Abstract
In rod‐shaped bacteria, the proper placement of the division septum at the midcell relies, at least partially, on the proteins of the Min system as an inhibitor of cell division. The main principle of Min system function involves the formation of an inhibitor gradient along the cell axis; however, the establishment of this gradient differs between two well‐studied gram‐negative and gram‐positive bacteria. While in gram‐negative Escherichia coli, the Min system undergoes pole‐to‐pole oscillation, in gram‐positive Bacillus subtilis, proper spatial inhibition is achieved by the preferential attraction of the Min proteins to the cell poles. Nevertheless, when E.coli Min proteins are inserted into B.subtilis cells, they still oscillate, which negatively affects asymmetric septation during sporulation in this organism. Interestingly, homologs of both Min systems were found to be present in various combinations in the genomes of anaerobic and endospore‐forming Clostridia, including the pathogenic Clostridium difficile. Here, we have investigated the localization and behavior of C.difficile Min protein homologs and showed that MinDE proteins of C.difficile can oscillate when expressed together in B.subtilis cells. We have also investigated the effects of this oscillation on B.subtilis sporulation, and observed decreased sporulation efficiency in strains harboring the MinDE genes. Additionally, we have evaluated the effects of C.difficile Min protein expression on vegetative division in this heterologous host.
Keywords: Bacillus subtilis, bacterial cell division, Clostridium difficile, Min system oscillation, sporulation
Introduction
Gram‐positive Bacillus subtilis and gram‐negative Escherichia coli are the most common model organisms used for studying cell division in rod‐shaped bacteria. Bacterial cell division is a strictly controlled, binary fission process leading to the formation of two equal daughter cells. FtsZ, a tubulin‐like protein, forms a structure termed the Z‐ring, which marks the position of the future division septum and serves as a scaffold for downstream division proteins. The placement of the division septum at the midcell site is very precise and the details of how this is achieved are still unknown. Two different mechanisms which have a negative effect on Z‐ring assembly have been described: nucleoid occlusion and the Min system (reviewed in Barák and Wilkinson 2007; Wu and Errington 2011; Rowlett and Margolin 2015). Recently, positive regulators of Z‐ring placement have been reported – the SsgAB system found in Streptomyces coelicolor (Willemse and van Wezel 2009), PomZ in Myxococcus xanthus (Treuner‐Lange et al. 2013), and MapZ in Streptococcus pneumoniae (Fleurie et al. 2014). The existence of a similar mechanism in B.subtilis has also been proposed (Monahan et al. 2014). It seems that Z‐ring placement is controlled differently by different bacterial species; many of the proteins involved in these systems are not highly conserved.
The Min system efficiently blocks unwanted polar septation during vegetative growth by creating a concentration gradient along the cell axis and hence protecting the polar sites from Z‐ring formation. The key component of the Min system is the MinC protein, which prevents Z‐ring formation by preventing FtsZ polymerization and by inhibiting interactions between FtsZ protofilaments (reviewed in Adams and Errington 2009). MinC is recruited to the cytoplasmic membrane, thereby triggering its inhibitory activity, by interacting with MinD, which binds reversibly to organized groups of anionic phospholipids within the membrane (Hu and Lutkenhaus 2001; Hu et al. 2002; Barák et al. 2008). The specific action and localization pattern of the MinCD complex at polar sites depends on an interaction with a third component of the Min system, termed the topological determinant. It is MinE in E.coli and DivIVA/MinJ in B.subtilis.
The behavior of E.coli Min proteins is based on a finely tuned interaction between MinE and MinD, and is highly dynamic. Upon binding of MinE to MinD, the ATPase activity of MinD is stimulated, resulting in the dissociation of the MinCD complex from the membrane and its reassociation at an adjacent site. This is manifested as rapid oscillation of all three proteins from one pole to the other, creating a bipolar MinC gradient and leaving only one place at the midcell site for FtsZ polymerization (Hu and Lutkenhaus 1999; Raskin and de Boer 1999a,b). B.subtilis does not encode a MinE homolog, on the other hand, and the polar localization of MinCD is achieved by an interaction with MinJ, which links the MinCD complex to the DivIVA protein (Bramkamp et al. 2008; Patrick and Kearns 2008). DivIVA stably localizes at the sites of septation based on its ability to bind to negatively curved membranes (Cha and Stewart 1997; Edwards and Errington 1997; Lenarcic et al. 2009; Eswaramoorthy et al. 2011), and it also persists at the cell poles. The preferential attraction of MinCD to the newly forming cell poles under the influence of MinJ/DivIVA blocks polar division in B.subtilis. This system is not entirely static: fast membrane dissociation and reassociation of B.subtilis MinD (MinDBs), which retains its ATPase activity, has been observed (Barák et al. 2008), but MinDBs does not drive the rapid oscillation of MinC as E.coli MinD (MinDEc) does in the presence of MinEEc.
One of the possible paths of the B.subtilis cell cycle is sporulation, which begins with asymmetric cell division. During vegetative cell division, DivIVA helps position MinCD at the cell poles, but during sporulation it plays a role in the proper segregation of chromosomes (Thomaides et al. 2001) by attracting the RacA protein (Ben‐Yehuda et al. 2003). RacA recognizes the oriC region of elongated sister chromosomes and recruits these chromosomes to the cell poles (Thomaides et al. 2001). In addition, it was shown that DivIVA also interacts with SpoIIE, the most crucial protein for asymmetric cell division (Eswaramoorthy et al. 2014).
Min systems are not essential for cell viability, however, their absence has a clear effect on the cell phenotype. In min mutant strains, polar cell division produces mixtures of “mini” cells, which lack chromosomes, and extended rods containing multiple nucleoids (Adler et al. 1967; Reeve et al. 1973; de Boer et al. 1989). Furthermore, when E.coli MinD and MinE are introduced into B.subtilis, MinDEc oscillates just as it does in E.coli cells, and this oscillating system interferes with asymmetric septum formation during B.subtilis sporulation (Jamroškovič et al. 2012). Given the clear difference in the phospholipid composition of the E.coli and B.subtilis membranes (Kusters et al. 1991; López et al. 1998), this behavior was somewhat unexpected.
Many spore‐forming bacteria from the phylum Firmicutes, including the Clostridia, contain homologs from both MinCDE and MinCDJ/DivIVA systems. For example, the gram‐positive pathogenic spore‐former Clostridium difficile harbors homologs of MinC, MinD, MinE, and also DivIVA. Exactly which homologs are present varies according to the organism (Stragier 2002; Jamroškovič et al. 2012; this study) and it is not known whether they form a Min system which behaves as either of the two described. It is not even known whether all of these homologs are functional.
Because of the nature of C.difficile anaerobic lifestyle and its confined genetic toolbox, we have decided to address these questions by investigating the mechanism of action of the C.difficile Min proteins (MinCd) in a heterologous B.subtilis host. We found that the Min proteins of C.difficile are functional in a heterologous host B.subtilis and can affect its vegetative division. We also found that the C.difficile MinD and MinE proteins exhibit oscillation, meaning that oscillating Min proteins are not confined only to gram‐negative bacteria. Oscillation of a YFP‐MinDCd fusion protein was observed in B.subtilis cells in the presence of MinECd. The same behavior can also be seen by combining the MinD and MinE proteins from E.coli and C.difficile, which opens interesting questions about the evolution of Min systems and the origins of gram‐positive and gram‐negative bacteria. Finally, we noted that the sporulation efficiency of those strains where oscillation was observed was diminished, indicating that either MinCd proteins or their oscillation interferes with the process of sporulation in B.subtilis.
