectariferous plants (Baker and Baker, 1986). Amino acids in nectar play a role in attracting protectors to protect plant (LANZA, 1991). Some essential amino acids in nectar from a particular flower is a crucial reason that pollinators remain changeless despite the nectar robbers and low nectar volumes (Maloof and Inouye, 2000; Petanidou et al., 2006). Furthermore, higher concentrations of amino acids in the robbed flower nectar from damaged tissues may be another reason (Camargo et al., 1984).
Proteins have also been detected in nectar, and it is thought that these proteins supply nectar consumers with organic nitrogen (Heil, 2011). Previous studies have revealed two other primary functions of nectar proteins: defense against microorganisms (Carter et al., 1999; Carter and Thornburg, 2004a; Harper et al., 2010) and post-secretory hydrolysis of nectar sugars into fructose and glucose for protectors (Heil et al., 2005). Proteins in flower nectar of ornamental tobacco (Nicotiana langsdorffii ×N. sanderae) nectar appear to protect the nectar from microbial infestation through the nectar redox cycle (Carter and Thornburg, 2004a; Carter et al., 2006, 2007; Park and Thornburg, 2009). The pathogenesis-related proteins chitinase, glucanases, and thaumatin-like proteins have been identified in nectar (Heil, 2011), as have GDSL lipase and Nectarin IV, which have been suggested to have antimicrobial functions (Kram et al., 2008; Harper et al., 2010). Thus, most of the characterized nectar proteins seem to play a role in protecting nectar against microorganisms. Although it has been some time since the presence of proteins in nectar was reported, there have been few comprehensive analyses of nectar proteins (Nepi et al., 2012).
Liriodendron tulipifera, also called tulip tree or yellow poplar, is a relict plant and one of only two species in this genus. L. tulipifera has one of the largest natural ranges of tree species of the eastern United States (Harlow and Harrar, 1969; Little, 1979). L. tulipifera is considered a basal angiosperm, and its floral and other structural features put it at an ideal phylogenetic position for comparative studies of the evolution of biological processes in land plants (Hunt, 1998; De Craene et al., 2003). Moreover, L. tulipifera is also a nectariferous plant (Liang et al., 2007). In one season, an L. tulipifera tree less than 20 years old can yield 3.6 kg of nectar (Beck and Sims, 1983). However, the structure of the nectary and proteins in nectar of this species are poorly understood.
Proteomics is a large-scale analysis of proteins in cell and organism has become a very important analysis method for protein characterisation (Pandey and Mann, 2000). Since two-dimensional polyacrylamide gel electrophoresis was developed for separating proteins in O’Farrell (1975) and higher resolution protein separation technologies incorporated later (Chen and Harmon, 2006), plant proteomics has largely been used to characterize responses to abiotic stress (Kosova et al., 2011) and in developmental (Hochholdinger et al., 2006; Kaufmann et al., 2011) and secretion (Agrawal et al., 2010) processes. These proteome analyses have often focused on changes at the subcellular level, such as in plastids, mitochondria, the endoplasmic reticulum, and the cell wall and membranes (Mahon et al., 2000; Canovas et al., 2004; Lee et al., 2012).
In this study, scanning electron microscopy (SEM) was used to elucidate the mechanism by which L. tulipifera secretes nectar and to characterize the nectary structure. Two-dimensional (2-D) gel electrophoresis and nano LC-MS/MS were applied to identify proteins in L. tulipifera flower nectar. Bioinformatics revealed potential functions of the proteins in nectar, and we identified a rubber elongation factor (REF) protein that we speculate may play a role in defense against microorganisms.
Materials and Methods
Floral Nectar Collection, pH and Protein Content Determination
Raw nectar of L. tulipifera L. (South Carolina accession) was collected from nearly opened flowers (pre-pollination stage) of an adult tree with a sterile pipette in a provenance trial plantation located in Xiashu, Jiangsu province (119°13′20′′E, 32°7′8′′N) at 6:00–7:00 am in May of 2014. The floral nectar samples were immediately frozen in liquid nitrogen until use.
The pH and protein content of fresh L. tulipifera nectar were determined according to Zha (Zha et al., 2013) and Bradford (Bradford, 1976), using bovine serum albumin as the standard. The averages of triplicate total protein content measurements are presented.
Morphology Observation by SEM
The secreting flowers were fixed in 2.5% (v/v) glutaraldehyde buffered with 0.2 M sodium phosphate buffer (pH 7.2) for SEM. The samples were then post-fixed in 1% (w/v) osmium tetroxide for 1 h and then washed three times in the buffer. The samples were dehydrated in a graded alcohol series and then examined in a Quanta 200 environmental scanning electron microscope at an accelerating voltage of 20 kV (FEI, USA; Zimmermann et al., 2007).
L. tulipifera Nectar Proteins (Nectarines) Separated by 1-D Gel Electrophoresis
Because there were no previous data on nectar proteins of L. tulipifera, we first identified proteins in the L. tulipifera nectar samples. Before electrophoresis, the samples were concentrated and purified with Amicon Ultra 3K centrifugal filter devices (Millipore, USA). The concentrated L. tulipifera nectar (5 μl, ∼10 μg total protein) was boiled in 2× sample buffer for 5 min then analyzed by SDS-PAGE (12.5% acrylamide gel) as described by Laemmli (Laemmli, 1970) with protein molecular weight markers (Bio-Rad, USA). Proteins were visualized by staining with Coomassie Brilliant Blue R-250.
2-D Gel Electrophoresis of L. tulipifera Nectarines and In-Gel Trypsin Digestion
For further high-resolution identification of nectarines for which the isoelectric point (pI) had not been determined, 2-D gel electrophoresis was carried out with wide range (pH 3–10) linear gradient IPG strips. The concentrated and purified L. tulipifera nectar was mixed with rehydration buffer containing 8 M urea, 4% CHAPS, 0.5% pH 3–10 linear IPG buffer and 14 mM dithiothreitol, and the mixture was subjected to isoelectric focusing using Ettan IPGphor III and 13 cm ReadyStrip IPG Strips (GE Healthcare, USA) with an immobilized pH gradient of 3–10 using the following settings: 12 h at 0 V, 2 h at 100 V, 1.5 h at 250 V, 1 h at 500 V, 1.5 h at increasing voltage from 2000 to 8000 V and 1.5 h at 8000 V.
Prior to second-dimension separation, the focused IPG strips were first equilibrated in 10 ml of equilibration buffer [6 M urea, 30% v/v glycerol, 75 mM Tris-HCl (pH 8.8), 2% (w/v) SDS] containing 1% DTT, then 2.5% iodoacetamide instead of DTT in the second equilibration, 15 min each at room temperature on a shaker. The equilibrated strips were transferred to the wells of 12.5% polyacrylamide gels for second-dimension electrophoresis. The wells were sealed with 0.5% agarose. SDS-PAGE was performed using the Ettan DALT six unit (GE Healthcare, USA) until the bromophenol blue dye front reached the bottom of the gel. After SDS-PAGE separation, the gels were stained with silver nitrate and scanned with Image Scanner (GE Healthcare, USA).
The excised protein spots were washed twice with ultrapure water and destained with 30 mM K3Fe(CN)6/100 mM NaS2O3 (1:1, v/v) for 20 min. Gel fragments were then dehydrated with 50% acetonitrile followed with 100% acetonitrile. The dried gel fragments were incubated with trypsin (20 μg/μl, Promega, USA) for 5 min at 4°C then covered and incubated with 25 mM NH4HCO3 in 10% acetonitrile at 37°C overnight (∼14 h) then extracted twice with 50 μl of 50% acetonitrile/5% trifluoroacetic acid (v/v) for 1 h. The extracted peptides were dried by freeze dryer for nanoLC-MS/MS.
LC-MS/MS Analysis
The dried peptides were redissolved in 0.1% formic acid (v/v) and analyzed with a nano-HPLC-MS system (Easy-nLC 1000; Thermo Fisher Scientific, CA), which was coupled to an LTQ-Orbitrap XL instrument (Thermo Fisher Scientific, Bremen, Germany; LTQ, linear trap quadrupole) equipped with a nanoelectrospray interface operated in positive ion mode. All peptides were passed through a μ-Precolumn (C18, 3 μm, 100 Å, 75 μm × 2 cm) before being separated with a reverse phase column (C18, 3 μm, 100 Å, 50 μm × 15 cm, all nanoViper, Thermo Fisher Scientific, CA). The mobile phases consisted of a gradient of solvent A (0.1% formic acid) to solvent B (0.1% formic acid in acetonitrile) run for 107 min per gel spot as follows: 530% B (0–20 min); 30–90% B (20–25 min); 98% B (25–30 min); 90–5% B (30–35 min). Continuum mass spectra data were acquired on an ESI-LTQ-Orbitrap-XL MS (Thermo Scientific, Bremen, Germany; ESI, electrospray ionization) with spray voltage of 2.00 kV and heated capillary temperature set at 175°C in the data-dependent mode of acquisition to automatically switch between Orbitrap-MS and LTQ-MS/MS acquisition. The six most intense precursor ions were sequentially isolated for fragmentation in the LTQ with CID, and the normalized collision energy was set to 35% with activation time of 30 ms. Activation Q was 0.25. Dynamic exclusion settings were repeat counts 2, repeat duration 30 s and exclusion duration 90 s. Survey full scan MS spectra (from m/z 350 to 1800) were acquired in the Orbitrap.
Three independent experiments were carried out. The proteins identified were similar in the three experiments.
