Abstract
Burkholderia contaminans MS14 shows significant antimicrobial activities against plant and animal pathogenic fungi and bacteria. The antifungal agent occidiofungin produced by MS14 has great potential for development of biopesticides and pharmaceutical drugs. However, the use of Burkholderia species as biocontrol agent in agriculture is restricted due to the difficulties in distinguishing between plant growth‐promoting bacteria and the pathogenic bacteria. The complete MS14 genome was sequenced and analyzed to find what beneficial and virulence‐related genes it harbors. The phylogenetic relatedness of B. contaminans MS14 and other 17 Burkholderia species was also analyzed. To research MS14′s potential virulence, the gene regions related to the antibiotic production, antibiotic resistance, and virulence were compared between MS14 and other Burkholderia genomes. The genome of B. contaminans MS14 was sequenced and annotated. The genomic analyses reveal the presence of multiple gene sets for antimicrobial biosynthesis, which contribute to its antimicrobial activities. BLAST results indicate that the MS14 genome harbors a large number of unique regions. MS14 is closely related to another plant growth‐promoting Burkholderia strain B. lata 383 according to the average nucleotide identity data. Moreover, according to the phylogenetic analysis, plant growth‐promoting species isolated from soils and mammalian pathogenic species are clustered together, respectively. MS14 has multiple antimicrobial activity‐related genes identified from the genome, but it lacks key virulence‐related gene loci found in the pathogenic strains. Additionally, plant growth‐promoting Burkholderia species have one or more antimicrobial biosynthesis genes in their genomes as compared with nonplant growth‐promoting soil‐isolated Burkholderia species. On the other hand, pathogenic species harbor multiple virulence‐associated gene loci that are not present in nonpathogenic Burkholderia species. The MS14 genome as well as Burkholderia species genome show considerable diversity. Multiple antimicrobial agent biosynthesis genes were identified in the genome of plant growth‐promoting species of Burkholderia. In addition, by comparing to nonpathogenic Burkholderia species, pathogenic Burkholderia species have more characterized homologs of the gene loci known to contribute to pathogenicity and virulence to plant and animals.
Keywords: Antimicrobial, Burkholderia contaminans MS14, comparative genomics, virulence, whole genome sequencing.
Background
Burkholderia is a gram‐negative, rod‐shaped, motile, and nonspore‐forming bacterium that has been identified in many diverse ecological niches (Francis et al. 2013). Currently, 88 species have been recognized in the genus Burkholderia (De Meyer et al. 2013). The ecological versatility of these bacteria is likely due to their unusually large genomes, which are often comprised of one or multiple large replicons with plasmids (Lessie et al. 1996). The bacterium has the ability to use a large array of carbon sources to synthesize secondary metabolites (el ‐Banna and Winkelmann 1998; Parke and Gurian‐Sherman 2001).
Most Burkholderia species isolated from soil are associated with plants. Some species are related to the promotion of plant growth and are considered to be plant growth‐promoting bacteria (PGPB). For example, B. phenoliruptrix and B. phymatum are effective in nitrogen fixing (Elliott et al. 2007; Zuleta et al. 2014), while B. phytofirmans induces larger root systems (Sessitsch et al. 2005). Interests in the use of Burkholderia species or their secondary metabolites in agriculture have increased. For example, the use of Burkholderia cepacia AMMDR1 could yield as efficient control of “damping‐off” disease caused by Pythium species and Rhizoctonia solani as that of the fungicide Captan (Heungens and Parke 2000; Parke and Gurian‐Sherman 2001). The relatively large Burkholderia genome harbors a large variety of antimicrobial biosynthesis genes. Occidiofungin (Gu et al. 2011; Ellis et al. 2012) produced by B. contaminans MS14 has significant antifungal activity. Pyrrolnitrin (el ‐Banna and Winkelmann 1998) was first identified as an antifungal antibiotic produced by Pseudomonas species and later found to be synthesized by several Burkholderia cepacia species. Pyoluteorin (Birchall et al. 1970) and lipopeptide AFC‐BC11 (Kang et al. 1998) are other antibiotics produced by Burkholderia. Siderophores are bacteriostatic agents that can inhibit pathogenic microorganism's growth by depleting iron in the soil (Hider and Kong 2010). Pyochelin (Serino et al. 1997) and ornibactin (Meyer et al. 1995) are common siderophores produced by Burkholderia species. On the other hand, plant‐pathogenic species of Burkholderia have also been identified, such as B. gladioli and B. glumae, which infect rice and other horticultural plants (Stoyanova et al. 2007; Ham et al. 2011). The commonly produced plant‐toxic secondary metabolites by Burkholderia species include polysaccharides and other toxins, such as rice grain rot and wilt causal agent toxoflavin (Latuasan and Berends 1961) and exopolysaccharide toxin cepacian (Ferreira et al. 2010) that contribute to the overall pathogenicity and success of the bacterium as a plant pathogen.
The potent antifungal occidiofungin was first identified from B. contaminans MS14 (Gu et al. 2011). Burkholderia contaminans MS14 was isolated from soil in Mississippi, it has a broad range of antifungal activities to plant and human pathogens by producing an oligopeptide occidiofungin (Lu et al. 2009). It is a novel fungicide that can significantly inhibit the growth of pathogens by interfering with cell wall synthesis or triggering apoptosis (Ellis et al. 2012; Emrick et al. 2013; Ravichandran et al. 2013). Genetically, the whole length of the ocf gene cluster required for production of occidiofungin has been characterized, which is composed of 16 ORFs (Gu et al. 2009). Among the 16 members of this cluster, ocfD, ocfE, ocfF, ocfH, and ocfJ were predicted to encode nonribosomal peptide synthesis (NRPS) or NRPS‐polyketide synthase (PKS), which are directly related to the biosynthesis of the antifungal compound occidiofungin. The genes ocfA, ocfC, ocfK, ocfL, ocfM, and ocfN were predicted to be involved in the secretion and modification of occidiofungin.
Burkholderia cepacia complex (Bcc) is a group of Burkholderia species that some are opportunistic bacteria and could cause lung disease in immunocompromised individuals (Mahenthiralingam et al. 2005). The Bcc group composed of nine different genomovars and at least 18 different species. These species include B. cepacia, B. cenocepacia, B. multivorans, B. vietnamiensis, B. stabilis, B. ambifaria, B. dolosa, B. anthina, and B. pyrrocinia (Lipuma 2005). B. cepacia is a common environmental species, but is also an important human pathogen which can create respiratory complications for cystic fibrosis (CF) patients (Mahenthiralingam et al. 2005). B. cenocepacia is a major CF pathogen and is responsible for 70% of the cases of Bcc infection (Mahenthiralingam et al. 2005). B. multivorans is the second most common Bcc species in CF infection (Mahenthiralingam et al. 2002), and some species like B. dolosa strains are frequently isolated from the CF patents (Vermis et al. 2004). Bcc infections contribute to the overall poor health of CF patients (Mahenthiralingam et al. 2002). It is important to be able to distinguish virulent Bcc species, as well as some other reported virulent Burkholderia species such as B. pseudomallei, from the less‐virulent soil‐isolated Burkholderia species.
Resistance to multiple antibiotics and disinfectants is very common among Burkholderia species. This antibiotic resistance feature makes them hard to treat and is crucial for human pathogenicity of Bcc species. There are several mechanisms that contribute to antibiotic resistance of the Bcc strains. First, efflux pumps are responsible for exclusion of antibiotics from the cell. Secondly, some Burkholderia species could significantly lower antimicrobial susceptibility by forming biofilm (Sawasdidoln et al. 2010) or by entering a nonreplicating state (Hamad et al. 2011). In addition, enzymatic inactivation of antibiotics either by modification or cleavage is a common resistance mechanism found in Burkholderia species (Tribuddharat et al. 2003).
Several well‐documented bacterial virulence features for mammalian pathogenesis are found in Burkholderia species. Cable pili and the 22‐kilodalton (KDa) adhesin are virulence factors associated with cepacia syndrome. These are required for B. cenocepacia to bind and cross the squamous epithelium leading to an intensified chronic infection (Urban et al. 2005). The periplasmic located superoxide dismutase (SOD) protects bacteria from oxidation by exogenously generated superoxide or peroxide (Keith and Valvano 2007). A 31.7 kb B. cepacia virulence genomic island (GIs) has been identified to harbor B. cepacia epidemic strain marker (BCESM) and possesses both virulence and metabolism‐associated genes (Baldwin et al. 2004). VgrG‐5 is a Burkholderia type VI secretion system 5‐associated protein, which is required for the full virulence of type VI secretion system 5 to induce multinucleate giant cell formation and full mammalian virulence (Schwarz et al. 2014).
In this study, we analyzed the phylogenetic relatedness of MS14 to 17 other Burkholderia species including plant pathogen, cystic fibrosis opportunistic, plant growth‐promoting strains, and other soil isolates. Being able to distinguish between safe environmental isolates of Burkholderia and a human pathogen is a current problem (Mahenthiralingam et al. 2008). Our analyses provide a whole‐genome approach to identify less‐virulent environmental isolates by comparing antibiotic biosynthesis, antibiotic resistance, and virulence loci among Burkholderia species. The results provide important information for evaluating Burkholderia species virulence according to their secondary metabolites production and virulence markers.