Experimental Procedures
Culture conditions and bacterial strains
Strains were grown in LB (Luria‐Bertani) medium (Ausubel et al. 1987) or in DSM (Difco sporulation medium)(Schaeffer et al. 1965) at 37°C. DNA manipulations and E.coli transformations were performed according to Sambrook et al. (1989). The B.subtilis strains used in this work are derivatives of B.subtilis MO1099, and were prepared by transformation using plasmid or chromosomal DNA following the method of Harwood and Cutting (1990). All B.subtilis and E.coli strains used in this study are listed in Table 1 and details of their construction are in Table S1. When required, media were supplemented with ampicillin (100 μg mLˉ1), spectinomycin (100 μg mLˉ1), erythromycin (1 μg mLˉ1), lincomycin (25 μg mLˉ1), kanamycin (10 μg mLˉ1 or 30 μg mLˉ1), chloramphenicol (5 μg mLˉ1), and tetracycline (5 μg mLˉ1). To induce the expression of proteins under the control of the Pxyl and Phyperspank promoters, the media were supplemented, respectively, with xylose at t0 to a final concentration of 0.02–0.5% (w/v) and IPTG (isopropyl β‐D‐1‐thiogalactopyranoside) to a final concentration of 0.1–0.5 mmol/L.
Table 1.
Bacterial strains used in this study
| Strain or plasmid | Genotype or description | Construction | Reference or origin |
|---|---|---|---|
| B. subtilis | |||
| PY79 | Prototrophic derivative of B. subtilis 168 | Youngman et al. (1984) | |
| MO649 | thrC::cat | Guérout‐Fleury et al. (1996) | |
| MO1099 | amyE::erm | Guérout‐Fleury et al. (1996) | |
| IB220 | spo0A::kan | Schmeisser et al. (2000) | |
| IB1056 | minD Bs ::cat, amyE::erm | Barák et al. (2008) | |
| IB1059 | minD Bs ::cat, amy::P xyl ‐gfp‐minD Bs spc | Pavlendová et al. (2010) | |
| IB1111 | minD Bs ::cat, amyE::P hyperspank ‐yfp‐minD Ec spc | Pavlendová et al. (2010) | |
| IB1112 | minD Bs ::cat, divIVA::tet amyE::P hyperspank ‐yfp‐minD Ec spc | Pavlendová et al. (2010) | |
| IB1141 | minC Bs ::kan | Pavlendová et al. (2010) | |
| IB1242 | minD Bs ::cat, divIVA Bs ::tet, amyE::P hyperspank ‐yfp‐minD Ec spc, thrC::P xyl ‐minE Ec erm | Jamroškovič et al. (2012) | |
| IB1362 | minJ Bs ::kan | Jamroškovič et al. (2012) | |
| IB1371 | minCD Bs ::kan | Jamroškovič et al. (2012) | |
| IB1410 | thrC::P xyl ‐minE cd erm | MO649::pNP‐minECd | This study |
| IB1412 | minD Bs ::cat, amyE::P hyperspank ‐yfp‐minD Ec spc, thr::P xyl ‐minE Cd erm | IB1111::chr DNA IB1410 | This study |
| IB1413 | minD Bs ::cat, divIVA::tet, amyE::P hyperspank ‐yfp‐ minD Ec spc, thr::P xyl ‐minE Cd erm | IB1112::chr DNA IB1410 | This study |
| IB1415 | amyE::P hyperspank ‐yfp‐minD cd spc | MO1099::pED‐yfp‐minDCd | This study |
| IB1416 | minD Bs ::cat, amyE::P hyperspank ‐yfp‐minD Cd spc | IB1056::chr DNA IB1415 | This study |
| IB1553 | minJ Bs ::kan, amyE::P hyperspank ‐yfp‐minD Cd spc | IB1415::chr DNA IB1362 | This study |
| IB1417 | amyE::P hyperspank ‐yfp‐minD Cd spc, thrC::P xyl ‐minE Cd erm | IB1415::chr DNA IB1410 | This study |
| IB1418 | minD Bs ::cat, amyE::P hyperspank ‐yfp‐minD Cd spc, thrC::P xyl ‐minE Cd erm | IB1416::chr DNA IB1410 | This study |
| IB1419 | minD Bs ::cat, minJ Bs ::kan, amyE::P hyperspank ‐yfp‐minD Cd spc | IB1416::chr DNA IB1362 | This study |
| IB1545 | minD Bs ::cat, minJ Bs ::kan | IB1056::chr DNA IB1362 | This study |
| IB1546 | minD Bs ::cat, minJ Bs ::kan, amyE::P hyperspank ‐yfp‐minD Cd spc, thrC::P xyl ‐ minE Cd erm | IB1419::chr DNA IB1410 | This study |
| IB1549 | amyE::P xyl ‐minC Cd spc | MO1099::pSG‐minCCd | This study |
| IB1550 | minC::kan, amyE::P xyl ‐minC Cd spc | IB1549::chr DNA IB1141 | This study |
| IB1552 | amyE::P xyl ‐minE Cd ‐mgfp spc | MO1099::pSG‐minECd‐mGFP | This study |
| IB1562 | minD Bs ::cat, minJ Bs ::kan, amyE::P xyl ‐gfp‐minD Bs spc, thrC::P xyl ‐minE Cd erm | IB1059::chr DNA IB1362::chr DNA IB1410 | This study |
| E. coli | |||
| MM294 | F − endA1 hsdR17 (rk ‐ , mk) supE44 thi‐1 recA + | Meselson and Yuan (1968) | |
| BTH101 | F − cya‐99 araD139 galE15 galK16 rpsL1(Str R ) hsdR2 mcrA1 mcrB1 | Karimova et al.(1998) | |
| C. difficile | |||
| C. difficile 630 | kind gift from Prof. Neil Fairweather | ||
Construction of recombinant plasmids
Plasmids were constructed using standard methods and propagated in E.coli MM294; their construction is described in Table S1. Primers used in the study are listed in Table S2. All PCR fragments were amplified from the chromosomal DNA of the C.difficile 630 strain (kind gift from Prof. Neil Fairweather, Imperial College London). C.difficile has been recently renamed to Peptoclostridium difficile (Yutin and Galperin 2013), but we continue to use its traditional name here.
Sporulation efficiency
The sporulation efficiency assay was performed as described in Harwood and Cutting (1990). Sporulation was induced by nutrient exhaustion in liquid DSM supplemented with 0.5 mmol/L IPTG, 0.5% xylose (w/v), and half the dose of the appropriate antibiotics at 37°C for 24 h after inoculation. Afterward, aliquots of the culture were serially diluted and plated on to LB plates before and after heat treatment (85°C, 15 min). Colonies formed from nontreated samples contain viable cells, those formed from heat‐treated samples contain cells that were able to sporulate. The sporulation efficiency was defined in terms of colony‐forming units (CFU) formed from nontreated samples (viable cells) and heat‐treated samples (spores), and was normalized against the sporulation efficiency of the originating strain. Each strain was assayed at least three times. The agar plates for photography were prepared by resuspending a single colony in 100 μL of liquid DSM and applying 10 μL of this suspension to DSM plates supplemented with 0.1 mmol/L IPTG and 0.02% xylose (w/v). These plates were sealed and incubated for 14 days at room temperature.