Data Processing and Protein Annotation
Raw files were acquired with Xcalibur 2.1.0 and converted into MGF format by Thermo Proteome Discoverer software v. 1.1.0.263 (Thermo Scientific, CA). The exported MGF files were searched against the local database downloaded from UniProtKB (Taxonomy: Viridiplantae, containing 36,020 sequences) with the MASCOT software (version 2.3, installed on a local server). The search criteria were as follows: enzyme, trypsin; fixed modification, carbamidomethyl (cysteine); variable modification, oxidation (methionine); peptide tolerance, 10 ppm; fragment mass tolerance, ±0.6 Da; peptide charge state, 2+, 3+; instrument profile, ESI-Trap; and one max missed cleavage. Hits were considered high confidence if at least three peptides were matched with ion scores >25 or proteins were identified by one or two peptides with score ≥40.
Real-Time Quantitative PCR
Leaf buds, shoot and leaves were collected with the nectar, as well as petals, bract, stamen, and stigma of highly secreting flowers. The different stages of nectary development were designated S1–S4 (Figure 1). Stage 1 (S1): 10 days before nectar secretion, the nectary area was light green. Stage 2 (S2): 3 days before secreting nectar, nectary area beginning to yellow. Stage 3 (S3): flowers beginning to intumesce and secrete nectar, nectary area fully yellow. Stage 4 (S4): 20 h after stage 3, nectary area was bright orange. Total RNA of all samples was extracted using the Total RNA Purification kit (Norgen, Canada). Quality and concentration of isolated RNA were assessed with a Nanodrop 2000 (Thermo Fisher Scientific, USA) and agarose gel electrophoresis.
FIGURE 1.
Developmental stages of Liriodendron tulipifera flower buds and nectaries. (S1) Ten days before nectar secretion, the nectary area was light green; (S2) 3 days before secreting nectar, nectary area beginning to yellow; (S3) flowers beginning to intumesce and secrete nectar, nectary area fully yellow; (S4) 20 h after stage 3, nectary area was bright orange. Red rectangles delineate the nectary area on the petals. The white bar is 1 cm.
cDNA was synthesized from total RNA using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, USA). Specific primers were designed according to the open reading frames in the scaffolds (Supplementary Table S1) using MacVector 11.1.0 software (MacVector, USA). The primers are shown in Supplementary Table S2. According to preliminary experiments, the optimal amount of cDNA and the number of PCR cycles corresponding to the exponential phase of the reaction were determined. Constitutively expressed 18S rRNA was amplified (primers also shown in Supplementary Table S2) as a loading control.
To investigate the relative expression of genes in different organs and stages, real-time PCR were performed using the LightCycler 480 II Real-Time PCR System with the LightCycler 480 SYBR Green I Master mix (Roche, Switzerland) and the following program: 5 min at 95°C (1 cycle); 10 s at 95°C, 15 s at 60°C and 20 s at 72°C (45 cycles). Semi-quantitative RT PCR was then performed according to the protocol in the KOD FX Polymerase manual (Toyobo, Japan).
The experiments shown are representative of independent experiments with triplicate assays.
Bioinformatic Analysis
To facilitate analysis of the functions of L. tulipifera nectar proteins and also to provide a resource for the study of other nectarines, we used Blast2GO assignments to annotate the nectar proteins with gene ontology (GO) terms. All of the annotated proteins were submitted to GO analysis using Blast2GO 3.0.9 software (Conesa et al., 2005) with default parameters. The analysis was performed by searching (BLASTp) the translated protein sequences from the open reading frames in the scaffolds (unpublished data) against the SWISS-PROT database using the public NCBI BLAST service. Only the statistically significant alignments (<1.0 E–5) were considered.
To better understand the functions and interactions of the identified L. tulipifera nectar proteins, a protein–protein interaction network analysis was performed with the online analysis tool STRING1 (version 10) with a confidence level of 4. Because the protein annotation was based on different organisms in the SWISS-PROT Viridiplantae database, all identified proteins were searched against the Arabidopsis thaliana protein database to obtain annotated protein entries for the network analysis.
Protein hydrophobicity was analyzed with ProtScale, one of the ExPASy online tools, with the Kyte & Doolittle hydrophobicity scale for amino acids. The BioEdit software version 7.0.9.0 was used to predict amino acid frequencies. All parameters were the default.
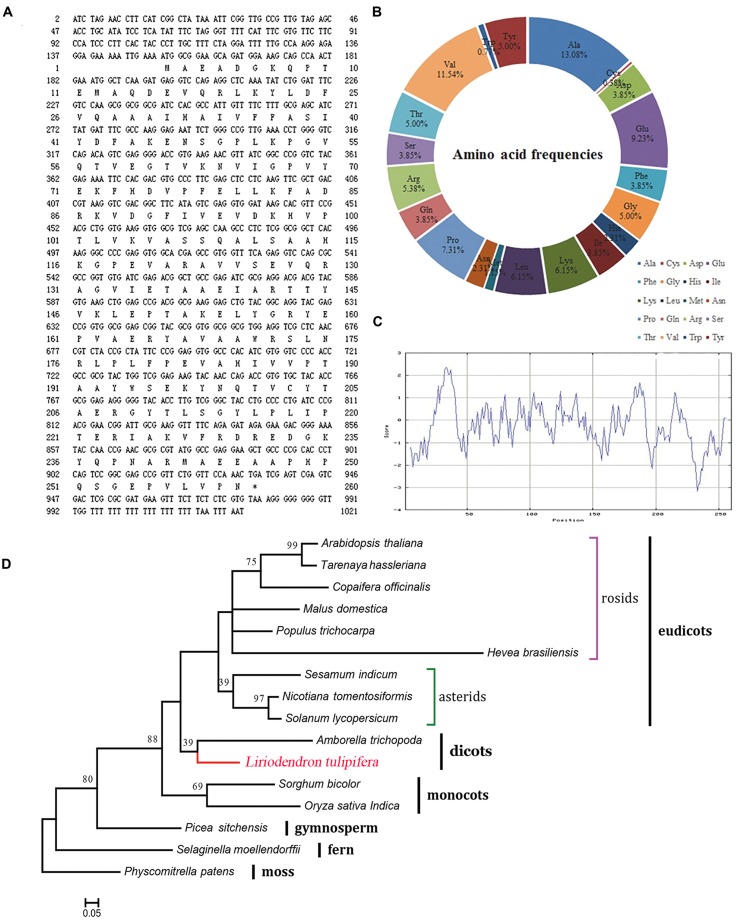
To examine the phylogenetic position of L. tulipifera REF sequence, we selected reported and predicted REF protein sequences of 15 species belonging to 15 different families representing order Eubryales to Brassicales. The multiple sequence alignment was trimmed using trimAL version 1.3 (Capella-Gutiérrez et al., 2009) and the phylogenetic analysis was performed with the MEGA 6 software (Tamura et al., 2013). The protein sequences were used to construct a maximum-likelihood phylogenetic tree (Figure 6D) and Physcomitrella patens was set as the outgroup. Initial trees for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using an LG model. A discrete Gamma distribution was used to model evolutionary rate differences among sites (G = 5, parameter = 1.4675). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The 15 amino acid sequences were downloaded from the NCBI Viridiplantae database (Supplementary Table S3).
FIGURE 6.
Characterization of the REF protein from L. tulipifera. (A) The nucleotide and predicted amino acid sequences. The asterisk marks the stop codon (TGA). (B) The amino acid frequencies. (C) ExPasy ProtScale hydrophobicity analysis with the Kyte & Doolittle scale and window size of 9. Positive values indicate hydrophobicity and negative values indicate hydrophilicity. (D) Maximum-likelihood phylogenetic tree of the homologous amino acid sequences. The scale bar indicates the branch length that corresponds to 0.05 substitutions per site. The red line shows L. tulipifera. The species and accession numbers are listed in Supplementary Table S3. Numbers below each node are bootstrap support values.
Results and Discussion
L. tulipifera General Floral Nectar Traits and Nectary Morphology
Liriodendron tulipifera nectar was first secreted when the flower buds began to intumesce (S3, Figure 1). In this stage, the anthers was still not fully mature but the gynoecium had been mature for fertilization. A lot of yellow stripes first occurred in the nectary area. Then the secreted nectar volume reached the maximum in the following 24 h (about 2∼4 h later after S4). Meanwhile, the petals began to unfold, while the anthers first became mature and the nectary area became orange–yellow to attract pollinators in the morning. This early secretion strategy would attract multiple insects or other organisms indiscriminately and provide ample opportunity for pollination. This may be a evidence that L. tulipifera is an entomophilous plant. Nearly opened flowers had accumulated ∼1600–2100 μl of raw nectar per flower (n = 20) before pollinators visit. Because of its extraordinary nectar-producing ability and wide distribution in North America, L. tulipifera is valued as a nectar source for honey production and as a source of wildlife food (Liang et al., 2011). L. tulipifera floral nectar was acidic, with pH of 4.8 ± 0.15 (mean ± SD, n = 15). The mean total protein content in pooled nectar samples was 130 ± 28 μg ml–1 (mean ± SD, n = 9) and exceeded the mean value of 100 μg ml–1 in floral nectar of most other species (Nicolson and Thornburg, 2007b).