Materials and Methods
Genome sequencing of B. contaminans MS14
B. contaminans MS14 was cultured in nutrient broth yeast (NBY) extract medium (Vidaver 1967) overnight in a shaker at 28°C. The genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA) and was used for library construction with Illumina Genomic DNA Sample Preparation Kit (Illumina, CA, USA). Twelve standard libraries (with an average insert size of 400~700 bp) and one mate pair library (with an average insert size of 7000 bp) were prepared and sequenced on the Illumina MiSeq and HiSeq instrument according to the manufacturer's instructions. The genome was de novo assembled similar as described by Tim Durfee, etc. (Durfee et al. 2008) using DNAStar Seqman NGen (Version 12, DNASTAR, Inc. Madison, WI U.S.). Briefly, a standard 400 bp insertion library and the mate pair library were selected as the input data for the assembling. Forty million short reads were scanned and extracted from the raw data files as input data. The short reads were preprocessed by Seqman NGen to trim adaptors and filter low‐quality reads. Automatic Mer size was chosen and a minimum match percentage of 98% was selected. Short reads (38.2 million) were assembled into 19 contigs that were ordered within SeqMan Pro (Version 12, DNASTAR, Inc. Madison, WI, USA) using mate pair data. The first round assembling was then used as a template for a complete reassembly. Independently de novo assembled sequence data from other libraries were incorporated to proof read the first assembly and to maximize coverage and quality. Adjacent contigs, if possible, were merged. The alignment resulted in a three‐contig scaffold spanning the whole genome. Gaps were filled, and three contig were bridged and circled by Polymerase Chain Reaction (PCR) and Sanger sequencing. No contigs remained unassembled which might correspond to plasmids. The annotated MS14 genome, which has three chromosomes, is available from NCBI under the accession numbers: CP009743, CP009744, and CP009745.
Other Burkholderia strains used for genome comparison
Seventeen previously sequenced Burkholderia strains were selected for comparative genome analysis based upon their characteristics and distinctive biological properties (Table 1). The selected species include MS14 and three PGPB B. lata 383, B. ambifaria AMMD, and B. phytofirmans PsJN (Coenye et al. 2001; Vanlaere et al. 2009; Weilharter et al. 2011), seven CF opportunistic pathogens and mammalian pathogens B. thailandensis E264, B. mallei ATCC 23344, B. cenocepacia J2315, B. multivorans ATCC 17616, B. pseudomallei 1026b, B. pseudomallei K96243, and B. oklahomensis EO147 (DeShazer et al. 2001; Biddick et al. 2003; Glass et al. 2006; Knappe et al. 2008; Holden et al. 2009). Plant‐pathogenic strains B. gladioli BSR3, which infects onions, rice, and iris (Seo et al. 2011), and B. glumae BGR1, which causes bacterial panicle blight on rice (Lim et al. 2009) were selected for comparison. Another five previously sequenced soil isolates including nitrogen‐fixing nodulator that were acquired from different habitats were also used: B. cepacia GG4, B. phenoliruptrix BR3459a, B. phymatum STM815, B. xenovorans LB400, and B. vietnamiensis G4 (Fries et al. 1997; Vandamme et al. 2002; Hong et al. 2012; de Oliveira Cunha et al. 2012).
Table 1.
List of strains used in comparative analysis
| Strain | Number of chromosomes | Number of plasmids | Size (Mb) | GC% | Gene | CDS | Source | Accession number |
|---|---|---|---|---|---|---|---|---|
| B. contaminans MS14 | 3 | – | 8.509 | 66.37 | 7582 | 7270 | Cotton field, MS, USA | CP009743,CP009744,CP009745 |
| B. lata 383 | 3 | – | 8.676 | 66.26 | 7823 | 7716 | Forest soil, Trinidad and Tobago | NC_007510.1, NC_007511.1, NC_007509.1 |
| B. ambifaria AMMD | 3 | 1 | 7.529 | 66.79 | 6717 | 6610 | Rhizosphere of healthy pea, Wisconsin, USA | NC_008390.1, NC_008391.1, NC_008392.1 |
| B. cenocepacia J2315 | 3 | 1 | 8.056 | 66.92 | 7365 | 7116 | CF patients, UK | NC_011000.1, NC_011001.1, NC_011002.1 |
| B. multivorans ATCC 17616 | 3 | 1 | 7.009 | 66.69 | 6372 | 6258 | Soil enriched with anthranilate, Berkeley, CA, USA | NC_010804.1, NC_010805.1, NC_010801.1 |
| B. gladioli BSR3 | 2 | 4 | 9.052 | 67.41 | 7757 | 7411 | Diseased rice sheath, South Korea. | NC_015381.1, NC_015376.1 |
| B. glumae BGR1 | 2 | 4 | 7.285 | 67.93 | 6302 | 5773 | Diseased Rice Grain, South Korea. | NC_012724.2, NC_012721.2 |
| B. mallei ATCC 23344 | 2 | – | 5.836 | 68.52 | 5506 | 5022 | Glanders patient, Myanmar | NC_006348.1, NC_006349.2 |
| B. oklahomensis EO147 | 2 | – | 7.314 | 66.94 | 6357 | 6264 | Wound infection, Georgia, USA | CP008726.1,CP008727.1 |
| B. phenoliruptrix BR3459a | 2 | 1 | 7.651 | 63.12 | 6605 | 6496 | Root nodule of Mimosa flocculosa, South America. | NC_018695.1, NC_018672.1 |
| B. phymatum STM815 | 2 | 2 | 8.677 | 62.28 | 7899 | 7496 | Root nodule of Machaerium lunatum, French Guiana. | NC_010622.1, NC_010623.1 |
| B. phytofirmans PsJN | 2 | 1 | 8.215 | 62.32 | 7484 | 7241 | Glomus vesiculiferum‐infected onion roots, plant‐beneficial bacterium | NC_010681.1, NC_010676.1 |
| B. pseudomallei 1026b | 2 | – | 7.231 | 68.16 | 6262 | 6070 | Septicemic melioidosis patient, Thailand | NC_017831.1, NC_017832.1 |
| B. pseudomallei K96243 | 2 | – | 7.248 | 68.05 | 5935 | 5727 | Septicemic melioidosis patient, Thailand | NC_006350.1, NC_006351.1 |
| B. thailandensis E264 | 2 | – | 6.724 | 67.65 | 5712 | 5632 | Rice field. Thailand. | NC_007651.1, NC_007650.1 |
| B. vietnamiensis G4 | 3 | 5 | 8.391 | 65.73 | 7861 | 7617 | Soil, California, USA | NC_009256.1, NC_009255.1, NC_009254.1 |
| B. xenovorans LB400 | 3 | – | 9.731 | 62.63 | 9043 | 8702 | Contaminated soil, New York, USA | NC_007951.1, NC_007952.1, NC_007953.1 |
| B. cepacia GG4 | 2 | – | 6.467 | 66.71 | 5903 | 5,825 | Ginger rhizosphere, Malaysia | NC_018513.1, NC_018514.1 |
Comparative analyses of Burkholderia genomes
Identification of putative protein‐encoding genes and annotation of B. contaminans MS14 whole genomes were performed by NCBI Prokaryotic Genome Automatic Annotation Pipeline (PGAAP) (Angiuoli et al. 2008), which was designed to annotate bacterial and archaeal genomes. Gene ontology (GO) analysis was performed by searching against protein database using BLAST2GO (Conesa et al. 2005), COG (http://www.ncbi.nlm.nih.gov/COG/) (Tatusov et al. 2003) and KEGG (Kyoto encyclopedia of genes and genomes; http://www.genome.jp/kegg/) (Kanehisa and Goto 2000). Progressive Mauve (Darling et al. 2010) was used to perform the multiple genome alignments. IslandViewer (Langille and Brinkman 2009) was used to predict and identify genomic islands (GIs) within the bacterial genomes with two different GIs prediction methods: SIGI‐HMM (Waack et al. 2006), which uses a hidden Markov model (HMM) (Baum and Petrie 1966) measuring codon usage for GIs prediction, and IslandPath‐DIMOB (Langille et al. 2008), which predicts GI based on identifying conserved regions across all genomes and unique regions to the query genome. Secondary metabolites‐ and antibiotics‐related genes were identified using antiSMASH (Medema et al. 2011). The average nucleotide identity (ANI) (Richter and Rossello‐Mora 2009) was calculated by a script developed by Kostas's lab (http://enve-omics.gatech.edu/), only the chromosome sequence of the Burkholderia strains were taken into account. Protein coding regions were visually compared using Easyfig (Sullivan et al. 2011) and genes were searched against the nonredundant database by BLASTx search. Protein sequence identity of 40% was used as the cutoff to distinguish between peptides of similar and nonsimilar structure (Rost 1999). BLASTn comparison of genomes was visualized by BRIG (Alikhan et al. 2011) and Circos (Krzywinski et al. 2009).
Identification and comparison of genetic loci associated with siderophores and antimicrobial production, virulence factors and antibiotic resistance
The gene clusters required for production of siderophores and antimicrobial secondary metabolites were analyzed among the selected Burkholderia genomes. The gene clusters responsible for antimicrobial product biosynthesis were analyzed including occidiofungin, pyrrolnitrin, pyoluteorin, and lipopeptide AFC‐BC11 as mentioned previously. Burkholderia rhizoxinica produced antitumor product rhizoxin (Partida‐Martinez and Hertweck 2007) and Burkholderia spp produced spliceostatin (Eustaquio et al. 2014) were also included. The compared siderophores include pyochelin and ornibactin.
Pathogenesis‐related factors of Burkholderia species to both plants and mammals were compared. Plant‐toxic exopolysaccharide cepacian and toxins toxoflavin (Suzuki et al. 2004), hydrogen cyanide (HCN) (Ryall et al. 2008), and 2‐heptyl‐3‐hydroxy‐4(1H)‐quinolone (Diggle et al. 2006) were compared within the studied genomes as plant‐pathogenic agents. CF‐related O‐antigen of lipopolysaccharides pathogenic to mammals (Ortega et al. 2005) was also compared as it is associated with transmissible infections in CF patients.