Fluorescence microscopy and cell length determination
Bacillus subtilis strains were inoculated from a fresh overnight plate to an OD600 of 0.1 and grown as liquid cultures in DSM to the desired phase. Protein expression was induced at t0 by the addition of IPTG and/or xylose to a final concentration of 0.1–0.5 mmol/L and 0.02–0.3% (w/v), respectively. A small amount of culture was examined microscopically on freshly prepared poly‐L‐lysine‐treated slides or transferred to microscope slides covered with a thin layer of 1% agarose in LB. If necessary, cells were concentrated by centrifugation (3 min at 2300 g) and resuspended in a small volume of supernatant prior to microscopic examination. Cells and septal membranes were visualized by staining the cell cultures with FM 4–64 (Molecular Probes) at a concentration of 1 mg mL−1. YFP, GFP, and FM 4–64 fluorescence was observed using an Olympus BX63 microscope equipped with a Hamamatsu Orca‐R2 camera and analysed by Olympus CellP imaging software. The length of the B.subtilis cells was measured as described previously (Pavlendová et al. 2010). Briefly, B.subtilis cultures were grown as for fluorescence microscopy. Prior to examination, cultures were stained with FM 4–64. The cell length was taken to be the axis length from one cell pole to the other as measured using ImageJ (http://imagej.nih.gov/ij/). The average cell length was determined for at least 500 cells from each sample. Minicells were not included in the calculations of the average cell lengths.
Bacterial two hybrid system
Fusions of MinDCd to the T18 and T25 fragments of adenylate cyclase were constructed for the bacterial adenylate cyclase two‐hybrid (BACTH) system (Karimova et al. 1998). The MinDCd sequence was PCR‐amplified using the respective primer pairs (Table S2) with the chromosomal DNA of C.difficile 630 as a template. These PCR fragments were then cloned into the BamHI/EcoRI sites of the pUT18, pUT18C, pKT25, and pKNT25 plasmids. Fusions of MinCBs, MinDBs, and MinJBs in the BACTH system had been previously prepared (Jamroškovič et al. 2012). To test protein–protein interactions, the adenylate cyclase‐deficient E.coli BTH101 strain was cotransformed with various plasmid combinations and plated onto LB plates supplemented with X‐gal (40 μg mL−1), IPTG (0.1 mmol/L), ampicillin (100 μg mL−1), and kanamycin (30 μg mL−1), and grown for 24–72 h at 30°C. Constructs were tested for autoinduction with the originating vectors containing only individual fragments of adenylate cyclase. The β‐galactosidase activity was measured as described by Miller (1972).
Bioinformatic analysis
The NCBI's PSI‐BLAST program (Altschul et al. 1997) was used to search for homologs using the default threshold of 0.005 and to evaluate identity and similarity of homologous sequences. The sequences of the following strains were used in queries and alignments: B.subtilis (Bacillus subtilis PY79; taxid: 1415167), C.difficile (Peptoclostridium difficile 630; taxid: 272563), and E. coli (E.coli str. K‐12 substr. MG1655; taxid: 511145). Specific strains of clostridia were selected based on the availability of their whole genome sequence. The positions amphipatic helices were predicted using AmphipaSeek (Sapay et al. 2006). Multiple alignment of protein sequences was done using ClustalW plugin of CLC Sequence Viewer 7.6 software (CLC Bio, Cambridge, MA).
Results
Clostridium difficile Min proteins can influence B.subtilis cell division
Our first question to address was whether the proteins of C.difficile are functional and could affect B.subtilis cell division. It was previously shown that higher expression of MinCBs and MinDBs in B.subtilis has a negative effect on bacterial cell division, resulting in elongation of the cells (Marston and Errington 1999). This effect was also observed when the E.coli MinCEc and MinDEc proteins were heterologously overexpressed in B.subtilis cells (Pavlendová et al. 2010). The average cell length of these elongated cells was 4 μm. To examine the effect of the C.difficile Min proteins on B.subtilis cells, we placed the corresponding genes under the control of inducible promoters. The resulting strains are listed in Table 1. Measurements of cell length were performed with no inducer and with both low and high concentrations of inducer (low = 0.1 mmol/L IPTG and/or 0.02% xylose; high = 0.5 mmol/L IPTG and/or 0.3% xylose); the results are summarized in Table S3 and are illustrated in Figure 1. Additionally, the average cell length of the wild‐type strain (MO1099) was measured with and without the addition of xylose, to exclude its effect on cell division (not shown).
Figure 1.
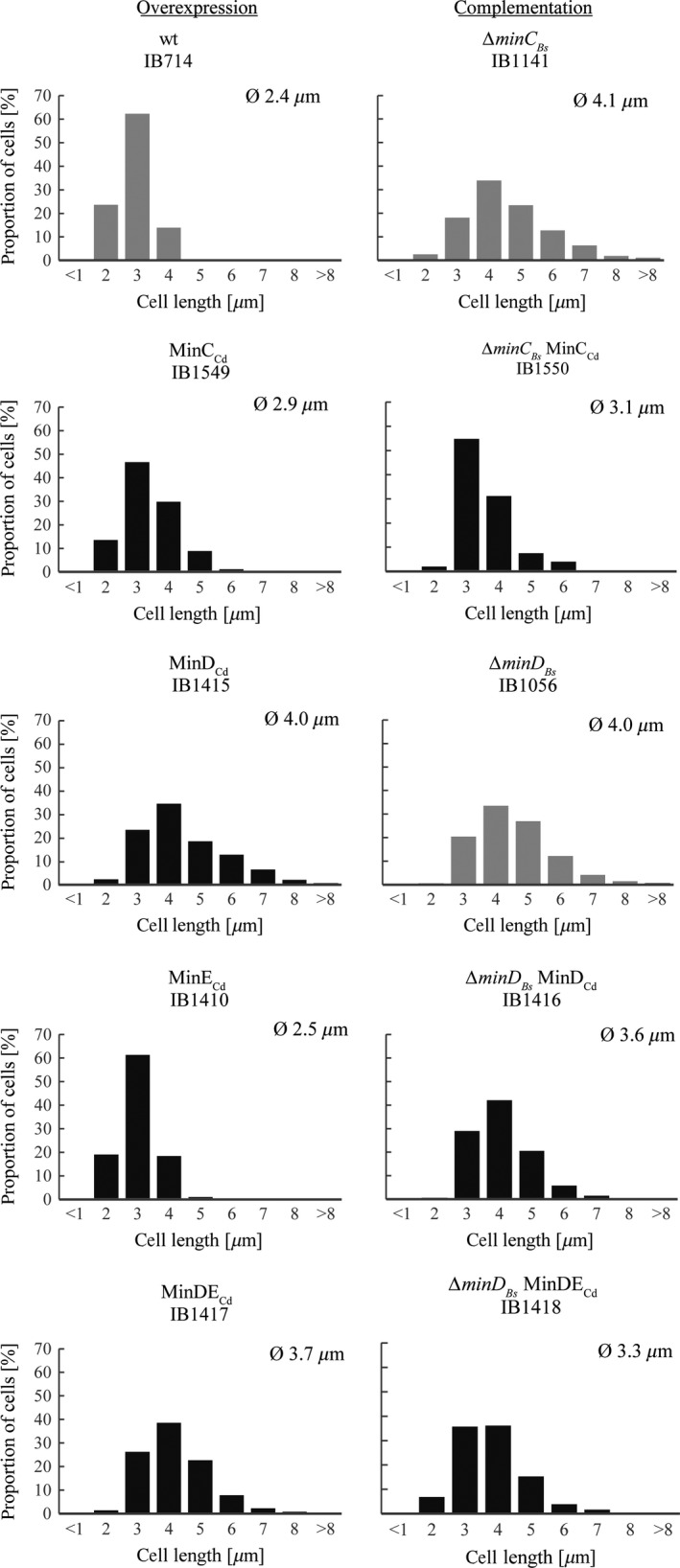
Cell length histograms. Left column: effects of overexpression of C. difficile Min proteins in wild‐type background. Expression was induced using 0.1 mM IPTG and/or 0.02% xylose, except for strain expressing MinCC d (IB1549), in which 0.3% xylose was used. Right column: complementation of MinBs proteins absence by MinCd proteins. Shown are induction conditions exhibiting the most notable complementation, that is 0.1 mM IPTG and/or 0.02% xylose except for strain ∆min DB s MinDEC d (IB1418), in which higher concentrations were used (0.5 mM IPTG and 0.3% xylose). Parental strains are in gray. Summary of all measurements can be found in Table S3.