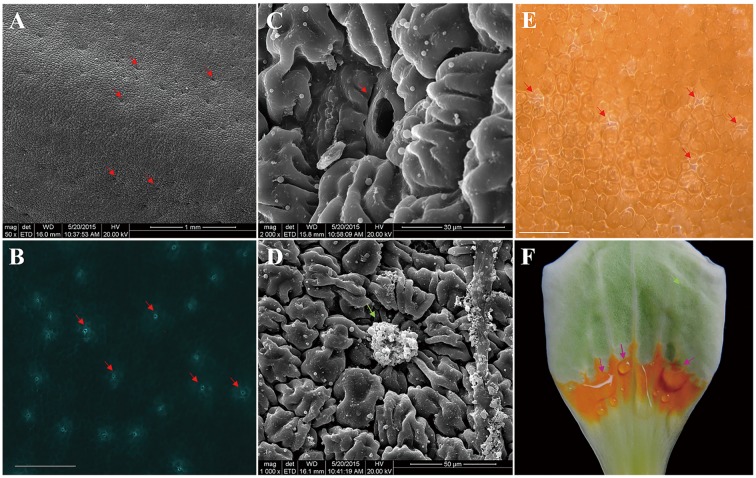
In stage S3, L. tulipifera nectar was secreted from the individual nectarostomata (Figures 2B,E), then formed liquid nectar drops onto the petal internal surface, where there was a yellow–orange fleshy ring (Figure 2F). In stage S4, the liquid nectar drops formed even bigger drops and flowed down to the base of petals. To further study the nectary structure, petals were examined by SEM. We observed that L. tulipifera nectar was exuded through numerous modified, sunken stomata in the epidermis of the lower glabrous part of the petals, which are made up of guard and epidermal cells (Figure 2C). The outer cuticular layer cells were more evident than guard cells before secretion, and they gradually collapsed after secretion (Figures 2A,D).
FIGURE 2.
Photographs of L. tulipifera nectary. (A) Glabrous internal surface of a petal showing the secretory stomata (red arrows). Magnification is 50×. (B) Autofluorescence image of Secretory stomata in GFP (red arrows). Magnification is 100×. Bar is 100 μm. (C) Higher magnification image of a secretory stoma of the nectary. Magnification is 2000×. (D) Higher magnification of a secretory stoma occluded with secretory material. Magnification is 1000×. (E) Secreting L. tulipifera nectar stomata (red arrows). Bar is 100 μm. (F) Isolated petal showing the orange ring and raw nectar (pink arrows). Bar is 1 cm.
Flowers can attract pollinating insects in several ways: they can offer beneficial shelter, they may have specialized color, odor, or pheromonal attractants or they may offer floral reward (Faegri and Van der Pijl, 1971; Miller, 1883). In fact, most flower, including those of L. tulipifera, rely primarily on the last category (Lundgren, 2009). L. tulipifera nectaries secrete thousands of μl of nectar per flower, whereas nectaries in most species generally produce <10 μl (Pacini et al., 2003). Furthermore, like the brightly colored flowers of others species, the colorful nectary of L. tulipifera attracts insects (Kevan, 1972). In conclusion, incorporation of these multiple strategies to attract insects can improve the fecundity of an entomophilous plant.
Nectary structure and position can differ among flowers to the point of being useful for taxonomic classification (Fahn, 1979). The nectary is typically composed of epidermis, with or without stomata, which normally mediates nectar release; parenchyma, which produces or stores substances that become dissolved in the nectar; and the vascular bundle, which conveys water or nutrients to the parenchyma (Pacini et al., 2003). Nectar secretion through stomata is the most common manner of nectar release (Bernardello, 2007; Nepi, 2007). Although L. tulipifera nectar is secreted through nectarostomata, the L. tulipifera flower nectary structure is very different than that of Brassicaceae flowers (Kram and Carter, 2009) and even that of Magnolia stellata in the same Magnoliaceae family (Erbar, 2014). As an apocarpous gynoecium flower, the nectary of L. tulipifera flowers was located on the modified orange–yellow part of petals, as in flowers of Helleborus and Symphyglossum (Vesprini et al., 1999; Stpiczynska and Davies, 2006), whereas in most species, it encircles the ovary (Brown, 1938; Zer and Fahn, 1992; Rivera, 2000; Konarska, 2010; Nocentini et al., 2012; Stpiczyñska et al., 2012; Lüttge, 2013; Stephens, 2013). Although this result was consistent with previous findings in plants in the Ranunculaceae family (Kosuge, 1994), it was different from what has been seen in M. stellata in the same family (Erbar and Leins, 2013). As the secreting petals produce a colorful ring, we hypothesize that the bright-colored and glabrous nectary tissues of L. tulipifera may be more favorable for attracting its pollinator.
L. tulipifera Nectarin Annotation by Gel Electrophoresis and LC-MS/MS
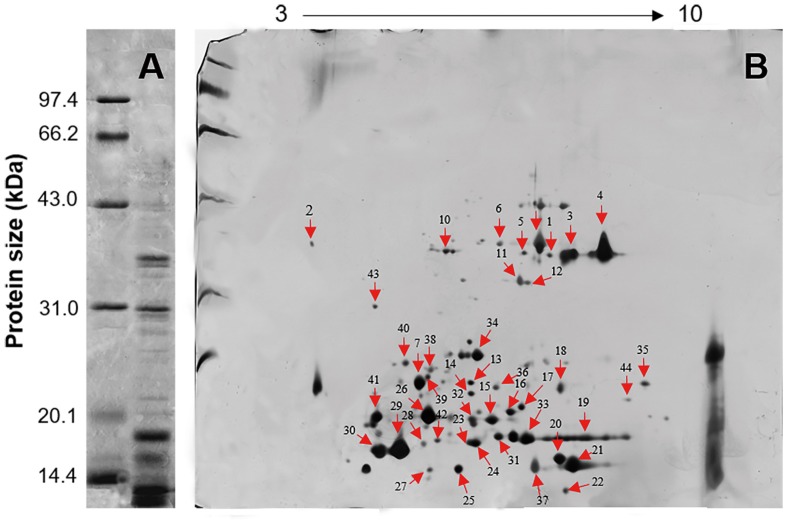
Five distinct main bands, ranging in size from 10 to 41 kDa, were yielded by 1-D SDS-PAGE and visualized by Coomassie Brilliant Blue R-250 staining under reducing conditions (Figure 3A). The 2-D gel electrophoresis showed that most of the L. tulipifera nectarines were acidic proteins ranging in molecular mass from 10 to 40 kDa, consistent with the patterns observed in the 1-D gel electrophoresis (Figure 3B).
FIGURE 3.
Gel electrophoresis of L. tulipifera nectar proteins. (A) 1D gel electrophoresis of L. tulipifera nectar proteins. Lane 1, reference proteins; lane 2, 10 μg of L. tulipifera nectar protein. (B) Identification of L. tulipifera nectar proteins (numbered arrows) separated by 2D gel electrophoresis using nanoLC-MS/MS analysis. Reference proteins were the same as in A and 120 μg of nectar protein was loaded. The protein identifications are listed in Table 1.
Individual L. tulipifera nectar proteins were further separated through 2-D gel electrophoresis, and 42 proteins were successfully annotated (Table 1) by searching against the SWISS-PROT database downloaded from UniProtKB. The proteins between 20 and 30 kDa were annotated as binding proteins, ribosylation factor, amino acid transferase and reductase (Figure 3B; Table 1). Those between 14 and 20 kDa were annotated as small ubiquitin-related modifier and carbonic anhydrase (Figure 3B; Table 1). The most abundant proteins below 14 kDa were ubiquitin-related proteins, REF, glutaredoxin and profilin (Figure 3B; Table 1). Interestingly, ubiquitin-related proteins and REF have not been observed in flower nectar of other plants, and no nectarin (NEC)-like proteins were found in L. tulipifera nectar (Carter et al., 1999; Ge et al., 2000; Carter and Thornburg, 2004b,c; Saglar Naqvi et al., 2005). However, carbonic anhydrase was detected in our survey and NEC3 in ornamental tobacco plants has been demonstrated to have the same carbonic anhydrase activity (Carter and Thornburg, 2004b). Therefore, some L. tulipifera nectar proteins may have analogous functions with known nectarines, but more information is needed to characterize their homology.
Table 1.
Floral nectar proteins of Liriodendron tulipifera Linn. (South Carolina accession) annotated by LC-MS/MS after 2-D gel electrophoresis.