The genes responsible for antibiotic resistance were analyzed within the researched genomes. First, four efflux pumps are compared: BpeAB‐OprB multidrug efflux pump that is responsible for the efflux of aminoglycosides gentamicin, streptomycin, and erythromycin (Chan et al. 2004), AmrAB–OprA efflux pump that is responsible for the extrude of aminoglycoside, macrolide, fluoroquinolones, and tetracyclines (Moore et al. 1999), BpeEF–OprC efflux pump that contributes to the resistance to chloramphenicol, fluoroquinolones, tetracyclines, and trimethoprim in clinical and environmental B. pseudomallei isolates (Podnecky et al. 2013), and a salicylate‐induced efflux pump that is associated with the resistance of chloramphenicol, trimethoprim, and ciprofloxacin (Nair et al. 2004). Biofilm formation and nonreplicating state alternation related genes were also compared: first, Cep Quorum‐sensing system‐related genes were selected for this system controls biofilm formation that could significantly reduce antimicrobial susceptibility (Huber et al. 2001). Second, arginine and pyruvate fermentation mechanism were analyzed for this mechanism are among those most highly induced in response to hypoxia under anaerobic conditions (Zuniga et al. 2002; Hamad et al. 2011), it is very likely that pathogenic Burkholderia bacteria utilize the same pathway for energy generation during respiratory stress in CF patients (Hamad et al. 2011). Moreover, beta‐lactamase‐induced enzymatic inactivation‐related genes were also analyzed due to their effects on ceftazidime and beta‐lactam drug clavulanic acid (Tribuddharat et al. 2003).
Virulence features associated genes that contributes to mammalian pathogenesis and opportunistic infections in CF were compared: Cable pili and the 22‐kilodalton adhesin biosnthesis genes that are required for B. cenocepacia binding and transmission, the SodC gene that encodes for a Cu2+ and Zn2+ containing periplasmic SOD that contributes to intracellular survival in CF patients, a zinc metalloprotease that may be involved in overall virulence of several Bcc strains(Corbett et al. 2003), the 31.7 kb Burkholderia cepacia virulence genomic island that harbors multiple virulence and metabolism‐associated genes, a melanin pigment that could aid the colonization and transmission of certain B. cepacia strains in CF patients (Zughaier et al. 1999), in addition, the VgrG‐5 protein that is required for type VI secretion system 5 to multinucleate giant cell formation (Schwarz et al. 2014).
Results
Genomic features of MS14
The complete genome of B. contaminans MS14 consists of three circular chromosomes of 3,522,585, 3,358,952, and 1,627,712 bp with a GC content of 66.89%, 66.30%, and 65.50%, respectively. A total of 7813 genes were identified from the genome, of which 231 were pseudogenes or partial genes. The three replicons encode 3,020, 2,943, and 1,307 predicted coding DNA sequences (CDSs), 1 noncoding RNAs (ncRNAs), 5 rRNA operons, and 65 tRNA loci. Forty‐nine genomic islands ranging from 4 kbp to 29 kbp were also identified by Islandviewer throughout the MS14 genome of which 22 genomic islands were identified from chromosome 3 (Table S1). Majority of the genomic islands genes encode hypothetical proteins. A summary of B. contaminans MS14 genome features are provided in Table 2 and the circular chromosomes are provided in Figure 1.
Table 2.
Chromosome statistics of Burkholderia contaminans MS14
| Feature | Chromosome 1 | Chromosome 2 | Chromosome 3 | Total |
|---|---|---|---|---|
| Size | 3,522,585 bp | 3,358,952 bp | 1,627,712 bp | 8,509,249 bp |
| Genes | 3130 | 3,049 | 1403 | 7582 |
| CDS | 3020 | 2,943 | 1307 | 7270 |
| Pseudogenes | 67 | 73 | 91 | 231 |
| rRNAs | 1 | 3 | 1 | 5 |
| tRNAs | 40 | 23 | 2 | 65 |
| ncRNA | 0 | 1 | 0 | 1 |
| G+C content | 66.89% | 66.30% | 65.50% | 66.40% |
Figure 1.
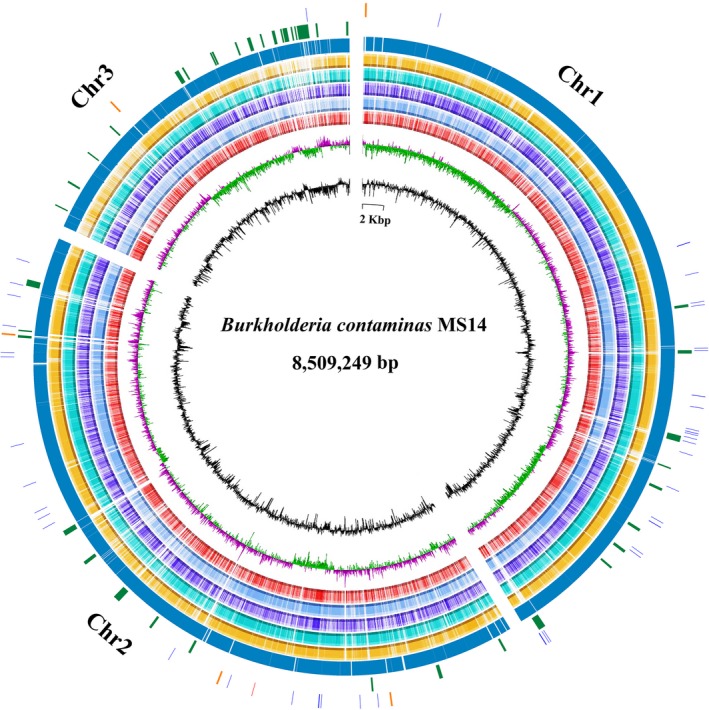
Circular representation of the B. contaminans MS14 genome in comparison with five sequenced Burkholderia whole genome. Rings from inside to outside: (1) GC content (black), (2) GC skew (purple and green), (3) BLAST comparison with B. mallei ATCC 23344 (red), (4) BLAST comparison with B. cenocepacia J2315 (aqua), (5) BLAST comparison with B. glumae BGR1 (slateblue), (6) BLAST comparison with B. ambifaria AMMD (cyan), (7) BLAST comparison with B. lata 383 (yellow), (8) Coding sequences of B. contaminans MS14 genome (dark blue), (9) Gene islands (dark green), (10) rRNA (yellow), tRNA (dark purple) and ncRNA (red). Figure generated by Circos with incorporated BLASTn result from BRIG (BLAST, Ring Image Generator).
An insight of B. contaminans MS14 genome indicated multiple predicted antibiotic‐ and antimicrobial‐related secondary metabolites biosynthetic gene loci by antiSMASH (Table S2), those include five terpene and two bacteriocin biosynthesis gene clusters. Two NRPS genes NL30_14890 and NL30_14895 on chromosome one share 91% and 90% nucleotide sequence similarity with orbJ and orbI NRPS genes in the ornibactin biosynthetic gene cluster (Agnoli et al. 2006), respectively. On the chromosome three, the genes NL30_32800, NL30_32795, NL30_32790, and NL30_32785 share an overall 90% nucleotide sequence similarity to the prnABCD genes in pyrrolnitrin biosynthetic gene cluster (Costa et al. 2009). In addition, two PKS genes NL30_36200 and NL30_36225 share a highest 69% and 66% amino acid sequence similarity to any published data. Strain MS14 is likely to produce a novel bioactive compound via those two PKS genes.
Blast research of B. contaminans MS14 genome against B. lata 383, B. ambifaria AMMD, B. glumae BGR1, B. cenocepacia J2315, and B. mallei ATCC 23344 genome revealed multiple unique gene regions, which were only found in the MS14 genome (Fig. 1). The BLASTn atlas showed that MS14 chromosome presents a large similarity with the PGPB B. lata 383, there is no surprise considering they belonged to taxon K of the Bcc prior to their reassignment as two different species. However, 5% coding genes present in MS14 chromosome did not share significant homology with any other species used in comparison. The BLASTn atlas also indicated that MS14 chromosome 3 is more diverse than the MS14 chromosomes 1 and 2, harboring more unique gene regions that do not share significant similarity with those in other genomes being compared. Moreover, MS14 chromosome 3 harbors majority of gene islands identified within its genome.
Gene ontology annotations were added to MS14 genes to conduct gene analysis in population according to analyzed transcripts (Ashburner et al. 2000). GO term distribution describing molecular function and biological process is shown in Figure 2. Besides the basic cell functions, GO terms connected to heterocyclic compound binding and organic cyclic compound binding are well represented in MS14 genome, that can selectively and noncovalently interact with heterocyclic compound and organic cyclic compound, of which many bioactive secondary metabolic are derived from. In addition, GO terms associated with ion binding, hydrolase activity, oxidoreductase, and transferase activity are also represented in large percentage in MS14 genome. On the other side, more than a quarter of the biological process GO terms are connected to the metabolic process. Those data indicate that, like some Burkholderia strains (Bartell et al. 2014), strain MS14 is very versatile from the metabolic standpoint.
Figure 2.