To explore the effect of C.difficile MinC (MinCCd) on B.subtilis cell division, we placed the gene under the control of a xylose‐inducible promoter (Pxyl) into an amyE locus. The cell length of the resulting IB1549 strain was measured without xylose and with of 0.3% xylose. This strain showed increased average length reaching 3 μm and occurrence of cells longer than 4 μm under both conditions up to 11% (Table S3). As the cell length also increased in the uninduced sample, it may be inferred that the cell division system of B.subtilis is so sensitive to MinCCd that even the low concentrations of it produced by a leaky Pxyl promoter (Vavrová et al. 2010) are enough to cause cell elongation. Leaky expression affected cell length in a previous Min system study as well (Pavlendová et al. 2010).
We also investigated the ability of MinDCd to interfere with the Min system of B.subtilis by introducing yfp‐minD Cd fusion under the control of an IPTG‐inducible promoter (Phyperspank) into an amyE locus, creating strain IB1415. We assumed that YFP‐MinDCd could functionally substitute for the native MinDCd, as both GFP‐MinDBs and GFP‐MinDEc are functional in their respective native organisms (Pavlendová et al. 2010; Raskin and de Boer 1999a). Cell length measurements were performed without IPTG and with two different IPTG concentrations, 0.1 mmol/L and 0.5 mmol/L. In the absence of inducer, the cell length of the strain harboring yfp‐minD Cd (IB1415) was unchanged relative to the parental MO1099 strain (2.4 μm; only 0.8% of cells are longer than 4 μm) and no minicells were observed, suggesting that even though Phyperspank is known to be leaky (Vavrová et al. 2010), the MinDCd amounts resulting from this leakiness are not sufficient to induce cell elongation. The addition of an inducer, regardless on the concentration, triggered elongation, with the average cell length reaching 4 μm and 40% of cells becoming longer than this (Table S3). Additionally, we determined the cell length of a strain harboring MinECd, to verify the effect of MinECd alone on cell division. To prepare a B.subtilis strain producing MinECd (IB1410), we placed the corresponding gene under the control of a xylose‐inducible promoter (Pxyl) into a thrC locus. We observed no notable change in average cell length (2.3–2.5 μm) regardless of the presence of inducer at either concentration (Table S3). This is the same behavior we observed for MinEEc in a previous study (Pavlendová et al. 2010). In E.coli, MinEEc overexpression is characterized by the production of minicells (de Boer et al. 1989), but neither MinECd nor MinEEc seemed to induce their formation when introduced into B.subtilis cells.
Finally, we assessed the effects of simultaneous MinDCdECd expression on the length of B.subtilis cells. A strain harboring both MinDCd and MinECd (yfp‐minD Cd minE Cd; IB1417) was prepared by transformation using chromosomal DNA as described in the Experimental procedures and Table 1. In the absence of inducers, IB1417 cells retained the same length as the parental wild‐type strain MO1099 (2.5 μm; Table S3). Both induction conditions lead to comparable elongation, with average cell length that exceeded 3.7 μm and 33% of cells were longer than 4 μm (Table S3). Apparently, increasing the inducer concentration, and thus the amounts of MinDCd and MinECd, does not further increase cell length. Taken together, these results show that MinCCd and MinDCd, but not MinECd, elongate cells and induce minicell formation when overexpressed in B.subtilis. Elongation was slightly less distinct when both MinDCd and MinECd were coexpressed at low inducer concentrations.
Complementation of the B.subtilis Min system with C.difficile Min proteins
It is known that the absence of MinC, MinD, or both in B.subtilis causes a slight cell elongation and the formation of minicells (Levin et al. 1992, 1998). There are several studies showing that the Min proteins of one organism can complement the function of the Min system of a different organism (Szeto et al. 2001; Tavva et al. 2006). For example, a functional exchange of Min systems between gram‐negative and gram‐positive bacteria showed that the expression of a heterologous E.coli MinDEc protein was able to partially rescue the ΔminD Bs phenotype of B.subtilis; however, the same could not be said for E.coli MinCEc, which failed to improve the ΔminC Bs phenotype (Pavlendová et al. 2010).
Here, we investigated whether the MinCCd and MinDCd proteins of the C.difficile Min system could restore defects caused by deleting their homologues in B.subtilis, and whether coexpressing MinDCd and MinECd together could restore defects caused by the absence of MinDBs. If MinCCd or MinDCd do complement MinCBs and MinDBs, the cells should become shorter and minicell formation should decrease. Previously utilized constructs with respective C.difficile genes under the control of inducible promoters were introduced into various B.subtilis mutant backgrounds. These strains and their complete genotypes are listed in Table 1. The average cell lengths of the resulting strains were measured in the presence of varying inducer concentrations and compared with those of their parental mutant strains. All of the following measurements are summarized in Table S3 and are illustrated in histogram in Figure 1. The length of parental strains B.subtilis ΔminD Bs (IB1056) and ΔminC Bs (IB1141) were both determined to be 4 μm on average, with 45% of cells being longer than that.
To investigate the ability of MinCCd to complement the absence of MinCBs, we created a strain producing MinCCd from a xylose‐inducible promoter (Pxyl) in a ΔminC Bs background (IB1550). The experiments were performed without xylose induction and with two different xylose concentrations (0.02% and 0.3%). Complementation effect was already observed in the absence of inducer, when leaky expression of MinCCd was sufficient to shorten the cells from 4.1 μm to 3.4 μm on average. Induced expression improved the phenotype even further, causing the percentage of cells longer than 4 μm drop from 45% to 12% (Table S3). Minicells were present in all samples of ΔminC Bs minC Cd.
Introducing a YFP‐MinDCd into a ΔminD Bs background (IB1056) yielded strain ΔminD Bs yfp‐minD Cd (IB1416). The experiments were carried out without IPTG and with two different IPTG concentrations, 0.1 mmol/L IPTG and 0.5 mmol/L IPTG. Measurements of strain IB1416 grown without inducer produced cells with an average length similar to that of the originating ΔminD Bs strain (Table S3). The low levels of MinDCd due to leaky expression therefore appeared to have no visible effect on cell length. Moderate expression of MinDCd (induction with 0.1 mmol/L IPTG) seemed to have a slight effect on cell division, as cell length decreased to 3.6 μm and the proportion of cells longer than 4 μm went down to 28%. Increasing the concentration of inducer to 0.5 mmol/L IPTG, however, lead to cell elongation (average of 4 μm and 43% of cells longer than 4 μm), just as seen during overexpression on a wild‐type background (IB1415) (Table S3). Minicells, which are a phenotype of both ΔminD Bs mutation and also, as we have shown here, MinDCd overexpression, were observed in all samples, but their frequency was not evaluated.