| Spot | Accession | Protein name | Score | Coverage % | MW/pI∗ | Hit Peptides | Species |
|---|---|---|---|---|---|---|---|
| 1 | A2Y7R5 | GTP-binding nuclear protein Ran-2 | 225 | 19 | 25.36/6.66 | LVIVGDGGTGK HLTGEFEK VCENIPIVLCGNK NLQYYEISAK |
Oryza sativa Indica Group |
| 2 | P15252 | Rubber elongation factor protein | 160 | 34 | 14.71/5.04 | SGPLQPGVDIIEGPVK NVAVPLYNR DASIQVVSAIR SLASSLPGQTK |
Hevea brasiliensis |
| 3 | A2Y7R5 | GTP-binding nuclear protein Ran-2 | 162 | 21 | 25.36/6.66 | HLTGEFEK LVIVGDGGTGK NLQYYEISAK VCENIPIVLCGNK |
Oryza sativa Indica Group |
| 4 | P15252 | Rubber elongation factor protein | 244 | 27 | 14.71/5.04 | SGPLQPGVDIIEGPVK DASIQVVSAIR SLASSLPGQTK |
Hevea brasiliensis |
| 5 | Q43209 | Protein-L-isoaspartate O-methyltransferase | 451 | 7 | 24.81/4.90 | VAEVMETIDR YVPLTSR |
Triticum aestivum |
| 6 | P11143 | Heat shock 70 kDa protein | 814 | 18 | 70.87/5.22 | VEIIANDQGNR TTPSYVAFTDTER QFAAEEISSMVLIK EIAEAYLGSTIK NAVVTVPAYFNDSQR DAGVIAGLNVMR IINEPTAAAIAYGLDK ATAGDTHLGGEDFDNR MVNHFVQEFK |
Zea mays |
| 7 | O82089 | Copper transport protein CCH | 255 | 12 | 13.08/4.91 | MEGVESFDIDIK MEGVESFDIDIKEQK |
Arabidopsis thaliana |
| 10 | Q9FRL8 | Glutathione S-transferase DHAR2 | 115 | 9 | 23.51/5.79 | VLLTLEEK NWSVPESLTSVR |
Arabidopsis thaliana |
| 11 | Q0JNR2 | Cysteine proteinase inhibitor 12 | 322 | 3 | 27.25/6.07 | ENALLEFVR | Oryza sativa Japonica Group |
| 12 | Q9SVD7 | Ubiquitin-conjugating enzyme E2 variant 1D | 264 | 34 | 16.69/6.20 | TLGSGGSSVVVPR LLEELER GIGDGTVSYGMDDGDDIYMR KLVQPPEGTCF |
Arabidopsis thaliana |
| 13 | P49310 | Glycine-rich RNA-binding protein GRP1A | 634 | 38 | 16.06/5.21 | ASPDVEYR CFVGGLAWATDDR DAIEGMNGQDLDGR SITVNEAQSR SGGGGGYGGGGGGYGGGGR |
Sinapis alba |
| 14 | A2XKU9 | Costars family protein | 173 | 27 | 9.63/5.78 | VTFGVIFNDDR CANIFEALVGTLR |
Oryza sativa Indica Group |
| 15 | P29449 | Thioredoxin H-type 1 | 2221 | 18 | 14.12/5.62 | KLVVVDFTASWCGPCR LVVVDFTASWCGPCR VDVDELK |
Nicotiana tabacum |
| 16 | P15252 | Rubber elongation factor protein | 378 | 52 | 14.71/5.04 | SGPLQPGVDIIEGPVK NVAVPLYNR FSYIPNGALK FVDSTVVASVTIIDR DASIQVVSAIR SLASSLPGQTK |
Hevea brasiliensis |
| P29449 | Thioredoxin H-type 1 | 217 | 18 | 14.12/5.62 | VDVDELK LVVVDFTASWCGPCR KLVVVDFTASWCGPCR |
Nicotiana tabacum | |
| 17 | Q84JT6 | Peptide methionine sulfoxide reductase B9 | 128 | 12 | 15.75/6.81 | AILSPEQFR HCVNSVSLK |
Arabidopsis thaliana |
| 18 | P35135 | Ubiquitin-conjugating enzyme E2-17 kDa | 237 | 47 | 16.68/7.71 | ELKDLQK VFHPNINSNGSICLDILK EQWSPALTISK VLLSICSLLTDPNPDDPLVPEIAHMYK AKYETTAR |
Solanum lycopersicum |
| Q03250 | Glycine-rich RNA-binding protein 7 | 633 | 21 | 16.94/5.85 | SITVNEAQSR CFVGGLAWATDDR DAIEGMNGQDLDGR |
Arabidopsis thaliana | |
| Q9FVI1 | Actin-depolymerizing factor 2 | 168 | 13 | 16.67/5.78 | MIYASSK IFFIAWSPDTAR |
Petunia × hybrida | |
| 19 | Q94A97 | Ubiquitin-conjugating enzyme E2 35 | 223 | 27 | 17.24/6.74 | LLSEPAPGISASPSEDNMR LELFLPEEYPMAAPK IYHPNIDK |
Arabidopsis thaliana |
| P55142 | Glutaredoxin-C6 | 61 | 30 | 11.94/5.77 | TVPNVFINGK AIELDGESDGSELQSALAEWTGQR |
Oryza sativa Japonica Group | |
| 20 | P69310 | Ubiquitin | 281 | 61 | 8.52/6.56 | TITLEVESSDTIDNVK IQDKEGIPPDQQR EGIPPDQQR TLADYNIQK ESTLHLVLR |
Avena sativa |
| 21 | P0CH10 | Ubiquitin-60S ribosomal protein L40 | 2014 | 54 | 14.92/9.94 | EGIPPDQQR TLADYNIQK ESTLHLVLR MQIFVKTLTGK LIFAGKQLEDGR IQDKEGIPPDQQR TITLEVESSDTIENVK TLADYNIQKESTLHLVLR |
Chlamydomonas reinhardtii |
| 22 | P84718 | Putative oxygen-evolving enhancer protein 1 | 141 | 24 | 12.27/4.35 | VINTWADIINR GGSTGYDNAVALPAGGR |
Pinus strobus |
| 23 | P69310 | Ubiquitin | 184 | 61 | 8.52/6.56 | TITLEVESSDTIDNVK IQDKEGIPPDQQR TLADYNIQK ESTLHLVLR |
Avena sativa |
| P15252 | Rubber elongation factor protein | 318 | 44 | 14.71/5.04 | NVAVPLYNR SLASSLPGQTK FSYIPNGALK SGPLQPGVDIIEGPVK FVDSTVVASVTIIDR |
Hevea brasiliensis | |
| 24 | P29449 | Thioredoxin H-type 1 | 285 | 18 | 14.12/5.62 | KLVVVDFTASWCGPCR LVVVDFTASWCGPCR VDVDELK |
Nicotiana tabacum |
| 25 | P0C030 | Ubiquitin-NEDD8-like protein RUB1 | 1629 | 62 | 17.12/5.77 | TITLEVESSDTIDNVK IQDKEGIPPDQQR EGIPPDQQR TLTGKEIEIDIEPTDTIDR EIEIDIEPTDTIDR VEEKEGIPPVQQR EGIPPVQQR DYNIEGGSVLHLVLALR |
Oryza sativa Japonica Group |
| A2XKU9 | Costars family protein | 181 | 27 | 9.63/5.78 | VTFGVIFNDDR CANIFEALVGTLR |
Oryza sativa Indica Group | |
| 26 | P29449 | Thioredoxin H-type 1 | 1314 | 25 | 14.12/5.62 | VDVDELK EVDRVVGAK LVVVDFTASWCGPCR KLVVVDFTASWCGPCR |
Nicotiana tabacum |
| Q03250 | Glycine-rich RNA-binding protein 7 | 675 | 21 | 16.94/5.85 | CFVGGLAWATDDR DAIEGMNGQDLDGR SITVNEAQSR |
Arabidopsis thaliana | |
| B4YYA9 | Costars family protein ST45-2 | 291 | 26 | 10.19/5.63 | VTFGVLFNDDR CANIFEALVGTLR |
Eutrema halophilum | |
| 27 | P0C030 | Ubiquitin-NEDD8-like protein RUB1 | 265 | 42 | 17.12/5.77 | TITLEVESSDTIDNVK TLADYNIQK EIEIDIEPTDTIDR EGIPPVQQR DYNIEGGSVLHLVLALR |
Oryza sativa Japonica Group |
| 28 | P15252 | Rubber elongation factor protein | 215 | 41 | 14.71/5.04 | SGPLQPGVDIIEGPVK NVAVPLYNR FSYIPNGALK DASIQVVSAIR SLASSLPGQTK |
Hevea brasiliensis |
| 29 | P15252 | Rubber elongation factor protein | 530 | 52 | 14.71/5.04 | SGPLQPGVDIIEGPVK NVAVPLYNR FSYIPNGALK FVDSTVVASVTIIDR DASIQVVSAIR |
Hevea brasiliensis |
| 30 | P15252 | Rubber elongation factor protein | 283 | 41 | 14.71/5.04 | SGPLQPGVDIIEGPVK NVAVPLYNR FSYIPNGALK DASIQVVSAIR SLASSLPGQTK |
Hevea brasiliensis |
| 31 | A4KA43 | Profilin-6 | 120 | 9 | 14.19/4.90 | YMVIQGEPGVVIR | Corylus avellana |
| P15252 | Rubber elongation factor protein | 111 | 44 | 14.71/5.04 | NVAVPLYNR SLASSLPGQTK DASIQVVSAIR SGPLQPGVDIIEGPVK FVDSTVVASVTIIDR |
Hevea brasiliensis | |
| 32 | A2YIW7 | Thioredoxin H-type | 112 | 5 | 13.32/5.16 | VDVDELK | Oryza sativa Indica Group |
| 33 | Q9C996 | GDSL esterase | 109 | 2 | 40.65/8.38 | CFGKMNVMAK | Arabidopsis thaliana |
| 34 | P0DH9 | ADP-ribosylation factor 2-B | 124 | 19 | 20.64/6.43 | ILMVGLDAAGK DAVLLVFANK QDLPNAMNAAEITDK |
Glycine soja |
| 35 | O49886 | Peptidyl-prolyl cis-trans isomerase | 98 | 12 | 18.51/8.36 | TAENFR FADENFIK FADENFIKK TEWLDGK |
Lupinus luteus |
| P49310 | Glycine-rich RNA-binding protein GRP1A | 204 | 33 | 16.06/5.21 | SITVNEAQSR CFVGGLAWATDDR DAIEGMNGQDLDGR |
Sinapis alba | |
| 36 | Q9FVI1 | Actin-depolymerizing factor 2 | 2168 | 13 | 16.67/5.78 | MIYASSK IFFIAWSPDTAR |
Petunia × hybrida |
| 37 | P0CG86 | Ubiquitin-40S ribosomal protein S27a | 158 | 28 | 17.87/9.83 | MQIFVK IQDKEGIPPDQQR TLADYNIQK ESTLHLVLR VDDATGKVTR |
Hordeum vulgare |
| 38 | P15252 | Rubber elongation factor protein | 439 | 52 | 14.71/5.04 | NVAVPLYNR SLASSLPGQTK FSYIPNGALK DASIQVVSAIR FVDSTVVASVTIIDR |
Hevea brasiliensis |
| Q9FVI1 | Actin-depolymerizing factor 2 | 765 | 18 | 16.67/5.78 | MIYASSK QKEVVVEK IFFIAWSPDTAR |
Petunia × hybrida | |
| 39 | Q03250 | Glycine-rich RNA-binding protein 7 | 326 | 21 | 16.94/5.85 | SITVNEAQSR CFVGGLAWATDDR DAIEGMNGQDLDGR |
Arabidopsis thaliana |
| 132270 | Rubber elongation factor protein | 338 | 52 | 14.71/5.04 | SGPLQPGVDIIEGPVK NVAVPLYNR FSYIPNGALK FVDSTVVASVTIIDR DASIQVVSAIR SLASSLPGQTK |
Hevea brasiliensis | |
| 40 | Q9FLP6 | Small ubiquitin-related modifier 2 | 439 | 35 | 11.76/5.35 | LMNAYCDR GQDGNEVFFR KLMNAYCDR VKGQDGNEVFFR QSVDFNSIAFLFDGR |
Arabidopsis thaliana |
| Q9FVI1 | Actin-depolymerizing factor 2 | 188 | 13 | 16.67/5.78 | MIYASSK IFFIAWSPDTAR |
Petunia × hybrida | |
| 41 | Q9SNW5 | Profilin-3 | 600 | 16 | 14.27/4.73 | KGSGGVTIK YMVIQGEPGAVIR |
Lilium longiflorum |
| 42 | A4KA43 | Profilin-6 | 169 | 9 | 14.19/4.90 | YMVIQGEPGVVIR | Corylus avellana |
| P15252 | Rubber elongation factor protein | 167 | 26 | 14.71/5.04 | SGPLQPGVDIIEGPVK NVAVPLYNR SLASSLPGQTK |
Hevea brasiliensis | |
| 43 | Q9ZSW9 | Translationally-controlled tumor protein homolog | 673 | 27 | 19.08/4.50 | QFVTYMK VVDIVDTFR LQEQPAFDK MLVYQDLLTGDELLSDSFPYK |
Hevea brasiliensis |
| Q0JNS6 | Calmodulin-1 | 181 | 37 | 16.88/4.11 | ELGTVMR HVMTNLGEK LTDEEVDEMIR DTDSEEELKEAFR VFDKDQNGFISAAELR |
Oryza sativa Japonica Group | |
| 44 | Q5Z9Z3 | Thioredoxin-like protein Clot | 219 | 7 | 15.34/4.88 | LTGVPTLIR FRLTGVPTLIR |
Oryza sativa Japonica Group |
∗The numerical value of Mw and pI just represent the accession, not the experimental value.