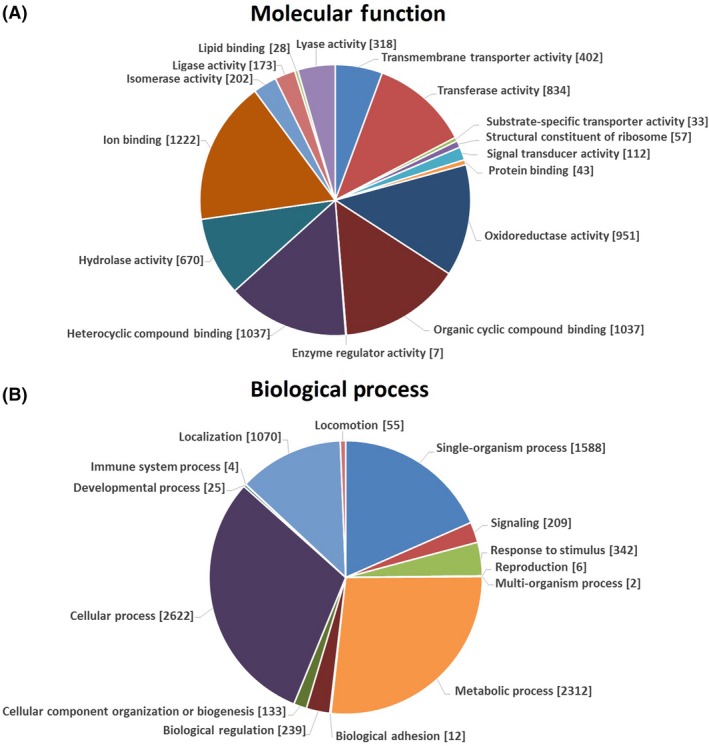
Gene ontology (GO) analyses of B. contaminans MS14 genome. GO analysis of B. contaminans MS14 genome corresponding to 7,582 genes as for their predicted involvement in molecular functions (A) and biological processes (B). Data are presented as level 3 GO categorization for molecular function and level 2 GO categorization for biological process. Classified gene objects are depicted as gene numbers (in brackets).
Phylogenetic relationships of sequenced Burkholderia species
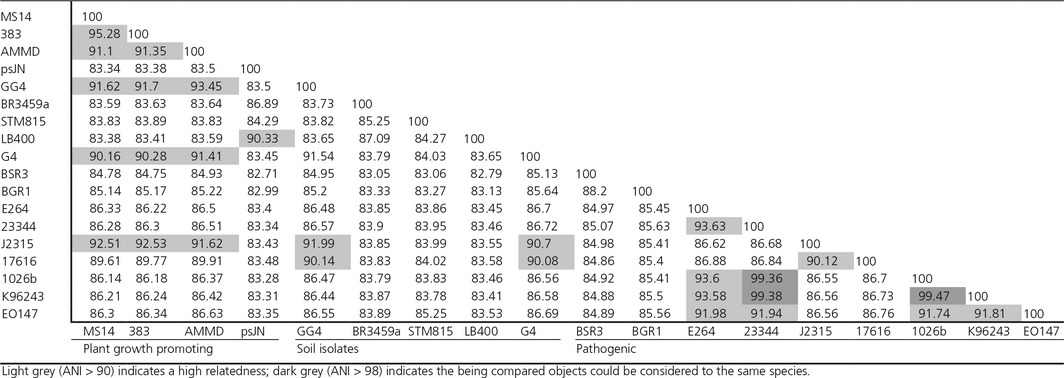
Phylogenetic relationships of sequenced Burkholderia species were analyzed. Strains with ANI values greater than 96%, which equate to a DNA–DNA hybridization value of 70%, are considered to be the same species (Richter and Rossello‐Mora 2009). Strains with ANI value greater than 90% are considered to have high genome relatedness. Strains with ANI value lower than 90% are considered to have divergent genomes (Konstantinidis et al. 2006). As shown in Table 3, strain MS14 has the greatest nucleotide identity similarity to B. lata 383, which is also a novel species within Burkholderia taxon K (Vanlaere et al. 2009). Soil isolate B. cepacia GG4 is most closely related to the PGPB strains B. ambifaria AMMD. The pathogenic B. cenocepacia J2315 is also closely related to PGPB strain MS14, 383, AMMD, soil isolates GG4 and G4 than to other pathogenic species. Soil isolates GG4 and G4 have high genome relatedness to pathogenic B. multivorans ATCC 17616. Plant‐pathogenic B. gladioli BSR3 and B. glumae BGR1 only showed limited gene relatedness between them due to the 2 Mb genome size difference, even though they share a similar host range. These ANI data suggest that genome sequences of the PGPB isolates are different from some virulent strains such as B. pseudomallei 1026b, B. pseudomallei K96243, and B. mallei ATCC 23344, which have very close phylogenetic relationship between them (ANI > 99%).
Table 3.
Average Nucleotide Identity (ANI) pairwise comparisons among sequenced Burkholderia strains
Production of siderophores and antimicrobial compounds
The comparison of antibiotics production‐related gene loci among the Burkholderia species are shown in Table 4. Only the core biosynthesis genes are being compared, as indicated in the table. Intact occidiofungin biosynthesis gene is only present in the AMMD and MS14. Several Burkholderia cepacia species were reported to produce pyrrolnitrin. As expected, the PGPB MS14, 383, AMMD, and B. oklahomensis EO147 all harbor the intact biosynthesis gene prnABCD as a gene cluster (Fig. 3). However, the pyrrolnitrin biosynthesis locus was not found in B.phytofirmans PsJN. Rhizoxin's biosynthesis gene RhiABCDE and spliceostatin biosynthesis gene clusters were not found in the Burkholderia genomes. Antifungal compound pyoluteorin biosynthesis genes pltB and pltC were not found in the analyzed genomes. Intact siderophore pyochelin biosynthesis pch gene cluster and ornibactin biosynthesis orb gene orbI and orbJ are widely possessed among the studied genomes compared to the lipopeptide AFC‐BC11, whose biosynthesis genes afcBACD are only harbored by MS14, 383, AMMD, and pathogenic B. cenocepacia J2315. None of the antimicrobial biosynthesis genes being studied were identified from soil isolate B. phenoliruptrix BR3459a and plant‐pathogen bacteria B. gladioli BSR3, and B. glumae BGR1.
Table 4.
Distribution of antibiotic and virulence compound symbiotic loci of Burkholderia strains
| Name | Biosynthesis Gene Homologs | PGPB | Soil isolates | Pathogenic | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 383 | MS14 | AMMD | psJN | GG4 | BR3459a | STM815 | LB400 | G4 | BSR3 | BGR1 | E264 | 23344 | J2315 | 17616 | 1026b | K96243 | EO147 | |||
| Antibiotic and siderophore | Occidiofungin | ocfD‐ocfJ | ‐ | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Pyrrolnitrin | prnA‐prnD | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | |
| Rhizoxin | rhiA‐rhiF | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ‐ | −− | − | |
| Spliceostatin | fr9C‐fr9I | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Pyoluteorin | pltB, pltC | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Pyochelin | pchR, pchD‐pchA | + | − | − | − | − | − | − | − | − | − | − | + | − | + | − | + | + | − | |
| Ornibactin | orbE, orbI, orbJ | + | + | + | + | + | − | + | + | + | − | − | + | + | + | + | + | + | + | |
| AFC‐BC11 | afcB, afcA, afcC, afcD | + | + | + | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | |
| Virulence metabolics | Cepacian | bceA‐bceK, bceN‐bceT | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Toxoflavin | toxR, toxA‐toxE | − | − | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | |
| Hydrogen Cyanide | hcnA‐hcnC | − | − | + | − | + | − | + | − | + | + | + | + | + | + | + | + | + | + | |
| 2‐heptyl‐3‐hydroxy‐4(1H)‐quinolone | pqsA‐pqsE | − | − | − | − | − | − | − | − | − | − | − | + | + | − | + | + | + | + | |
Figure 3.
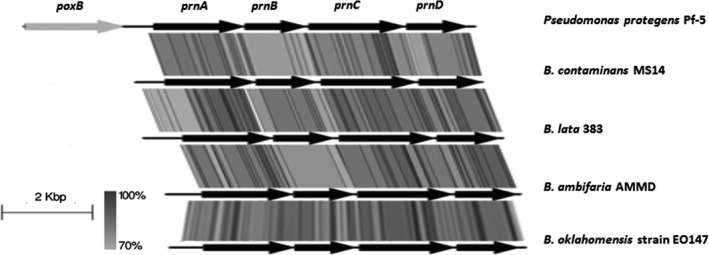
Pyrrolnitrin biosynthesis locus genetics. Figure showing the Pyrrolnitrin biosynthesis locus identified in analyzed Burkholderia genomes. The CDS marked in black are the prnABCD operon, which is highly conserved in the four Burkholderia strains showing in the figure. Figure was generated by Easyfig.
Virulent secondary metabolites production
Plant‐virulent secondary metabolites‐related loci are compared and shown in Table 4. The cepacian biosynthesis genes are possessed among all Burkholderia genomes. Only the two plant‐pathogen BSR3 and BGR1 harbors toxoflavin biosynthesis gene toxR and toxA‐toxE homologs. Conversely, HCN and 2‐heptyl‐3‐hydroxy‐4(1H)‐quinolone biosynthesis gene pqsA‐pqsE homologs are more commonly possessed by the studied genomes. AMMD has hydrogen cyanide gene cluster identified, which is not present in the PGPB strains MS14 and 383. The overall data show MS14, 383, AMMD, and other environmental isolates have very few toxin biosynthesis genes in their genome compared to those of the genomes of the plant‐pathogenic species.