We also investigated changes in cell length when MinDCd is expressed together with MinECd in a strain lacking MinDBs (ΔminD Bs minD Cd minE Cd; IB1418). In this strain, only the induced expression of MinDCdECd decreased both cell length and the proportion of cells longer than 4–3.5 μm and 29%, respectively, when using a lower induction level (0.1 mmol/L IPTG, 0.02% xylose), and to 3.3 μm and 21% when using a higher induction level (0.5 mmol/L IPTG, 0.3% xylose) (Table S3).
In conclusion, our results suggest that MinCCd is able to complement for the absence of MinCBs. MinDCd alone can only partially substitute for a missing MinDBs, but when coexpressed with MinECd, considerably enhanced complementation is observed.
Localization of C.difficile MinD and MinE in B.subtilis
As observation using fluorescent proteins is not yet commonly feasible in the anaerobic C.difficile, we explored the localization of its Min proteins in a heterologous B.subtilis system, which has previously proven to be a suitable environment for the study of Min proteins. We introduced a YFP‐MinDCd fusion into B.subtilis cells under the control of an IPTG‐inducible promoter at the amyE locus. This fusion was introduced into wild‐type, ΔminD Bs and ΔminJ Bs mutant backgrounds creating strains IB1415, IB1417, and IB1553, respectively. As expected from the similarity of MinDCd to MinDBs, MinDCd localized to the cell membrane and often formed foci (Figs. 2A–C). The localization pattern between the wild‐type and mutant strains did not differ and we often observed short helical‐like structures resembling those seen previously with B.subtilis MinD (Barák et al. 2008). In many instances, we observed localization to the sites of vegetative and asymmetric septa as well as the polar sites (Figs. 2A and B).
Figure 2.
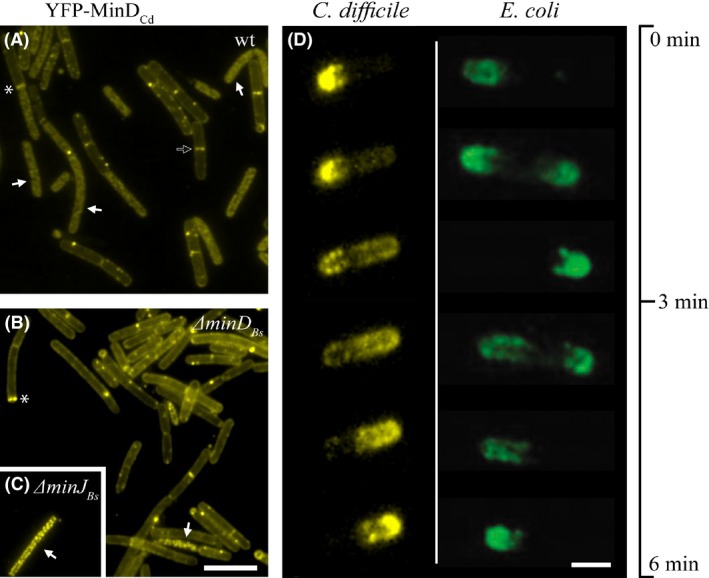
Localization and oscillation of C.difficile Min proteins. (A)–(C) Localization of YFP‐tagged MinDC d expressed from Phyperspank in wild‐type and mutant B. subtilis backgrounds. (A) wt (IB1415), (B) ∆min DB s (IB1416), (C) ∆min JB s (IB1553). Full arrows point to examples of cells where the fine‐structure signal resembles the localization pattern of the native MinDB s along lipid spirals; the empty arrow indicates an example of localization to the vegetative septum, and the asterisk, to the asymmetric septum. Expression was induced with 0.1 mM IPTG; the scale bar represents 5 μm. (D) Time‐lapse images recorded over a period of 6 min showing oscillation of MinDC dMinEC d in a B.subtilis ∆min DB s background (IB1418), compared to oscillation of E.coli proteins in a B.subtilis ∆min DB s background (Jamroškovič et al. 2012). Expression of MinDC d was induced with 0.1 mM IPTG and MinEC d with 0.02% xylose; scale bar represents 1 μm. Available also as Video S1 in Supporting Information.
The localization of MinECd was examined using a MinECd‐GFP fusion placed under the control of a xylose‐inducible promoter in a wild‐type background (IB1552). The observed signal was dispersed throughout the cytoplasm (not shown), which is similar to the localization of E.coli MinEEc‐GFP in B.subtilis observed previously (Pavlendová et al. 2010).
Oscillation of C.difficile MinDE proteins in B.subtilis
We have previously shown that the oscillation of the E.coli Min system can be reproduced in B.subtilis (Jamroškovič et al. 2012). Because manipulation of an anaerobic pathogenic bacteria poses a number of complications, we decided to explore the behavior of the C.difficile Min proteins in B.subtilis cells. We introduced MinECd, under the control of a xylose‐inducible promoter at the thrC locus, into strains already harboring a YFP‐MinDCd fusion at the amyE locus. This gave rise to strains carrying the C.difficile genes MinD Cd and MinE Cd in wild‐type, ΔminD Bs and ΔminD Bs ΔminJ Bs , mutant backgrounds (strains IB1417, IB1418 and IB1546). When these proteins were coexpressed using 0.1 mmol/L IPTG and 0.02% xylose on the wild‐type background (IB1417), we observed oscillation of YFP‐MinDCd from one pole to the other in a small number of cells (roughly 3% of 250 cells). This is in contrast with the behavior of the corresponding E.coli proteins in B.subtilis, which showed no oscillation at all in the wild‐type. This effect was probably due to an interaction between MinDBs and MinDEc (Jamroškovič et al. 2012).
In E.coli, Min oscillation cycle period is 20–50 sec (Raskin and de Boer 1999a; Touhami et al. 2006); the oscillation of the E.coli proteins in B.subtilis is somewhat slower, with a period of 1.5–3 min (recorded against a ∆minD Bs ∆divIVA Bs background; IB1242), and increasing the temperature to 30°C or 37°C does not affect the oscillation speed (Jamroškovič et al. 2012). Strain ΔminD Bs ΔdivIVA Bs yfp‐minD Ec minE Ec (IB1242) was used as a control strain in this study, to ensure our conditions are properly set, as it showed the most extensive oscillation of E.coli Min proteins in B.subtilis.
The oscillation of the C.difficile proteins observed against a wild‐type B.subtilis background (IB1417) was even slower than the E.coli ones, at a pace of about 3–5 min per cycle at room temperature. The oscillation period in strains depleted of MinDBs or both MinDBs and MinJBs did not change and remained at 3–5 min per cycle. However, the absence of these components seemed to increase the proportion of cells in which oscillation was observed. In a strain lacking MinDBs (IB1418, Fig. 2D, Video S1), the effect was similar to the wild‐type strain (4% of 50 cells), but when both MinDBs and MinJBs were absent (IB1546), the oscillation was observed in up to 50% of the cells (Video S2). Regardless of the strain observed, the oscillation often stopped after 10 min and the YFP signal became dispersed throughout the cell. The speed and extent of oscillation for various organisms and heterologous systems is summarized in Table 2.
Table 2.