Although it has been known that nectar contains proteins for some time, few nectarines have been characterized in detail (Peumans et al., 1997; Carter and Thornburg, 2000, 2004b,c; Thornburg et al., 2003; Kram et al., 2008; Harper et al., 2010; Hillwig et al., 2010, 2011; Nepi et al., 2011; Zha et al., 2013). During our proteomics survey, we detected isomerase, transferase, carbonic anhydrase, short-chain dehydrogenase reductase, ATPase, diphosphate kinase, and GDSL esterase in L. tulipifera floral nectar. One of GDSL esterase/lipases identified in our survey (spot 33) has been reported to have antimicrobial activities in both Jacaranda mimosifolia (Kram et al., 2008) and Arabidopsis (Oh et al., 2005) nectar. A diversity of defense proteins, like NADPH oxidase, endochitinase, β-1, 3-glucanases and xylosidase have been shown to defense against microorganisms in floral and extrafloral nectar (Carter and Thornburg, 2000, 2004a; Saglar Naqvi et al., 2005; Carter et al., 2007; Kram et al., 2008; González-Teuber et al., 2009, 2010; Hillwig et al., 2011; Nepi et al., 2011). Thus, we speculate that the enzymes identified in L. tulipifera nectar may also play an important role in the interaction of nectar with the biotic environment. The functions of other enzymes in the nectar are still unclear and may be explored in future studies.
The ubiquitin-related proteins are involved in many biological processes in almost all organisms. The presence of these proteins in secreted nectar may indicate contamination of the secreted floral nectar with cellular proteins arising from natural cellular degradation occurring during the nectar secretion process. This cellular degradation has previously been observed in the ornamental tobacco nectary (Carter et al., 2007).
Classification of L. tulipifera Nectar Proteins
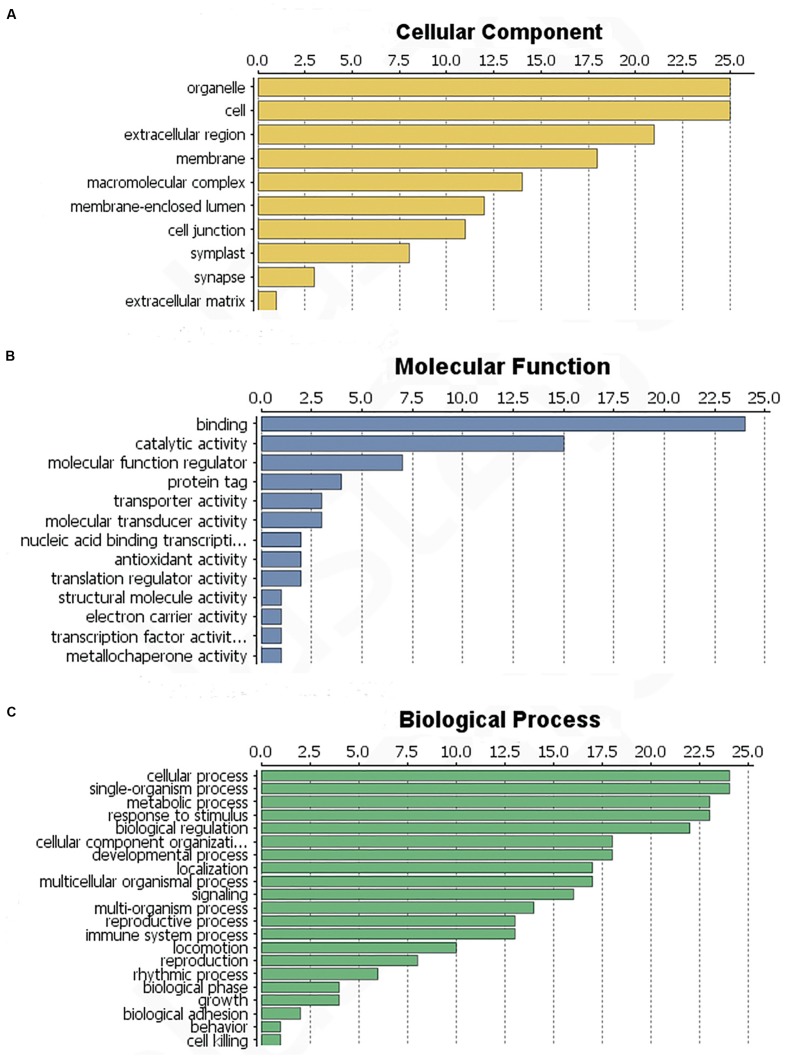
The identified proteins were categorized according to the three main GO categories: cellular component, molecular function, and biological process. In terms of cellular component, the largest group of proteins corresponded to the cell and organelle subcategories, followed by extracellular region, membranes, macromolecular complexes and the membrane-enclosed lumen (Figure 4A). In molecular function category of the identified proteins, the most prominent was binding, catalytic activity and molecular function regulation. Antioxidant activity was also found, but in a low component (Figure 4B). For biological process category, single-organism processes, response to stimulus, cellular processes and metabolic process were top terms (Figure 4C). In summary, most of the identified proteins were involved in catalytic activity, binding and antioxidant activity, response to stimulus and immune system processes.
FIGURE 4.
Functional classification of the identified L. tulipifera floral nectar proteins. (A) Cellular component profile. (B) Molecular function profile. (C) Biological process distribution. Numbers on the x-axes indicate the number of proteins.
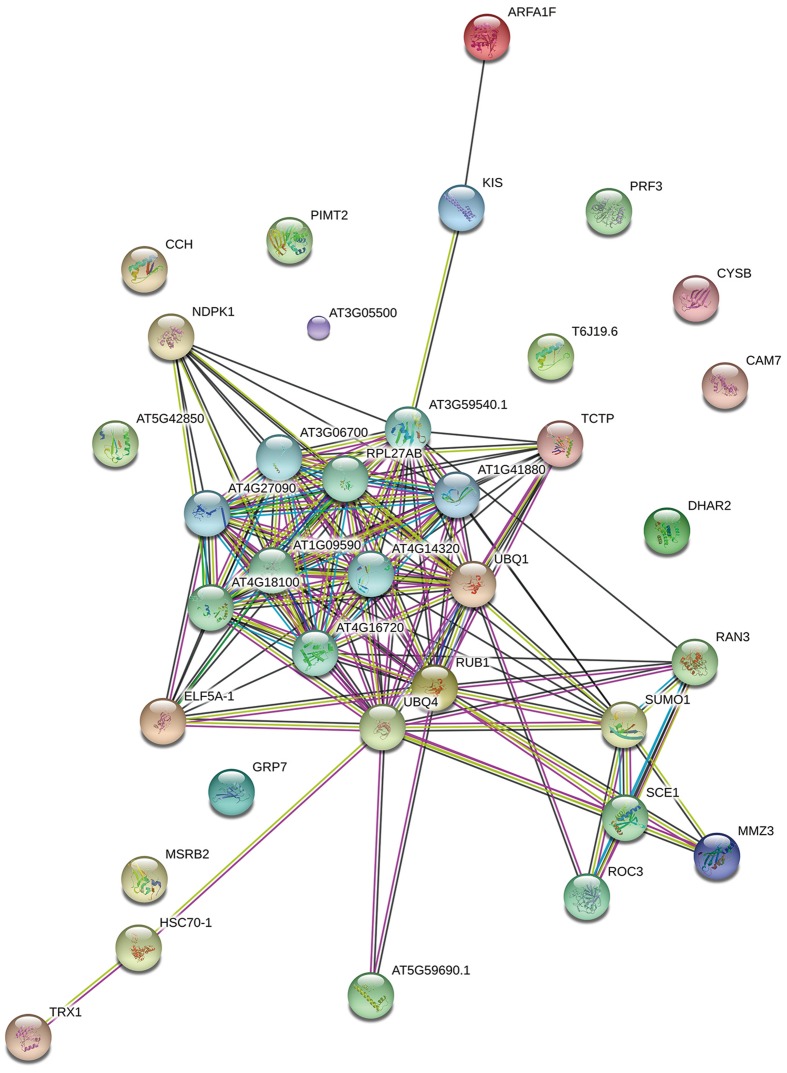
To determine the function of proteins involved in the various biological processes and the protein–protein interactions, the identified protein sequences were submitted to STRING online. The protein–protein interactions are shown in Figure 5. Many ubiquitination-related proteins and kinases were involved in the predicted interactions. Ubiquitination is an important post-translational protein modification and regulates a wide range of cellular processes, including responses to hormones, light, sucrose, development signals, and pathogens (Ellis et al., 2002). Previous studies revealed that ubiquitination may also play an important role in plant defense against pathogens. (Xie et al., 1998; Kim and Delaney, 2002; Devoto et al., 2003; Lee et al., 2007). This suggests that L. tulipifera flower nectar may have defense functions. This new protein database will be a valuable resource for further studies on L. tulipifera flower nectar proteins.