Human pathogenic CF‐related O‐antigen biosynthesis gene cluster of selected Burkholderia species were compared, the O‐antigen biosynthesis locus genetics are shown in Figure 4. The 29 kb gene region of B. cenocepacia J2315 containing 24 genes responsible for O‐antigen biosynthesis and lipid A‐core component was compared to the other 17 Burkholderia species. The homologs of first 13 genes, from waaA to wbiF that are required for the biosynthesis of lipid A‐core component, assembly initiation of the O antigen subunits and translocation of O‐antigen subunit across membrane, were identified from the genomes of all the Bcc species analyzed in this study. The homologs of the wbxCDEF genes that are also required for O‐antigen biosynthesis were identified from K96243, 1026b, AMMD, 17616, GG4, and EO147 genome (not shown in the BLAST region), but not identified from strains E264 and 23344. Possessing large amount of genes within O‐antigen cluster indicates a high possibility of a strain's virulence (Ortega et al. 2005).
Figure 4.
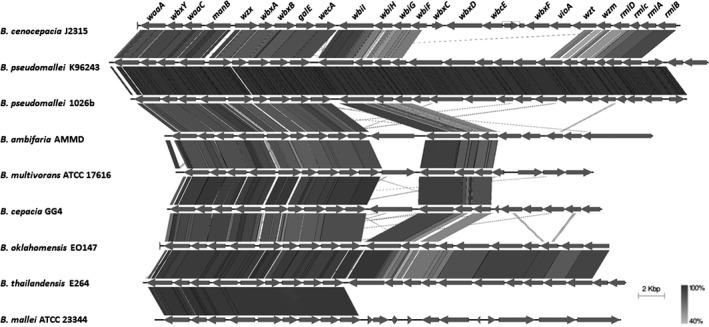
Cystic fibrosis (CF)‐related O‐antigen biosynthesis locus genetics. Figure showing a 29 kb gene region of B. cenocepacia J2315 containing 24 genes responsible for O‐antigen biosynthesis and lipid A‐core component.
Virulence features
Pathogenic Burkholderia species including CF opportunistic isolates have more key virulent genes identified than nonpathogenic species (Table 5). Strain J2315 harbors intact cable pili and the 22‐kilodalton adhesin biosynthesis gene, strain 17,616 harbors intact adhesin biosynthesis gene cluster, but lacks the cable pili biosynthesis gene cluster. Moreover, lack of cable pili biosynthesis genes in other Burkholderia species indicates their inability to attach to the host cell to initiate infection. The periplasmic superoxide dismutase SodC gene (Keith and Valvano 2007) is commonly present among Bcc species indicating their self‐protection ability. Zinc metalloprotease biosynthesis genes were found in the PGPB genomes; however, there is no evidence showing a direct relationship between the production of zinc metalloprotease and virulence. The 31.7 kb Burkholderia cepacia virulence genomic island is only harbored by strain J2315. The lack of this genomic island indicates a significant decrease in the pathogenicity potential in other Burkholderia species. The biosynthesis gene of melanin had been identified from all analyzed Burkholderia genomes. VgrG‐5 protein biosynthesis gene is present in the pathogenic strains E264, 23344, 1026b, K96243, EO147.
Table 5.
Distribution of antibiotic resistance and pathogenic symbiotic loci of Burkholderia strains
| Name | PGPB | Soil strains | Pathogenic strains | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 383 | MS14 | AMMD | psJN | GG4 | BR3459a | STM815 | LB400 | G4 | BSR3 | BGR1 | E264 | 23344 | J2315 | 17616 | 1026b | K96243 | EO147 | ||
| Antibiotic resistance | BpeAB‐OprB Multidrug Efflux Pump | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| AmrAB–OprA Multidrug Efflux Pump | + | + | + | − | + | − | + | + | + | + | + | + | + | + | + | + | + | + | |
| BpeEF‐OprC Multidrug Efflux Pump | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| Salicylate‐Induced Antibiotic Efflux Pump | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| Cep Quorum‐sensing System | + | + | + | − | + | − | − | − | + | + | + | + | + | + | + | + | + | + | |
| Arginine and Pyruvate Fermentation | − | − | − | + | − | − | + | + | − | − | − | + | + | − | − | + | + | + | |
| Beta‐Lactamase | + | + | + | − | + | − | + | − | + | + | + | + | + | + | + | + | + | + | |
| Porins | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| Pathogenic symbiotic | Cable Pili | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − |
| 22‐Kilodalton Adhesin | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | − | |
| Periplasmic Superoxide Dismutase SodC | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| Zinc Metalloprotease | + | + | + | − | − | − | − | − | − | − | − | + | + | + | − | + | + | + | |
| Burkholderia cepacia Virulence Genomic Island | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | |
| Melanin | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| Type VI Secretion System‐Exported Protein VgrG‐5 | − | − | − | − | − | − | − | − | − | − | − | + | + | − | − | + | + | + | |
Antibiotic resistance
Antibiotic resistance is a common feature among studied Burkholderia (Table 5). First, our results show that the four well‐studied efflux pumps (AmrAB–OprA efflux pump, BpeAB‐OprB multidrug efflux pump, BpeEF–OprC efflux pump, and salicylate‐induced efflux pump) are commonly present among the sequenced genomes, however, the AmrAB–OprA efflux pump was not identified from genome of strains psJN and BR3459a. Cep quorum‐sensing system is also commonly identified in the sequenced genomes. This data indicated that the majority of the sequenced Burkholderia species being analyzed have biofilm formation potential. However, the exceptions were psJN and the soil isolates BR3459a, STM815, and LB400. Arginine and pyruvate fermentation‐related genes were identified from PGPB strain psJN, soil isolates STM815, LB400 and pathogenic strains E264, 23344, 1026b, K96243, and EO147. The biosynthesis of beta‐lactamase gene was not identified from psJN, BR3459a, and GG4. Taking all into account, the results provide genetic evidence to understand Burkholderia species possess multiple antibiotic resistance mechanisms to adapt and to survival different the environment. Overall, pathogenic Burkholderia species possess a majority of both antibiotic‐resistant genes and virulence‐related genes. The PGPB strains, on the other hand, have the majority of the antibiotic‐resistant‐related genes, but lack the key virulence‐related genes like the biosynthesis genes for cable pili, adhesin, and the VgrG‐5 protein.
Discussion
B. contaminans MS14 is a versatile environmental isolate that shows significant antifungal and antibacterial activity. By investigating MS14′s genome, we found loci contributing to biosynthesis of several antimicrobial agents, which may partially explain its plant growth‐promoting activity. There are multiple gene regions related to antimicrobial production or antagonistic activity. The wide spectrum of antibiotic biosynthesis genes possessed by MS14 may contribute to its survivability when competing with other soil‐born microorganisms.
Genome comparative analysis shows significant horizontal gene transfers occurred during the evolution of MS14. The MS14 genome contains 49 genomic islands that are absent in other studied B. contaminans genomes. One genomic island on chromosome 3 encodes a putative‐exported protein that has a 58% amino acid identify to the type VI secretion protein Rhs of Ralstonia sp. GA3‐3 (NCBI Reference Sequence: WP_037024668.1) (Pearce et al. 2013). This protein is predicted to facilitate the exclusion of molecules from the cell wall and is possibly involved in the antibiotic resistance features (Filloux et al. 2008). The acquisition and exchange of mobile genetic elements in addition to GIs has contributed to the overall genome plasticity as well as MS14 antagonistic activities. Presumably, the integration of new biosynthetic pathways has made the bacterial strain more adaptated to various ecological niches. The contribution of the other GIs to survival of MS14 remains unknown, since many of the coding genes in the GIs code for hypothetical proteins.
The average nucleotide identity is one of the most robust measurements of genomic relatedness between strains (Kim et al. 2014). The ANI data show that the nonpathogenic and pathogenic isolates are clustered together, respectively. These results confirm that distinct lineages exist among Burkholderia species (Angus et al. 2014). The ANI values between B. pseudomallei 1026b, B. pseudomallei K96243, and B. mallei ATCC 23344 were previously shown to be all above 99%. Therefore, these three isolates can be considered the same species according to Konstantinidis et al. (Konstantinidis et al. 2006). However, progressive Mauve results on chromosome 1 indicate significant gene shuffling and rearrangement between these three species, suggesting a limited gene synteny between the isolates that have very high ANI values (Fig. S1). What is learned from these observations is that considerable gene rearrangement can occur between two species that have more than 99% ANI. Interestingly, pathogenic B. cenocepacia J2315 has a high genome relatedness to PGPB MS14, 383, and AMMD, even the following genomic analysis shows J2315 harbors almost all key virulent‐related genes, which are not present in the PGPB species. Burkholderia species can have very high flexibility and mosaicism among their genomes. These results support the notion that conventional classification of Burkholderia species is not able to distinguish between pathogenic and nonpathogenic species and that a whole genome comparison as was conducted with MS14 is needed to make this distinction.
MS14, PGPB 383 and AMMD have a wide range of common antimicrobial biosynthesis gene clusters within their genomes, supporting the possible production of multiple antimicrobial agents that contribute to their plant growth‐promoting activity by inhibiting soil‐borne plant pathogens, including pathogenic fungi and bacteria. Those antimicrobial agents include bactericidal agents like pyrrolnitrin and bacteriostatic siderophores. Occidiofungin is highly effective in killing pathogenic fungi by altering cell wall integrity or by inducing apoptosis (Lu et al. 2009; Emrick et al. 2013), however, its biosynthesis gene cluster was not found in the closely related 383. Alternatively, another pyochelin biosynthesis genes were found in 383 that is not present in MS14 genome. These results show that the PGPB strains have multiple mechanisms for inhibiting competing microorganism that increase their survivability.