Comparison of oscillation times and efficiency between Min systems
| System | Organism | Genotype | Oscillation efficiency [%] | Oscillation period | Reference |
|---|---|---|---|---|---|
| E.coli | E.coli | – | 100 | 20–50 sec | Raskin and de Boer 1999a; Touhami et al. 2006 |
| E.coli | B.subtilis | YFP‐minD Ec minE Ec | 0 | – | Jamroškovič et al. 2012 |
| E.coli | B.subtilis | ∆minD Bs ∆divIVA Bs YFP‐minD Ec minE Ec | ~100 | 1.5–3 min | Jamroškovič et al. 2012 |
| C.difficile | B.subtilis | YFP‐minD Cd minE Cd | 3 | 3–5 min | This study |
| C.difficile | B.subtilis | ∆minD Bs YFP‐minD Cd minE Cd | 4 | 3–5 min | This study |
| C.difficile | B.subtilis | ∆minD Bs ∆minJ Bs YFP‐minD Cd minE Cd | 50 | 3–5 min | This study |
To determine if C.difficile MinE could drive oscillation of MinD of B.subtilis, we prepared a strain carrying a combination of GFP‐tagged MinDBs and MinECd in the B.subtilis ΔminD Bs ΔminJ Bs background which showed the most efficient oscillation of MinDCdECd. Fluorescence microscopy of this strain (ΔminD Bs ΔminJ Bs gfp‐minD Bs minE Cd; IB1562) revealed that no oscillation or movement of foci could be observed after induction with 0.04% xylose (not shown). The GFP signal was distributed as random foci throughout the cell and along the helical structures which are characteristic of MinDBs (Barák et al. 2008).
Swap of Min system oscillating components between C.difficile and E.coli
To investigate the interchangeability of the Min proteins from the gram‐positive C.difficile and the gram‐negative E.coli and their ability to oscillate together, we prepared B.subtilis strains ΔminD Bs yfp‐minD Ec minE Cd (IB1412) and ΔminD Bs ΔdivIVA Bs yfp‐minD Ec minE Cd (IB1413). After inducing the expression of YFP‐MinDEc and MinECd, we observed oscillation of YFP‐MinDEc with a period of 3–5 min, similar to that observed before for strain expressing MinDE originating from C.difficile (IB1417). This oscillation was only observed in a small portion of the ΔminD Bs cells (IB1412) and improved in cells with a ΔminD Bs ΔdivIVA Bs background (IB1413; not quantified statistically). These results show that there is some compatibility between the oscillating systems of these two evolutionarily distant gram‐positive and gram‐negative species.
Oscillating Min system of C.difficile interferes with B.subtilis sporulation
In our previous study, we showed that the oscillating E. coli Min system blocks sporulation at the asymmetric septum formation step (Jamroškovič et al. 2012). An intriguing question is therefore whether spore‐forming C.difficile also possesses an oscillating Min system that interferes with its sporulation. We assessed the sporulation efficiency of various B.subtilis strains in the presence of inducers (0.5 mmol/L IPTG, 0.5% xylose). The sporulation efficiency of both the wild‐type and a ΔminD Bs strain harboring the oscillating C.difficile MinDE proteins (IB1417 and IB1418) dropped to 32% and 45%, respectively (Fig. 3). A dramatic decrease in sporulation efficiency, down to 0.03%, was observed in the strain which lacked both MinDBs and MinJBs (IB1546). This was also the strain with the most effective oscillation. This drop in sporulation efficiency seems to be related to the proportion of cells in which oscillation is observed.
Figure 3.
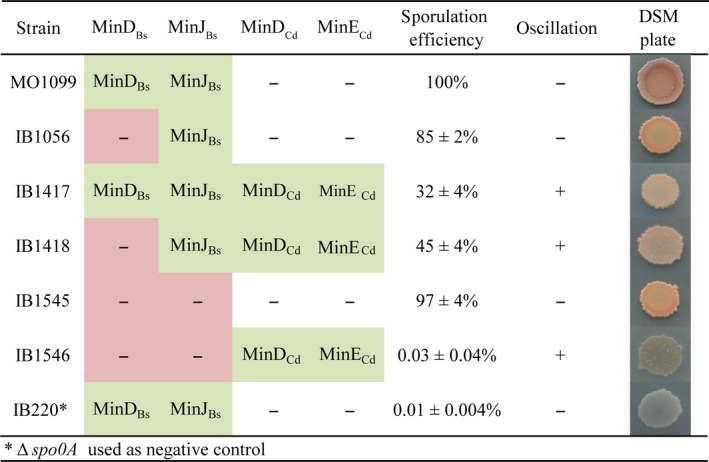
Sporulation efficiency of B.subtilis strains. Sporulation efficiency is given as the mean ± SD of at least three independent assays, each normalized against a wild‐type control. Sporulating colonies develop brown color, while nonsporulating light are brown to translucent, as seen in the ∆spo0A negative control (IB220).
Protein–protein interactions between C.difficile MinD and B.subtilis Min proteins
To improve our understanding of the behavior of the C.difficile Min proteins in B.subtilis, we looked for interactions between the C.difficile and B.subtilis proteins using a bacterial two‐hybrid system (BACTH). The strength of these interactions was quantified using a β‐galactosidase assay. A very strong interaction was detected between MinDCd and MinCBs, while a weaker one was found between MinDCd and MinJBs (Fig. 4). It is possible that the lower affinity of MinDCd for MinJBs might explain why only partial complementation was observed when MinDCd was expressed against a ΔminD Bs background (IB1416), as the MinD–MinJ interaction would clearly be a limiting factor. The MinD proteins from the two organisms seem to interact with each other strongly as well. The interaction observed between MinDCd and MinCBs confirms that the MinDCd/MinECd system can indeed utilize the host B.subtilis MinC, as suggested by the complementation experiments in the ΔminD Bs minD Cd minE Cd (IB1418) strain.
Figure 4.
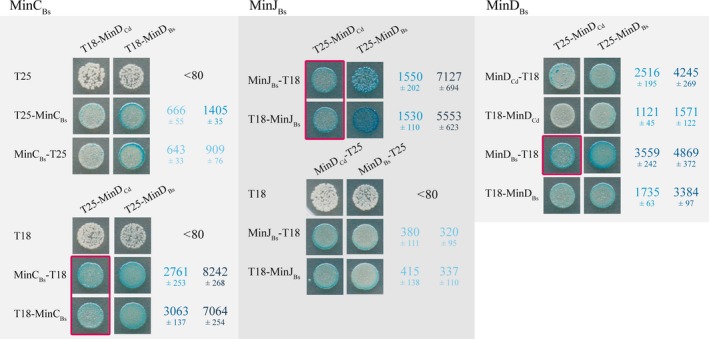
Protein–protein interactions between MinDC d and the B.subtilis Min proteins compared alongside interactions among the B.subtilis Min proteins as detected by bacterial two‐hybrid system BACTH. Interactions were quantified using a β‐galactosidase assay and are expressed in Miller units as mean values ± SD of at least three independent measurements. The color intensity corresponds to the strength of the interaction; red boxes highlight strong positive interactions between heterologous proteins. Negative controls were all below 80 MU.
Discussion
Clostridium difficile is an important human pathogen, causing serious problems in hospitals and medical facilities (reviewed in Burke and Lamont 2014). Because of its strictly anaerobic life style and its demanding transformation procedures, we have still only a limited knowledge of some of its basic processes, including cell division. For example, commonly used reporter genes, such as fluorescent proteins or luciferase, require oxygen for protein folding or enzyme activity (Heim et al. 1994; Hastings and Gibson 1967). In spite of ongoing efforts, methods and reporter assays suitable for anaerobic or low‐oxygen conditions are only now starting to emerge (Drepper et al. 2007; Edwards et al. 2015; Buckley et al. 2015; Ransom et al. 2015). Because of these problems, we decided to investigate the mechanism of action of C.difficile Min proteins and their effects on vegetative cell division and sporulation in a heterologous B.subtilis host. This, the first study focused on the C.difficile Min system, may help us to understand the role of its Min proteins in the asymmetric division and spore formation of this medically significant bacterium.