FIGURE 5.
Functional networks of the identified L. tulipifera floral nectar proteins. Different line colors represent the types of evidence used in predicting the associations: gene fusion (red), neighborhood (green), co-occurrence across genomes (blue), co-expression (black), experimental (purple), association in curated databases (light blue), text mining (yellow), homology (light purple).
Sequence and Expression Analysis of L. tulipifera REF
Among the proteins that were identified by our approach, we found L. tulipifera REF (spot 4), which had not been detected in previous nectar research. REF, a member of the REF/SRPP-like protein family, is the major allergen in latex (Czuppon et al., 1993), and it has also been shown to be involved in resistance mechanisms (Ko et al., 2003).
The open reading frame is 1021 bp in length, encoding a putative 260-amino acid protein (Figure 6A). The theoretical isoelectric point is 5.93 and the molecular weight is 28.82 kDa. To verify the conserved domain we submitted the protein sequence to the SMART server and aligned with Hevea brasiliensis REF sequence (Hev b1, P15252). The protein sequence contains a 216-aa conserved domain and is highly consistent with the H. brasiliensis sequence (Supplementary Figure S1). The frequencies of amino acids were deduced with the BioEdit software. Notably, alanine and cysteine were the most (13.08%) and least (0.38%) frequently coded amino acids (Figure 6B), respectively, and the peptide contained all 20 amino acids. The protein was predicted to be hydrophilic, with an overall average hydrophobicity score of -0.238 (Figure 6C).
Phylogenetically, L. tulipifera REF fell into the clade of dicots between monocots and eudicots, and it was closely related to Amborella trichopoda. This phylogenetic topology was congruent with previous phylogenetic analyses (Berthelot et al., 2014b); further supporting that L. tulipifera REF is a REF protein.
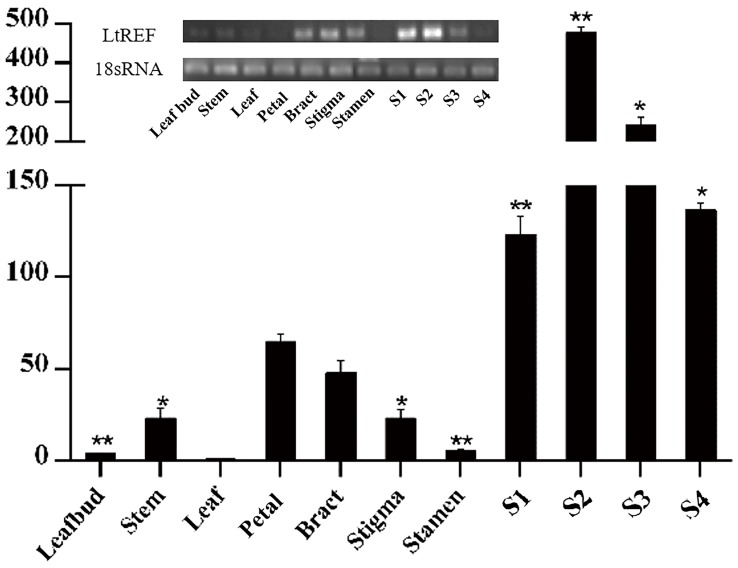
To further analyze the L. tulipifera REF, we performed real-time PCR to detect expression of the L. tulipifera REF gene in different tissues and, in particular, in different stages of nectary development. REF’s expression was scarcely detectable in leaves, leaf bud, and stem, and was higher in petals and bracts. By contrast, the expression was dramatically higher in the nectary. To be more specific, the expression was the highest in S2 and steadily decreased in S3 and S4 (Figure 7). In summary, the expression in reproductive organs was higher than that in vegetative organs. As a whole, the tendencies observed in the real-time PCR were consistent with the semi-quantitative RT PCR results (Figure 7). The increased expression of L. tulipifera REF in S2 may be in preparation for protein secretion in S3.
FIGURE 7.
Quantitation of expression of the REF gene in different tissues of L. tulipifera. S1–S4 correspond to the nectary tissue developmental stages in Figure 1 (∗∗P < 0.01, ∗P < 0.05).
We found that L. tulipifera REF, like MuSI in sweet potato (Seo et al., 2010; Kim et al., 2011), contains a REF domain and is highly similar to H. brasiliensis REF. REF is also able to interact with the membranes of yeasts and erythrocytes, leading to their agglutination (Berthelot et al., 2014a). However, antifungal or antimicrobial activities of REF have not been clearly demonstrated (Kanokwiroon, 2007). It is thus possible that REF has antimicrobial or other activities, and, given the homology, L. tulipifera REF may play a defensive role in L. tulipifera nectar.
Conclusion
In our investigation, we first characterized the nectary structure of L. tulipifera by morphological observation. The L. tulipifera nectar was secreted from modificatory stoma as most other species. Nonetheless, the nectary of L. tulipifera was positioned in a more colorful and accessible area on the petals and secreted considerably more nectar, which was distinctly different from other genus in Magnoliaceae. These differences were vitally advantageous to attract insects for pollination.
More importantly, we applied proteomic and bioinformatic approaches to obtain a proteomic description of L. tulipifera nectar. Among the 42 identified proteins in the nectar of L. tulipifera, most of them are involved in catalytic activity, antioxidant activity, response to stimulus, biological regulation and immune system processes. In addition, a REF protein was also detected in nectar for the first time. Further bioinformation and expression analysis suggested that L. tulipifera REF have allergic characteristic and may play a defensive role against microorganisms in nectar. This research provides valuable information both on nectary structure and proteins in nectar, but further studies are necessary to fully elucidate the functions of all nectar proteins.
Author Contributions
JC and JS designed the experiment, prepared samples and drafted the manuscript. YZ, ML, FZ, HZ, and LY performed the experiment. YL and GW contributed to the reagents and performed the experiment. YZ analyzed, interpreted the LC-MS/MS data, and drafted the manuscript. All authors contributed to and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by National High Technology Research and Development Program of China (863 Program, 2013AA102705), Specialized Research Fund for the Doctoral Program of Higher Education (SRFDP, 20113204130002), National Science Foundation of China (No. 31170619), Program for New Century Excellent Talents in University, Talent project by the Ministry of Science and Technology, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, and Priority Academic Program Development of Jiangsu Higher Education Institutions.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00826
References
- Agrawal G. K., Jwa N. S., Lebrun M. H., Job D., Rakwal R. (2010). Plant secretome: unlocking secrets of the secreted proteins. Proteomics 10 799–827. 10.1002/pmic.200900514 [DOI] [PubMed] [Google Scholar]
- Baker H., Baker I. (1986). The occurrence and significance of amino acids in floral nectar. Plant Syst. Evol. 151 175–186. 10.1007/BF02430273 [DOI] [Google Scholar]
- Baker H. G., Baker I. (1975). Studies of nectar-constitution and pollinator-plant coevolution. Coevol. Anim. Plants 100 591–600. [Google Scholar]
- Beck D. E., Sims D. H. (1983). “Yellow-poplar,” in Silvicultural Systems for the Major Forest Types of the United States, Agriculture Handbook Vol. 445 ed. Burns R. M. (Washington, DC: U.S. Department of Agriculture, Forest Service; ), 180–182. [Google Scholar]
- Bernardello G. (2007). “A systematic survey of floral nectaries,” in Nectaries and Nectar, eds Nicolson S. W., Nepi M., Pacini E. (Dordrecht: Springer; ), 19–128. [Google Scholar]
- Berthelot K., Lecomte S., Estevez Y., Coulary-Salin B., Peruch F. (2014a). Homologous Hevea brasiliensis REF (Hevb1) and SRPP (Hevb3) present different auto-assembling. Biochim. Biophys. Acta 1844 473–485. 10.1016/j.bbapap.2013.10.017 [DOI] [PubMed] [Google Scholar]
- Berthelot K., Lecomte S., Estevez Y., Peruch F. (2014b). Hevea brasiliensis REF (Hev b 1) and SRPP (Hev b 3): an overview on rubber particle proteins. Biochimie 106 1–9. 10.1016/j.biochi.2014.07.002 [DOI] [PubMed] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- Brown W. H. (1938). The bearing of nectaries on the phylogeny of flowering plants. Proc. Am. Philos. Soc. 79 549–595. [Google Scholar]
- Búrquez A., Corbet S. (1991). Do flowers reabsorb nectar? Funct. Ecol. 5 369–379. 10.2307/2389808 [DOI] [Google Scholar]
- Camargo J. D., Gottsberger G., Silberbauer-Gottsberger I. (1984). On the phenology and flower visiting behavior of Oxaea flavescens (Klug)(Oxaeinae, Andrenidae, Hymenoptera) in São Paulo, Brazil. Beitr. Biol. Pflanzen. 59 159–179. [Google Scholar]
- Canovas F. M., Dumas-Gaudot E., Recorbet G., Jorrin J., Mock H. P., Rossignol M. (2004). Plant proteome analysis. Proteomics 4 285–298. 10.1002/pmic.200300602 [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C., Graham R. A., Thornburg R. W. (1999). Nectarin I is a novel, soluble germin-like protein expressed in the nectar of Nicotiana sp. Plant Mol. Biol. 41 207–216. 10.1023/A:1006363508648 [DOI] [PubMed] [Google Scholar]