Euan L.S. Thomson and Jonathan J. Dennis reported that B. vietnamiensis DBO1 harbors an occidiofungin gene cluster homolog that is related to hemolytic activity and virulence (Thomson and Dennis 2012). However, two key polyketide biosynthesis genes 6477 and 6478 presented in DBO1 that are required for full virulence were not found in the MS14 genome. According to the author, the disruption of either of those two polyketide synthase genes led to a complete loss of human erythrocytes hemolytic activity. Conversely, hemolytic activity can be utilized in beneficial way. For example, hemolytic bacterium Pseudomonas entomophila had been utilized as entomopathogenic bacterium in killing insects to protect plants (Vallet‐Gely et al. 2010). Moreover, hemolytic activity has not been reported to increase a strain's chance of being an opportunistic microorganism. According to the same research data, only one out of 30 CF Burkholderia isolates have detectable hemolytic activity, yet regular soil/water Burkholderia isolates have shown moderate to high hemolytic activity. This observation indicates that hemolytic activity is not an important determinant for virulence of Burkholderia species and that the potential for hemolytic activity does not increase the possibility of MS14's chance of being a CF opportunistic bacterium.
Toxin biosynthesis clusters were mainly identified from pathogenic Burkholderia species, as expected. For example, toxoflavin biosynthesis gene cluster was identified from the plant‐pathogen species BSR3 and BGR1, this result further confirmed their pathogenicity in inducing bacterial blight in many field crops (Lim et al. 2009; Seo et al. 2011). The HCN is extremely toxic to both plants and human beings (http://pubchem.ncbi.nlm.nih.gov/compound/hydrogen_cyanide#section=Top; http://www.cyanidecode.org/cyanide-facts/environmental-health-effects). The biosynthesis genes for HCN production are present in all pathogenic species. Additionally, all pathogenic species in this study have the O‐antigen biosynthesis‐related genes associated with CF patient infection. It is not surprising that pathogenic isolates have more toxin‐producing genes. However, PGPB AMMD strain also has the CF‐related O‐antigen biosynthesis genes and hydrogen cyanide biosynthesis genes. This observation shows that the Bcc strain AMMD, which is effective at controlling plant‐pathogenic fungi, may contribute to CF infections. Given the presence of such virulence factors should raise concern for its use as a PGPB in agriculture. Whereas, MS14 and 383 only have the cepacian bce‐I gene cluster. Since cepacian is not required for the initiation of biofilm formation and only facilitates bacterial persistence (Cunha et al. 2004; Ferreira et al. 2010), the production of cepacian is not a major virulence determinant.
Antibiotic resistance is a very common feature among Burkholderia species. Possessing multiple efflux pumps facilitates viability of bacterial cells in different ecological niches. MS14 and 383 do not have arginine and pyruvate fermentation mechanism‐related genes that are crucial in cell energy generation under low oxygen conditions. The species that possess arginine and pyruvate fermentation are more capable of surviving in low oxygen environments, such as the environment found in mucus of CF patients' respiratory system. The cells remain latent in low oxygen environments making them more difficult to be killed by antibiotics (Hamad et al. 2011). Lack of arginine and pyruvate fermentation‐related genes in MS14 and 383 genomes indicates that both of these strains are not likely chronic opportunistic microorganisms.
PGPB and environmental isolates lack the key pathogenic features, such as cable pili formation and adhesin secretion activity. These are necessary for initiating and causing a CF opportunistic infection. Type VI secretion systems that have been reported to be related to animal's melioidosis (Burtnick et al. 2011) have been identified from all researched Burkholderia genomes. However, the VgrG‐5 protein, which is required for type VI secretion system 5 to induce multinucleate giant cell formation and full virulence, has its biosynthesis genes only identified from most pathogenic species. Melanin aids in environmental survival, however, the relationship between the production of melanin and the increase in virulence is unknown (Keith et al. 2007). The virulence feature comparison shows that PGPB and environmental isolates have less virulence‐related genes in their genomes, which indicates that PGPB characterized in this study pose less risk of causing an infection when compared to the pathogenic and CF opportunistic species.
The studied Burkholderia species can be separated into a virulent group and a less‐virulent group based on the data summarized above. The first group includes opportunistic pathogens, mammalian pathogens, and plant pathogens. This group is highly problematic in causing human or plant disease due to multiple virulence‐related genes identified from their genomes. The less‐virulent group includes plant growth‐promoting species, nitrogen‐fixing nodulators, and other environmental isolates that were used in the genomic analysis. The analysis of virulence traits in the genomes of the PGPB further confirmed that the plant‐associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis as described by Angus et al. (Angus et al. 2014). Those studied Burkholderia species are highly unlikely to be pathogenic to either plants or mammals based on the functional gene analysis. Whole genomic analysis approach as described in our study, should be prompted to identify the virulence level of a Burkholderia isolate.
Conclusion
B. contaminans MS14 has three chromosomes with a genome size of 8.5 Mb. Multiple antibiotic and bacteriocin biosynthesis genes contribute to its overall antimicrobial activity. BLASTn result of MS14 against other Burkholderia genomes reveals 362 unique coding genes that are not present in other researched genomes. Fifty‐one percent of the unique genes encode protein products with unknown function, 18% of the unique genes encode for metabolic and secretion system‐related proteins. ANI data indicates MS14 is closely related to B. lata 383 with a 95.28% ANI score, and the ANI data also demonstrates the diversity of Burkholderia species genomes. However, significant syntenic gene rearrangement can occur between two very closely related genomes, for example, between B. pseudomallei K96243 and B. pseudomallei 1026b, which have a more than 99% ANI score. The secondary metabolites biosynthesis genes and virulence‐associated loci were compared among Burkholderia species. These comparisons provide an insight of distinguishing between pathogenic and nonpathogenic Burkholderia species.
MS14 has biosynthesis gene cluster identified for antibiotic occidiofungin, pyrrolnitrin, ornibactin, and AFC‐BC11. Other strains, such as PGPB 383 and AMMD also have similar antibiotic biosynthesis gene clusters identified from their genomes. Conversely, MS14 does not have gene homologs for the production of virulent secondary metabolite toxoflavin, hydrogen cyanide, or 2‐heptyl‐3‐hydroxy‐4(1H)‐quinolone, which are commonly produced by plant‐pathogenic Burkholderia species. Besides, MS14 does not harbor gene loci that are related to key virulence features possessed by human pathogenic Bcc species. For example, MS14 does not have biosynthesis gene homologs for cable pili, 22‐Kilodalton adhesion, virulent protein VgrG‐5, or the intact Burkholderia cepacia virulence genomic island that harbors BCESM. These results indicate that MS14 has a low virulent potential and could be used to separate MS14 from the pathogenic Burkholderia species. Consequently, soil isolates could also be distinguished from pathogenic strains by the analysis of established virulence factors and feature genes.
Conflict of Interests
The authors declare no competing interests.
Ethics Statement
This research did not involve any human or animal subjects, materials, or data and therefore did not require any ethics oversight or approval in these respects.
Supporting information
Table S1. Gene islands of Burkholderia contaminans MS14.
Table S2. Secondary metabolites and antibiotics prediction of Burkholderia contaminans MS14.
Figure S1. Synteny of B. mallei ATCC 23344, B. pseudomallei K96243 and B. pseudomallei 1026b chromosome 1.
Acknowledgment
We thank Richard Baird, Sead Sabanadzovic, and Justin Thornton for helpful discussion, Chuan‐Yu Hsu for preparing one standard DNA‐Seq library. This research was funded by USDA NIFA to SL (MIS‐401170).