Our analysis of MinCCd and MinDCd in vegetatively growing cells shows that these proteins are able to affect cell division in B.subtilis. We found that both MinCCd and MinDCd can complement for the missing B.subtilis counterparts. Interestingly, the same could not be said for MinCEc, which was previously shown to fail in MinCBs complementation (Pavlendová et al. 2010), and which has higher similarity to MinCBs (35/58% identity/similarity based on BLAST alignment) than MinCCd has (29/51% identity/similarity; Fig. 5A, Fig. S1). As MinDBs is more similar to MinDCd than it is to MinDEc (64/81% identity/similarity compared to 44/67%; Fig. 5A, Fig. S1), we might expect MinDCd to better complement a MinDBs deletion than MinDEc (Pavlendová et al. 2010), however, this is not what we observed. The B.subtilis Min system is finely tuned, with relatively small changes having clearly notable effects, so it is not surprising that substituting one of the components with a replacement that has different binding affinities for all the other elements involved with the system leads to divergent effects, a feature which might not be reflected or predicted solely by the sequence similarity. More importantly, the complementation of MinDBs absence by MinDCd/MinECd coexpression revealed that this oscillating Min system can still aid in proper septum placement when the native Min system is disturbed, provided that these proteins can engage the native system's MinC.
Figure 5.
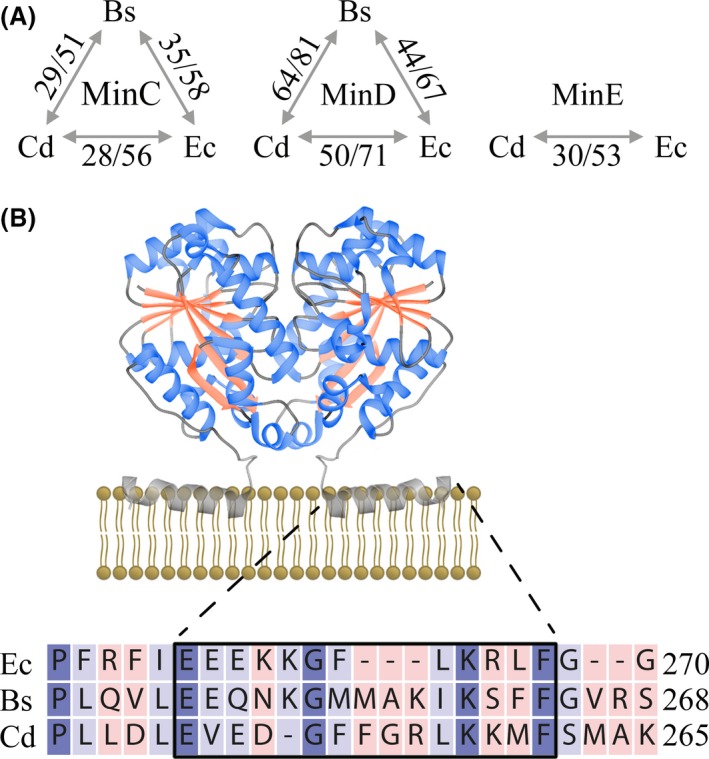
Bioinformatic analysis of MinCd proteins. (A) Percent sequence identity (same residues)/similarity (same residues + positive substitutions) between the Min proteins of B.subtilis (Bs), C.difficile (Cd) and E.coli (Ec) based on data from BLAST search (Altschul et al. 1997). (B) Model of the attachment of a MinDE c dimer to the membrane interface through its amphipathic helix. This model is based on the MinDE c crystal structure (PDB ID: 3Q9L), which contains residues 1–260, and thus lacks some of the residues involved in helix formation. Below: Alignment of the C‐terminal region of MinD containing the amphipathic helix. The consensus region is boxed, identical residues are violet, and conservation is indicated by color intensity.
The observed localization of MinDCd along helical structures suggests that this protein recognizes the anionic phospholipids organized in a helical manner in the B.subtilis membrane. The C‐terminal region of MinD from various organisms, including E.coli and B.subtilis, contains a consensus amphipathic helical region that anchors it to the membrane (Szeto et al. 2002). This consensus sequence can also be found in C.difficile MinD (Fig. 5B) and an amphipathic helix was also predicted at the C‐terminus by AmphipaSeek (not shown; Sapay et al. 2006). This helix would then be responsible for the membrane localization of MinDCd in B.subtilis, which is similar to that observed for MinDEc in the same organism.
Bacillus subtilis and C.difficile have very different membrane compositions, In fact, Clostridium species display distinct variations in their major polar lipid contents and C.difficile has an exceptionally variable membrane lipid composition, even for different isolates of the same strain (Korachi et al. 2002). Phosphatidylglycerol (PG) and cardiolipin (CL) have been identified, but interestingly, phosphatidylethanolamine is completely absent and other lipids have been proposed to balance the negative charge of PG and CL (Drucker et al. 1996; Korachi et al. 2002; Guan et al. 2014). Phosphatidylglycerol and CL represent 24% and 4% of the membrane phospholipids in E.coli (Kusters et al. 1991), 40% and 20% in B.subtilis (López et al. 1998), and 30% and 16% in C.difficile (Guan et al. 2014).
The ability to oscillate is an intrinsic characteristic of the Min proteins and emerges whenever some minimal criteria are met (Loose et al. 2008). Thus, a different membrane composition does not pose any obstacle for oscillation, as long as enough negatively charged lipids are present, although the resulting charge density does affect oscillation parameters such as wavelength and velocity (Vecchiarelli et al. 2014; Zieske and Schwille 2014). It has been suggested that these differences arise from the differences between the mechanisms of membrane binding by MinDEc and MinEEc (Vecchiarelli et al. 2014). Previous successful reconstitutions of oscillation in heterologous systems (Ramirez‐Arcos et al. 2002; Jamroškovič et al. 2012) suggest that, in the complex environment of cell, the most limiting factor is the interaction between the heterologous Min proteins within the host organism. In our case, oscillation markedly improved when the MinCd system was introduced into a ΔminD Bs ΔminJ Bs background (IB1546), which allowed oscillation of the heterologous Min system.
All B.subtilis strains expressing an oscillating MinCd system exhibited disturbed sporulation. The severity of the sporulation defect seems to be correlated with improved oscillation efficiency. Two important questions remain: first, what is the underlying cause of this failed sporulation, and second, does it affect the sporulation of C.difficile at all? It is possible that the differences in sporulation and sporulation regulation between B. subtilis and C.difficile cause oscillation to be inhibitory in the heterologous organism, but not in the native one. Another possibility is that an oscillating MinCd system only inhibits polar division during particular parts of the cell life cycle, such as vegetative growth, and is shut down or modulated during sporulation. Yet, a third possibility is that this system does, in fact, cause C.difficile to sporulate less efficiently than B.subtilis, but provides additional advantage for its different lifestyle and environmental niche.
Although complex transcriptional data for C.difficile are still lacking, we can still make some inferences from the work of Saujet et al. (2013). They found that the ftsZ, minC, minD, minE, and divIVA genes were all positively controlled by SigH, the key regulator of the transition phase in C.difficile, which is of comparable importance to Spo0A. SigH is also involved in the expression of ftsZ, minC, and minD in B. subtilis as well (Britton et al. 2002). These results suggest that the MinCDE proteins are present in the early stages of sporulation in C.difficile. The genome of C.difficile also harbors a DivIVA homolog (Table. S4), which in B.subtilis has a role in sporulation, but the question of whether it serves as a polar tether for the Min system as it does in B.subtilis remains open, as we were not able to identify a MinJ homologue. It is still possible that some other protein fills the role of MinJ in connecting the MinCD system to DivIVA.