- Carter C., Healy R., Nicole M., Naqvi S. S., Ren G., Park S., et al. (2007). Tobacco nectaries express a novel NADPH oxidase implicated in the defense of floral reproductive tissues against microorganisms. Plant Physiol. 143 389–399. 10.1104/pp.106.089326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C., Shafir S., Yehonatan L., Palmer R. G., Thornburg R. (2006). A novel role for proline in plant floral nectars. Naturwissenschaften 93 72–79. 10.1007/s00114-005-0062-1 [DOI] [PubMed] [Google Scholar]
- Carter C., Thornburg R. W. (2000). Tobacco nectarin I purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defense of floral reproductive tissues. J. Biol. Chem. 275 36726–36733. 10.1074/jbc.M006461200 [DOI] [PubMed] [Google Scholar]
- Carter C., Thornburg R. W. (2004a). Is the nectar redox cycle a floral defense against microbial attack? Trends Plant Sci. 9 320–324. 10.1016/j.tplants.2004.05.008 [DOI] [PubMed] [Google Scholar]
- Carter C. J., Thornburg R. W. (2004b). Tobacco Nectarin III is a bifunctional enzyme with monodehydroascorbate reductase and carbonic anhydrase activities. Plant Mol. Biol. 54 415–425. 10.1023/B:PLAN.0000036373.84579.13 [DOI] [PubMed] [Google Scholar]
- Carter C. J., Thornburg R. W. (2004c). Tobacco nectarin V is a flavin-containing berberine bridge enzyme-like protein with glucose oxidase activity. Plant Physiol. 134 460–469. 10.1104/pp.103.027482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Harmon A. C. (2006). Advances in plant proteomics. Proteomics 6 5504–5516. 10.1002/pmic.200600143 [DOI] [PubMed] [Google Scholar]
- Conesa A., Götz S., García-Gómez J. M., Terol J., Talón M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- Czuppon A. B., Chen Z., Rennert S., Engelke T., Meyer H. E., Heber M., et al. (1993). The rubber elongation factor of rubber trees (Hevea brasiliensis) is the major allergen in latex. J. Allergy Clin. Immunol. 92 690–697. 10.1016/0091-6749(93)90012-5 [DOI] [PubMed] [Google Scholar]
- De Craene L. P. R., Soltis P. S., Soltis D. E. (2003). Evolution of floral structures in basal angiosperms. Int. J. Plant Sci. 164 S329–S363. 10.1086/377063 [DOI] [Google Scholar]
- Deinzer M., Thomson P., Burgett D., Isaacson D. (1977). Pyrrolizidine alkaloids: their occurrence in honey from tansy ragwort (Senecio jacobaea L.). Science 195 497–499. 10.1126/science.835011 [DOI] [PubMed] [Google Scholar]
- Devoto A., Muskett P. R., Shirasu K. (2003). Role of ubiquitination in the regulation of plant defence against pathogens. Curr. Opin. Plant Biol. 6 307–311. 10.1016/s1369-5266(03)00060-8 [DOI] [PubMed] [Google Scholar]
- Ellis C., Turner J. G., Devoto A. (2002). Protein complexes mediate signalling in plant responses to hormones, light, sucrose and pathogens. Plant Mol. Biol. 50 971–980. 10.1023/A:1021291522243 [DOI] [PubMed] [Google Scholar]
- Erbar C. (2014). Nectar secretion and nectaries in basal angiosperms, magnoliids and non-core eudicots and a comparison with core eudicots. Plant Divers. Evol. 131 63–143. 10.1127/1869-6155/2014/0131-0075 [DOI] [Google Scholar]
- Erbar C., Leins P. (2013). Nectar production in the pollen flower of Anemone nemorosa in comparison with other Ranunculaceae and Magnolia (Magnoliaceae). Organ. Divers. Evol. 13 287–300. 10.1007/s13127-013-0131-9 [DOI] [Google Scholar]
- Faegri K., Van der Pijl L. (1971). The Principles of Pollination Ecology. NewYork, NY: Pergamon Press. [Google Scholar]
- Fahn A. (1979). Secretory Tissues in Plants. Cambridge: Academic Press. [Google Scholar]
- Ge Y. X., Angenent G. C., Wittich P. E., Peters J., Franken J., Busscher M., et al. (2000). NEC1 a novel gene, highly expressed in nectary tissue of Petunia hybrida. Plant J. 24 725–734. 10.1111/j.1365-313X.2000.00926.x [DOI] [PubMed] [Google Scholar]
- González-Teuber M., Pozo M. J., Muck A., Svatos A., Adame-Álvarez R. M., Heil M. (2010). Glucanases and chitinases as causal agents in the protection of Acacia extrafloral nectar from infestation by phytopathogens. Plant Physiol. 152 1705–1715. 10.1104/pp.109.148478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Teuber M., Eilmus S., Muck A., Svatos A., Heil M. (2009). Pathogenesis-related proteins protect extrafloral nectar from microbial infestation. Plant J. 58 464–473. 10.1111/j.1365-313X.2009.03790.x [DOI] [PubMed] [Google Scholar]
- Griebel C., Hess G. (1940). The vitamin C content of flower nectar of certain Labiatae. Zeit. Untersuch. Lebensmitt. 79 168–171. 10.1007/BF01662427 [DOI] [Google Scholar]
- Harlow W., Harrar E. S. (1969). Textbook of Dendrology, Vol. 5 New York, NY: McGraw-Hill Book Company, 12; 10.2307/1930600 [DOI] [Google Scholar]
- Harper A. D., Stalnaker S. H., Wells L., Darvill A., Thornburg R., York W. S. (2010). Interaction of Nectarin 4 with a fungal protein triggers a microbial surveillance and defense mechanism in nectar. Phytochemistry 71 1963–1969. 10.1016/j.phytochem.2010.09.009 [DOI] [PubMed] [Google Scholar]
- Heil M. (2011). Nectar: generation, regulation and ecological functions. Trends Plant Sci. 16 191–200. 10.1016/j.tplants.2011.01.003 [DOI] [PubMed] [Google Scholar]
- Heil M., Rattke J., Boland W. (2005). Postsecretory hydrolysis of nectar sucrose and specialization in ant/plant mutualism. Science 308 560–563. 10.1126/science.1107536 [DOI] [PubMed] [Google Scholar]
- Hillwig M. S., Kanobe C., Thornburg R. W., Macintosh G. C. (2011). Identification of S-RNase and peroxidase in petunia nectar. J. Plant Physiol. 168 734–738. 10.1016/j.jplph.2010.10.002 [DOI] [PubMed] [Google Scholar]
- Hillwig M. S., Liu X., Liu G., Thornburg R. W., MacIntosh G. C. (2010). Petunia nectar proteins have ribonuclease activity. J. Exp. Bot. 61 2951–2965. 10.1093/jxb/erq119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F., Sauer M., Dembinsky D., Hoecker N., Muthreich N., Saleem M., et al. (2006). Proteomic dissection of plant development. Proteomics 6 4076–4083. 10.1002/pmic.200500851 [DOI] [PubMed] [Google Scholar]
- Hunt D. (1998). Magnolias and their Allies (Milborne Port: International Dendrology Society and the Magnolia Society; ). [Google Scholar]
- Kanokwiroon K. (2007). Antimicrobials from Latex of Hevea brasiliensis. Ph.D. thesis, Prince of Songkla University, Hat Yai. [Google Scholar]
- Kaufmann K., Smaczniak C., de Vries S., Angenent G. C., Karlova R. (2011). Proteomics insights into plant signaling and development. Proteomics 11 744–755. 10.1002/pmic.201000418 [DOI] [PubMed] [Google Scholar]
- Kevan P. G. (1972). Floral colors in the high arctic with reference to insect-flower relations and pollination. Can. J. Bot. 50 2289–2316. 10.1139/b72-298 [DOI] [Google Scholar]
- Kim H. S., Delaney T. P. (2002). Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell 14 1469–1482. 10.1105/tpc.001867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-N., Kim J.-S., Seo S.-G., Lee Y., Baek S.-W., Kim I.-S., et al. (2011). Cadmium resistance in tobacco plants expressing the MuSI gene. Plant Biotechnol. Rep. 5 323–329. 10.1007/s11816-011-0186-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.-H., Chow K.-S., Han K.-H. (2003). Transcriptome analysis reveals novel features of the molecular events occurring in the laticifers of Hevea brasiliensis (para rubber tree). Plant Mol. Biol. 53 479–492. 10.1023/B:PLAN.0000019119.66643.5d [DOI] [PubMed] [Google Scholar]
- Konarska A. (2010). Flower nectary structure in Cornus alba L. Plant Syst. Evol. 291 1–6. 10.1007/s00606-010-0364-4 [DOI] [Google Scholar]
- Kosova K., Vitamvas P., Prasil I. T., Renaut J. (2011). Plant proteome changes under abiotic stress–contribution of proteomics studies to understanding plant stress response. J. Proteomics 74 1301–1322. 10.1016/j.jprot.2011.02.006 [DOI] [PubMed] [Google Scholar]
- Kosuge K. (1994). Petal Evolution in Ranunculaceae. New York, NY: Springer. [Google Scholar]
- Kram B. W., Bainbridge E. A., Perera M. A. D., Carter C. (2008). Identification, cloning and characterization of a GDSL lipase secreted into the nectar of Jacaranda mimosifolia. Plant Mol. Biol. 68 173–183. 10.1007/s11103-008-9361-1 [DOI] [PubMed] [Google Scholar]
- Kram B. W., Carter C. J. (2009). Arabidopsis thaliana as a model for functional nectary analysis. Sex. Plant Reprod. 22 235–246. 10.1007/s00497-009-0112-5 [DOI] [PubMed] [Google Scholar]