MicrobiologyOpen 2016; 5(3): 353–369
Reference
- Agnoli, K. , Lowe C. A., Farmer K. L., Husnain S. I., and Thomas M. S.. 2006. The ornibactin biosynthesis and transport genes of Burkholderia cenocepacia are regulated by an extracytoplasmic function sigma factor which is a part of the Fur regulon. J. Bacteriol. 188:3631–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhan, N. F. , Petty N. K., Ben Zakour N. L., and Beatson S. A.. 2011. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genom. 12:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiuoli, S. V. , Gussman A., Klimke W., Cochrane G., D. Field , Garrity G., et al. 2008. Toward an online repository of standard operating procedures (SOPs) for (meta)genomic annotation. OMICS 12:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus, A. A. , Agapakis C. M., Fong S., Yerrapragada S., Estrada‐de los Santos P., Yang P., et al. 2014. Plant‐associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS ONE 9:e83779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M. , Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., et al. 2000. Gene Ontology: tool for the unification of biology. Nat. Genet. 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, A. , Sokol P. A., Parkhill J., and Mahenthiralingam E.. 2004. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism‐associated genes in Burkholderia cenocepacia. Infect. Immun. 72:1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el ‐Banna, N. , and Winkelmann G.. 1998. Pyrrolnitrin from Burkholderia cepacia: antibiotic activity against fungi and novel activities against streptomycetes. J. Appl. Microbiol. 85:69–78. [DOI] [PubMed] [Google Scholar]
- Bartell, J. A. , Yen P., Varga J. J., Goldberg J. B., and Papin J. A.. 2014. Comparative metabolic systems analysis of pathogenic Burkholderia. J. Bacteriol. 196:210–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, L. E. , and Petrie T.. 1966. Statistical inference for probabilistic functions of finite state markov chains. Ann. Math. Stat. 37:1554–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddick, R. , Spilker T., Martin A., and LiPuma J. J.. 2003. Evidence of transmission of Burkholderia cepacia, Burkholderia multivorans and Burkholderia dolosa among persons with cystic fibrosis. FEMS Microbiol. Lett. 228:57–62. [DOI] [PubMed] [Google Scholar]
- Birchall, G. R. , Hughes C. G., and Rees A. H.. 1970. Newer synthes of the pyoluteorin antibiotics. Tetrahedron Lett. 11:4879–4882. [DOI] [PubMed] [Google Scholar]
- Burtnick, M. N. , Brett P. J., Harding S. V., Ngugi S. A., Ribot W. J., Chantratita N., et al. 2011. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79:1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, Y. Y. , Tan T. M., Ong Y. M., and Chua K. L.. 2004. BpeAB‐OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 48:1128–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye, T. , Mahenthiralingam E., Henry D., LiPuma J. J., Laevens S., Gillis M., et al. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis‐related isolates. Int. J. Syst. Evol. Microbiol. 51:1481–1490. [DOI] [PubMed] [Google Scholar]
- Conesa, A. , Götz S., García‐Gómez J. M., Terol J., Talón M., and Robles M.. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Corbett, C. R. , Burtnick M. N., Kooi C., Woods D. E., and Sokol P. A.. 2003. An extracellular zinc metalloprotease gene of Burkholderia cepacia. Microbiology 149:2263–2271. [DOI] [PubMed] [Google Scholar]
- Costa, R. , van Aarle I. M., Mendes R., and van Elsas J. D.. 2009. Genomics of pyrrolnitrin biosynthetic loci: evidence for conservation and whole‐operon mobility within gram‐negative bacteria. Environ. Microbiol. 11:159–175. [DOI] [PubMed] [Google Scholar]
- Cunha, M. V. , Sousa S. A., Leitao J. H., Moreira L. M., Videira P. A., and Sa‐Correia I.. 2004. Studies on the involvement of the exopolysaccharide produced by cystic fibrosis‐associated isolates of the Burkholderia cepacia complex in biofilm formation and in persistence of respiratory infections. J. Clin. Microbiol. 42:3052–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling, A. E. , Mau B., and Perna N. T.. 2010. ProgressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer, S. E. , Cnockaert M., Ardley J. K., Maker G., Yates R., Howieson J. G., et al. 2013. Burkholderia sprentiae sp. nov., isolated from Lebeckia ambigua root nodules. Int. J. Syst. Evol. Microbiol. 63:3950–3957. [DOI] [PubMed] [Google Scholar]
- DeShazer, D. , Waag D. M., Fritz D. L., and Woods D. E.. 2001. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microb. Pathog. 30:253–269. [DOI] [PubMed] [Google Scholar]
- Diggle, S. P. , Lumjiaktase P., Dipilato F., Winzer K., Kunakorn M., Barrett D. A., et al. 2006. Functional genetic analysis reveals a 2‐Alkyl‐4‐quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem. Biol. 13:701–710. [DOI] [PubMed] [Google Scholar]
- Durfee, T. , Nelson R., Baldwin S., Plunkett G. 3rd, Burland V., Mau B., et al. 2008. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J. Bacteriol. 190:2597–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, G. N. , Chen W. M., Chou J. H., Wang H. C., Sheu S. Y., Perin L., et al. 2007. Burkholderia phymatum is a highly effective nitrogen‐fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 173:168–180. [DOI] [PubMed] [Google Scholar]
- Ellis, D. , Gosai J., Emrick C., Heintz R., Romans L., Gordon D., et al. 2012. Occidiofungin's chemical stability and in vitro potency against Candida species. Antimicrob. Agents Chemother. 56:765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrick, D. , Ravichandran A., Gosai J., Lu S., Gordon D. M., and Smith L.. 2013. The antifungal occidiofungin triggers an apoptotic mechanism of cell death in yeast. J. Nat. Prod. 76:829–838. [DOI] [PubMed] [Google Scholar]
- Eustaquio, A. S. , Janso J. E., Ratnayake A. S., O'Donnell C. J., and Koehn F. E.. 2014. Spliceostatin hemiketal biosynthesis in Burkholderia spp. is catalyzed by an iron/alpha‐ketoglutarate‐dependent dioxygenase. Proc. Natl Acad. Sci. USA 111:E3376–E3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, A. S. , Leitao J. H., Silva I. N., Pinheiro P. F., Sousa S. A., Ramos C. G., et al. 2010. Distribution of cepacian biosynthesis genes among environmental and clinical Burkholderia strains and role of cepacian exopolysaccharide in resistance to stress conditions. Appl. Environ. Microbiol. 76:441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux, A. , Hachani A., and Bleves S.. 2008. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154:1570–1583. [DOI] [PubMed] [Google Scholar]
- Francis, F. , Kim J., Ramaraj T., Farmer A., Rush M. C., and Ham J. H.. 2013. Comparative genomic analysis of two Burkholderia glumae strains from different geographic origins reveals a high degree of plasticity in genome structure associated with genomic islands. Mol. Genet. Genomics 288:195–203. [DOI] [PubMed] [Google Scholar]
- Fries, M. R. , Forney L. J., and Tiedje J. M.. 1997. Phenol‐ and toluene‐degrading microbial populations from an aquifer in which successful trichloroethene cometabolism occurred. Appl. Environ. Microbiol. 63:1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass, M. B. , Steigerwalt A. G., Jordan J. G., Wilkins P. P., and Gee J. E.. 2006. Burkholderia oklahomensis sp. nov., a Burkholderia pseudomallei‐like species formerly known as the Oklahoma strain of Pseudomonas pseudomallei. Int. J. Syst. Evol. Microbiol. 56:2171–2176. [DOI] [PubMed] [Google Scholar]
- Gu, G. , Smith L., Wang N., Wang H., and Lu S. E.. 2009. Biosynthesis of an antifungal oligopeptide in Burkholderia contaminans strain MS14. Biochem. Biophys. Res. Commun. 380:328–332. [DOI] [PubMed] [Google Scholar]
- Gu, G. , Smith L., Liu A., and Lu S. E.. 2011. Genetic and biochemical map for the biosynthesis of occidiofungin, an antifungal produced by Burkholderia contaminans strain MS14. Appl. Environ. Microbiol. 77:6189–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham, J. H. , Melanson R. A., and Rush M. C.. 2011. Burkholderia glumae: next major pathogen of rice? Mol. Plant. Pathol. 12:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamad, M. A. , Austin C. R., Stewart A. L., Higgins M., Vazquez‐Torres A., and Voskuil M. I.. 2011. Adaptation and antibiotic tolerance of anaerobic Burkholderia pseudomallei. Antimicrob. Agents Chemother. 55:3313–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heungens, K. , and Parke J. L.. 2000. Zoospore homing and infection events: effects of the biocontrol bacterium Burkholderia cepacia AMMDR1 on two oomycete pathogens of pea (Pisum sativum L.). Appl. Environ. Microbiol. 66:5192–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hider, R. C. , and Kong X.. 2010. Chemistry and biology of siderophores. Nat. Prod. Rep. 27:637–657. [DOI] [PubMed] [Google Scholar]
- Holden, M. T. , Seth‐Smith H. M., Crossman L. C., Sebaihia M., Bentley S. D., Cerdeno‐Tarraga A. M., et al. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 191:261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, K. W. , Koh C. L., Sam C. K., Yin W. F., and Chan K. G.. 2012. Complete genome sequence of Burkholderia sp. Strain GG4, a betaproteobacterium that reduces 3‐oxo‐N‐acylhomoserine lactones and produces different N‐acylhomoserine lactones. J. Bacteriol. 194:6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, B. , Riedel K., Hentzer M., Heydorn A., Gotschlich A., Givskov M., et al. 2001. The cep quorum‐sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517–2528. [DOI] [PubMed] [Google Scholar]
- Kanehisa, M. , and Goto S.. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Y. , Carlson R., Tharpe W., and Schell M. A.. 1998. Characterization of genes involved in biosynthesis of a novel antibiotic from Burkholderia cepacia BC11 and their role in biological control of Rhizoctonia solani. Appl. Environ. Microbiol. 64:3939–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, K. E. , and Valvano M. A.. 2007. Characterization of SodC, a periplasmic superoxide dismutase from Burkholderia cenocepacia. Infect. Immun. 75:2451–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, K. E. , Killip L., He P., Moran G. R., and Valvano M. A.. 2007. Burkholderia cenocepacia C5424 produces a pigment with antioxidant properties using a homogentisate intermediate. J. Bacteriol. 189:9057–9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. , Oh H. S., Park S. C., and Chun J.. 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64:346–351. [DOI] [PubMed] [Google Scholar]
- Knappe, T. A. , Linne U., Zirah S., Rebuffat S., Xie X., and Marahiel M. A.. 2008. Isolation and structural characterization of capistruin, a lasso peptide predicted from the genome sequence of Burkholderia thailandensis E264. J. Am. Chem. Soc. 130:11446–11454. [DOI] [PubMed] [Google Scholar]
- Konstantinidis, K. T. , Ramette A., and Tiedje J. M.. 2006. The bacterial species definition in the genomic era. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1929–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski, M. , Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille, M. G. , and Brinkman F. S.. 2009. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 25:664–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille, M. G. , Hsiao W. W., and Brinkman F. S.. 2008. Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinformatics 9:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latuasan, H. E. , and Berends W.. 1961. On the origin of the toxicity of toxoflavin. Biochim. Biophys. Acta 52:502–508. [DOI] [PubMed] [Google Scholar]
- Lessie, T. G. , Hendrickson W., Manning B. D., and Devereux R.. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117–128. [DOI] [PubMed] [Google Scholar]
- Lim, J. , Lee T. H., Nahm B. H., Choi Y. D., Kim M., and Hwang I.. 2009. Complete genome sequence of Burkholderia glumae BGR1. J. Bacteriol. 191:3758–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipuma, J. J. 2005. Update on the Burkholderia cepacia complex. Curr. Opin. Pulm. Med. 11:528–533. [DOI] [PubMed] [Google Scholar]
- Lu, S. E. , Novak J., Austin F. W., Gu G., Ellis D., Kirk M., et al. 2009. Occidiofungin, a unique antifungal glycopeptide produced by a strain of Burkholderia contaminans . Biochemistry 48:8312–8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam, E. , Baldwin A., and Vandamme P.. 2002. Burkholderia cepacia complex infection in patients with cystic fibrosis. J. Med. Microbiol. 51:533–538. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam, E. , Urban T. A., and Goldberg J. B.. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144–156. [DOI] [PubMed] [Google Scholar]
- Mahenthiralingam, E. , Baldwin A., and Dowson C. G.. 2008. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104:1539–1551. [DOI] [PubMed] [Google Scholar]
- Medema, M. H. , Blin K., Cimermancic P., de Jager V., Zakrzewski P., Fischbach M. A., et al. 2011. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39:W339–W346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, J. M. , Van V. T., Stintzi A., Berge O., and Winkelmann G.. 1995. Ornibactin production and transport properties in strains of Burkholderia vietnamiensis and Burkholderia cepacia (formerly Pseudomonas cepacia). Biometals 8:309–317. [DOI] [PubMed] [Google Scholar]
- Moore, R. A. , DeShazer D., Reckseidler S., Weissman A., and Woods D. E.. 1999. Efflux‐mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43:465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, B. M. , Cheung K. J. Jr, Griffith A., Burns J. L.. 2004. Salicylate induces an antibiotic efflux pump in Burkholderia cepacia complex genomovar III (B. cenocepacia). J. Clin. Invest. 113:464–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Cunha, C. , Zuleta Goda L. F., Paula de Almeida L. G., Prioli Ciapina L., Prioli Ciapina L., Lustrino Borges W., et al. 2012. Complete genome sequence of Burkholderia phenoliruptrix BR3459a (CLA1), a heat‐tolerant, nitrogen‐fixing symbiont of Mimosa flocculosa. J. Bacteriol. 194:6675–6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega, X. , Hunt T. A., Loutet S., Vinion‐Dubiel A. D., Datta A., Choudhury B., et al. 2005. Reconstitution of O‐specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J. Bacteriol. 187:1324–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parke, J. L. , and Gurian‐Sherman D.. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39:225–258. [DOI] [PubMed] [Google Scholar]
- Partida‐Martinez, L. P. , and Hertweck C.. 2007. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. ChemBioChem 8:41–45. [DOI] [PubMed] [Google Scholar]
- Pearce, S. L. , Pushiri H., Oakeshott J. G., Russell R. J., and Pandey G.. 2013. Draft genome sequence of ralstonia sp. strain ga3‐3, isolated from Australian suburban soil. Genome. Announc. 1:e00414–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podnecky, N. L. , Wuthiekanun V., Peacock S. J., and Schweizer H. P.. 2013. The BpeEF‐OprC efflux pump is responsible for widespread trimethoprim resistance in clinical and environmental Burkholderia pseudomallei isolates. Antimicrob. Agents Chemother. 57:4381–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran, A. , Gu G., Escano J., Lu S. E., and Smith L.. 2013. The presence of two cyclase thioesterases expands the conformational freedom of the cyclic Peptide occidiofungin. J. Nat. Prod. 76:150–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, M. , and Rossello‐Mora R.. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl Acad. Sci. USA 106:19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost, B. 1999. Twilight zone of protein sequence alignments. Protein Eng. 12:85–94. [DOI] [PubMed] [Google Scholar]
- Ryall, B. , Lee X., Zlosnik J. E., Hoshino S., and Williams H. D.. 2008. Bacteria of the Burkholderia cepacia complex are cyanogenic under biofilm and colonial growth conditions. BMC Microbiol. 8:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawasdidoln, C. , Taweechaisupapong S., Sermswan R. W., U. Tattawasart , Tungpradabkul S., and Wongratanacheewin S.. 2010. Growing Burkholderia pseudomallei in biofilm stimulating conditions significantly induces antimicrobial resistance. PLoS ONE 5:e9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, S. , Singh P., Robertson J. D., LeRoux M., Skerrett S. J., Goodlett D. R., et al. 2014. VgrG‐5 is a Burkholderia type VI secretion system‐exported protein required for multinucleated giant cell formation and virulence. Infect. Immun. 82:1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, Y. S. , Lim J., Choi B. S., Kim H., Goo E., Lee B., et al. 2011. Complete genome sequence of Burkholderia gladioli BSR3. J. Bacteriol. 193:3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino, L. , Reimmann C., Visca P., Beyeler M., Chiesa V. D., and Haas D.. 1997. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron‐regulated pchDCBA operon in Pseudomonas aeruginosa. J. Bacteriol. 179:248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessitsch, A. , Coenye T., Sturz A. V., Vandamme P., Barka E. A., Salles J. F., et al. 2005. Burkholderia phytofirmans sp. nov., a novel plant‐associated bacterium with plant‐beneficial properties. Int. J. Syst. Evol. Microbiol. 55:1187–1192. [DOI] [PubMed] [Google Scholar]
- Stoyanova, M. , Pavlina I., Moncheva P., and Bogatzevska N.. 2007. Biodiversity and Incidence of Burkholderia Species. Biotechnol. Biotechnol. Equip. 21:306–310. [Google Scholar]
- Sullivan, M. J. , Petty N. K., and Beatson S. A.. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, F. , Sawada H., Azegami K., and Tsuchiya K.. 2004. Molecular characterization of the tox operon involved in toxoflavin biosynthesis of Burkholderia glumae. J. Gen. Plant Pathol. 70:97–107. [Google Scholar]
- Tatusov, R. L. , Fedorova N. D., Jackson J. D., Jacobs A. R., Kiryutin B., Koonin E. V., et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, E. L. , and Dennis J. J.. 2012. A Burkholderia cepacia complex non‐ribosomal peptide‐synthesized toxin is hemolytic and required for full virulence. Virulence 3:286–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribuddharat, C. , Moore R. A., Baker P., and Woods D. E.. 2003. Burkholderia pseudomallei class a beta‐lactamase mutations that confer selective resistance against ceftazidime or clavulanic acid inhibition. Antimicrob. Agents Chemother. 47:2082–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, T. A. , Goldberg J. B., Forstner J. F., and Sajjan U. S.. 2005. Cable pili and the 22‐kilodalton adhesin are required for Burkholderia cenocepacia binding to and transmigration across the squamous epithelium. Infect. Immun. 73:5426–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet‐Gely, I. , Novikov A., Augusto L., Liehl P., Bolbach G., Pechy‐Tarr M., et al. 2010. Association of hemolytic activity of Pseudomonas entomophila, a versatile soil bacterium, with cyclic lipopeptide production. Appl. Environ. Microbiol. 76:910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme, P. , Goris J., Chen W. M., de Vos P., and Willems A.. 2002. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25:507–512. [DOI] [PubMed] [Google Scholar]
- Vanlaere, E. , Baldwin A., Gevers D., Henry D., De Brandt E., LiPuma J. J., et al. 2009. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59:102–111. [DOI] [PubMed] [Google Scholar]
- Vermis, K. , Coenye T., LiPuma J. J., Mahenthiralingam E., Nelis H. J., and Vandamme P.. 2004. Proposal to accommodate Burkholderia cepacia genomovar VI as Burkholderia dolosa sp. nov. Int. J. Syst. Evol. Microbiol. 54:689–691. [DOI] [PubMed] [Google Scholar]
- Vidaver, A. K. 1967. Synthetic and complex media for the rapid detection of fluorescence of phytopathogenic pseudomonads: effect of the carbon source. Appl. Microbiol. 15:1523–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waack, S. , Keller O., Asper R., Brodag T., Damm C., Fricke W. F., et al. 2006. Score‐based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics 7:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilharter, A. , Mitter B., Shin M. V., Chain P. S., Nowak J., and Sessitsch A.. 2011. Complete genome sequence of the plant growth‐promoting endophyte Burkholderia phytofirmans strain PsJN. J. Bacteriol. 193:3383–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zughaier, S. M. , Ryley H. C., and Jackson S. K.. 1999. A melanin pigment purified from an epidemic strain of Burkholderia cepacia attenuates monocyte respiratory burst activity by scavenging superoxide anion. Infect. Immun. 67:908–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuleta, L. F. , Cunha Cde. O., de Carvalho F. M., Ciapina L. P., Souza R. C., Mercante F. M., et al. 2014. The complete genome of Burkholderia phenoliruptrix strain BR3459a, a symbiont of Mimosa flocculosa: highlighting the coexistence of symbiotic and pathogenic genes. BMC Genom. 15:535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga, M. , Perez G., and Gonzalez‐Candelas F.. 2002. Evolution of arginine deiminase (ADI) pathway genes. Mol. Phylogenet. Evol. 25:429–444. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Gene islands of Burkholderia contaminans MS14.
Table S2. Secondary metabolites and antibiotics prediction of Burkholderia contaminans MS14.
Figure S1. Synteny of B. mallei ATCC 23344, B. pseudomallei K96243 and B. pseudomallei 1026b chromosome 1.