- Laemmli U. (1970). Most commonly used discontinuous buffer system for SDS electrophoresis. Nature 227 680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- LANZA J. (1991). Response of fire ants (Formicidae: Solenopsis invicta and S. gerninata) to artificial nectars with amino acids. Ecol. Entomol. 16 203–210. 10.1111/j.1365-2311.1991.tb00210.x [DOI] [Google Scholar]
- Lee C. P., Eubel H., Solheim C., Millar A. H. (2012). Mitochondrial proteome heterogeneity between tissues from the vegetative and reproductive stages of Arabidopsis thaliana development. J. Proteome Res. 11 3326–3343. 10.1021/pr3001157 [DOI] [PubMed] [Google Scholar]
- Lee J., Nam J., Park H. C., Na G., Miura K., Jin J. B., et al. (2007). Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 49 79–90. 10.1111/j.1365-313X.2006.02947.x [DOI] [PubMed] [Google Scholar]
- Liang H., Ayyampalayam S., Wickett N., Barakat A., Xu Y., Landherr L., et al. (2011). Generation of a large-scale genomic resource for functional and comparative genomics in Liriodendron tulipifera L. Tree Genet. Genomes 7 941–954. 10.1007/s11295-011-0386-2 [DOI] [Google Scholar]
- Liang H., Fang E. G., Tomkins J. P., Luo M., Kudrna D., Kim H. R., et al. (2007). Development of a BAC library for yellow-poplar (Liriodendron tulipifera) and the identification of genes associated with flower development and lignin biosynthesis. Tree Genet. Genomes 3 215–225. 10.1007/s11295-006-0057-x [DOI] [Google Scholar]
- Little E. L., Jr. (1979). Checklist of United States trees (native and naturalized). Agric. Handb. 541 375. [Google Scholar]
- Lundgren J. G. (2009). Relationships of Natural Enemies and Non-Prey Foods. New York, NY: Springer Science & Business Media. [Google Scholar]
- Lüttge U. (2013). Green nectaries: the role of photosynthesis in secretion. Bot. J. Linn. Soc. 173 1–11. 10.1111/boj.12066 [DOI] [Google Scholar]
- Mahon P., Packman L. C., Dupree P. (2000). A proteomic analysis of organelles from Arabidopsis thaliana. Electrophoresis 21 3488–3499. [DOI] [PubMed] [Google Scholar]
- Maloof J. E., Inouye D. W. (2000). Are nectar robbers cheaters or mutualists? Ecology 81 2651–2661. 10.1890/0012-9658(2000)081[2651:ANRCOM]2.0.CO;2 [DOI] [Google Scholar]
- Miller H. (1883). Die Befruchtung der Blumen durch Insekten (English trans. 1883 by D. Thompson ‘The fertilisation of flowers,’ London). (London: Macmillan, Co.). [Google Scholar]
- Nepi M. (2007). “Nectary structure and ultrastructure,” in Nectaries and Nectar, eds Nicolson S. W., Nepi M., Pacini E. (Dordrecht: Springer; ), 129–166. [Google Scholar]
- Nepi M., Bini L., Bianchi L., Puglia M., Abate M., Cai G. (2011). Xylan-degrading enzymes in male and female flower nectar of Cucurbita pepo. Ann. Bot. 108 521–527. 10.1093/aob/mcr165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepi M., Soligo C., Nocentini D., Abate M., Guarnieri M., Cai G., et al. (2012). Amino acids and protein profile in floral nectar: much more than a simple reward. Flora 207 475–481. 10.1016/j.flora.2012.06.002 [DOI] [Google Scholar]
- Nicolson S. W., Thornburg R. W. (2007a). “Nectar chemistry,” in Nectaries and Nectar, eds Nicolson S., Nepi M., Pacini E. (Dordrecht: Springer; ), 289–342. [Google Scholar]
- Nicolson S. W., Thornburg R. W. (2007b). “Nectar chemistry,” in Nectaries and Nectar, eds Nicolson S., Nepi M., Pacini E. (Dordrecht: Springer; ), 215–264. [Google Scholar]
- Nocentini D., Pacini E., Guarnieri M., Nepi M. (2012). Flower morphology, nectar traits and pollinators of Cerinthe major (Boraginaceae-Lithospermeae). Flora 207 186–196. 10.1016/j.flora.2012.01.004 [DOI] [Google Scholar]
- O’Farrell P. H. (1975). High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250 4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Oh I. S., Park A. R., Bae M. S., Kwon S. J., Kim Y. S., Lee J. E., et al. (2005). Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 17 2832–2847. 10.1105/tpc.105.034819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacini E., Nepi M., Vesprini J. (2003). Nectar biodiversity: a short review. Plant Syst. Evol. 238 7–21. 10.1007/s00606-002-0277-y [DOI] [Google Scholar]
- Pandey A., Mann M. (2000). Proteomics to study genes and genomes. Nature 405 837–846. 10.1038/35015709 [DOI] [PubMed] [Google Scholar]
- Park S., Thornburg R. W. (2009). Biochemistry of nectar proteins. J. Plant Biol. 52 27–34. 10.1007/s12374-008-9007-5 [DOI] [Google Scholar]
- Petanidou T., Van Laere A., Ellis W. N., Smets E. (2006). What shapes amino acid and sugar composition in Mediterranean floral nectars? Oikos 115 155–169. 10.1111/j.2006.0030-1299.14487.x [DOI] [Google Scholar]
- Peumans W. J., Smeets K., Van Nerum K., Van Leuven F., Van Damme E. J. (1997). Lectin and alliinase are the predominant proteins in nectar from leek (Allium porrum L.) flowers. Planta 201 298–302. 10.1007/s004250050070 [DOI] [PubMed] [Google Scholar]
- Rivera G. L. (2000). Nuptial nectary structure of Bignoniaceae from Argentina. Darwiniana 38 227–239. [Google Scholar]
- Rodriguez-Arce A., Diaz N. (1992). The stability of beta-carotene in mango nectar. J. Agric. Univ. PR 76 101–102. [Google Scholar]
- Roshchina V. A. V., Roshchina V. D. (1993). Excretory Function of Higher Plants. Berlin: Springer. [Google Scholar]
- Saglar Naqvi S. M., Harper A., Carter C., Ren G., Guirgis A., York W., et al. (2005). Tobacco Nectarin IV is a specific inhibitor of fungal xylosidases secreted into the nectar of ornamental tobacco plants. Plant Physiol. 139 1389–1400. 10.1104/pp.105.065227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S.-G., Kim J.-S., Yang Y.-S., Jun B.-K., Kang S.-W., Lee G.-P., et al. (2010). Cloning and characterization of the new multiple stress responsible gene I (MuSI) from sweet potato. Genes Genomics 32 544–552. 10.1007/s13258-010-0093-7 [DOI] [Google Scholar]
- Stephens D. (2013). Pollination Ecology and the Floral Reward of Vaccinium myrtilloides and V. vitis-idaea (Ericaceae). MSc thesis, University of Saskatchewan, Saskatoon, SK. [Google Scholar]
- Stpiczynska M., Davies K. L. (2006). Nectary structure in Symphyglossum sanguineum [Rchb. f.] Schltr.[Orchidaceae]. Acta Agrobot. 59 7–16. 10.5586/aa.2006.001 [DOI] [Google Scholar]
- Stpiczyñska M., Kamiñska M., Zych M. (2012). Nectary structure in Dichogamous flowers of Polemonium caeruleum L. (Polemoniaceae). Acta Biol. Crac. Ser. Bot. 54 61–68. 10.2478/v10182-012-0019-6 [DOI] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg R., Carter C., Powell A., Mittler R., Rizhsky L., Horner H. (2003). A major function of the tobacco floral nectary is defense against microbial attack. Plant Syst. Evol. 238 211–218. 10.1007/s00606-003-0282-9 [DOI] [Google Scholar]
- Vesprini J., Nepi M., Pacini E. (1999). Nectary structure, nectar secretion patterns and nectar composition in two Helleborus species. Plant Biol. 1 560–568. 10.1111/j.1438-8677.1999.tb00784.x [DOI] [Google Scholar]
- Xie D.-X., Feys B. F., James S., Nieto-Rostro M., Turner J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094. 10.1126/science.280.5366.1091 [DOI] [PubMed] [Google Scholar]
- Zer H., Fahn A. (1992). Floral nectaries of Rosmarinus officinalis L. Structure, ultrastructure and nectar secretion. Ann. Bot. 70 391–397. [Google Scholar]
- Zha H.-G., Flowers V. L., Yang M., Chen L.-Y., Sun H. (2012). Acidic α-galactosidase is the most abundant nectarin in floral nectar of common tobacco (Nicotiana tabacum). Ann. Bot. 109 735–745. 10.1093/aob/mcr321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha H.-G., Liu T., Zhou J.-J., Sun H. (2013). MS-desi, a desiccation-related protein in the floral nectar of the evergreen velvet bean (Mucuna sempervirens Hemsl): molecular identification and characterization. Planta 1 77–89. 10.1007/s00425-013-1876-2 [DOI] [PubMed] [Google Scholar]
- Zimmermann F., Ebert M., Worringen A., Schütz L., Weinbruch S. (2007). Environmental scanning electron microscopy (ESEM) as a new technique to determine the ice nucleation capability of individual atmospheric aerosol particles. Atmos. Environ. 41 8219–8227. 10.1016/j.atmosenv.2007.06.023 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.