The Clostridia are a diverse group of bacteria, and, despite their common historical designation as gram‐positive, a number of them have been found to have a membrane organization more characteristic of gram‐negative bacteria (and were thus moved into a separate class, Negativicutes), together with the ability to form endospores (Yutin and Galperin 2013). Acetonema longum is a distant relative of Clostridium spp. and a lesser known member of Negativicutes. A study of the sporulation and germination of this organism revealed a remarkable inversion of the inner membrane of the mother cell, to become the outer membrane of the germinating cell (Tocheva et al. 2011). This brings us to the question of evolution of gram‐positive and gram‐negative bacteria, an exciting topic on which many opposing theories exist. The work of Tocheva et al. 2011suggests how the outer membrane of gram‐negative bacteria might have evolved, and more broadly, how gram‐negatives could have arisen from gram‐positives. A.longum could therefore represent a missing link between the two groups. Our analysis of some Clostridia and Negativicutes members' genomes shows that many possess Min proteins from both systems (Table. S4), suggesting that the two systems might have evolved in a gram‐positive bacterium. Whether and how these systems could coexist in clostridia remains to be resolved by future studies. Until convenient methods for directly studying clostridia are developed, B.subtilis could serve as host system for these studies.
Conflict of Interest
Authors declare no conflict of interest.
Supporting information
Figure S1: Multiple sequence alignment of Min proteins.
Video S1: Oscillation of YFP‐tagged MinDCd in the presence of MinECd, recorded in B.subtilis ΔminD Bs minD Cd minE Cd (IB1418). Scale bar represents 1 μm.
Video S2: Oscillation of YFP‐tagged MinDCd in the presence of MinECd, recorded in B.subtilis ΔminD Bs ΔminJ Bs minD Cd minE Cd (IB1546). Scale bar represents 5 μm.
Table S1: Plasmids used in this study and their construction.
Table S2: Primers used in this study.
Table S3: Cell length measurements.
Table S4: The sequence similarity/identity of Min proteins of selected members of Clostridia and Negativicutes compared with their counterparts in E.coli and B.subtilis. Similarity and identity values are derived from a BLAST query (Altschul et al. 1997). For MinC and MinD, the sequence of the B.subtilis proteins was used as reference, since these queries gave lower E‐values and higher query cover and identities than the E.coli sequences (not shown). The E.coli sequence was used as a reference for MinE, and B.subtilis sequences for a search of MinJ and DivIVA homologs. All listed organisms are endospore‐formers except E.coli.
Acknowledgments
The authors thank Emília Chovancová for technical assistance and all members of the laboratory for consultation and help. We thank Jacob Bauer for helpful comments on the manuscript. This work was supported by Grant 2/0009/13 from the Slovak Academy of Sciences, by grant APVV‐00335‐10, APVV‐14‐0181 and by the Research & Development Operational Programme funded by the ERDF (ITMS code: 26240220044). .
MicrobiologyOpen 2016; 5(3): 387–401
References
- Adams, D. W. , and Errington J.. 2009. Bacterial cell division, assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642–653. [DOI] [PubMed] [Google Scholar]
- Adler, H. I. , Fisher W. D., Cohen A., and Hardigree A. A.. 1967. Miniature Escherichia coli cells deficient in DNA. Proc. Natl Acad. Sci. USA 57:321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F. , Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., et al. 1997. Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F. M. , Brent R., Kingston R. E., Moore D. O., Seidmann J. S., Smith J., et al. 1987. Current Protocols in Molecular Biology. Wiley, New York. [Google Scholar]
- Barák, I. , and Wilkinson A. J.. 2007. Division site recognition in Escherichia coli and Bacillus subtilis . FEMS Microbiol. Rev. 31:311–326. [DOI] [PubMed] [Google Scholar]
- Barák, I. , Muchová K., Wilkinson A. J., O'Toole P. J., and Pavlendová N.. 2008. Lipid spirals in Bacillus subtilis and their role in cell division. Mol. Microbiol. 68:1315–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Yehuda, S. , Rudner D. Z., and Losick R.. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532–536. [DOI] [PubMed] [Google Scholar]
- de Boer, P. A. J. , Crossley R. E., and Rothfield L. I.. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli . Cell 56:641–649. [DOI] [PubMed] [Google Scholar]
- Bramkamp, M. , Emmins R., Weston L., Donovan C., Daniel R. A., and Errington J.. 2008. A novel component of the division‐site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol. Microbiol. 70:1556–1569. [DOI] [PubMed] [Google Scholar]
- Britton, R. A. , Eichenberger P., Gonzalez‐Pastor J. E., Fawcett P., Monson R., Losick R., et al. 2002. Genome‐wide analysis of the stationary‐phase sigma factor sigma‐H. regulon of Bacillus subtilis . J. Bacteriol. 184:4881–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley, A. M. , Petersen J., Roe A. J., Douce G. R., and Christie J. M.. 2015. LOV‐based reporters for fluorescence imaging. Curr. Opin. Chem. Biol. 27:39–45. [DOI] [PubMed] [Google Scholar]
- Burke, K. E. , and Lamont J. T.. 2014. Clostridium difficile infection: a worldwide disease. Gut. Liv. 8:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha, J. H. , and Stewart G. C.. 1997. The divIVA minicell locus of Bacillus subtilis . J. Bacteriol. 179:1671–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drepper, T. , Eggert T., Circolone F., Heck A., Krauss U., Guterl J. K., et al. 2007. Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 25:443–445. [DOI] [PubMed] [Google Scholar]
- Drucker, D. B. , Wardle H. M., and Boote V.. 1996. Phospholipid profiles of Clostridium difficile . J. Bacteriol. 178:5844–5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, D. H. , and Errington J.. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24:905–915. [DOI] [PubMed] [Google Scholar]
- Edwards, A. N. , Pascual R. A., Childress K. O., Nawrocki K. L., Woods E. C., and McBride S. M.. 2015. An alkaline phosphatase reporter for use in Clostridium difficile . Anaerobe 32:98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy, P. , Erb M., Gregory J., and Silverman J.. 2011. Cellular architecture mediates DivIVA ultrastructure and regulates Min activity in Bacillus subtilis . MBio 2:e00257–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy, P. , Winter P. W., Wawrzusin P., York A. G., Shroff H., and Ramamurthi K. S.. 2014. Asymmetric division and differential gene expression during a bacterial developmental program requires DivIVA. PLoS Genet. 10:e1004526. doi:10.1371/journal.pgen.1004526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleurie, A. , Lesterlin C., Manuse S., Zhao C., Cluzel C., Lavergne J. P., et al. 2014. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae . Nature 516:259–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Z. , Katzianer D., Zhu J., and Goldfine H.. 2014. Clostridium difficile contains plasmalogen species of phospholipids and glycolipids. Biochim. Biophys. Acta 1841:1353–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout‐Fleury, A. M. , Frandsen N., and Stragier P.. 1996. Plasmids for ectopic integration in Bacillus subtilis . Gene 180:57–61. [DOI] [PubMed] [Google Scholar]
- Harwood, C. R. , and Cutting S. M.. 1990. Molecular Biological Methods for Bacillus. Wiley, Chichester, UK. [Google Scholar]
- Hastings, J. W. , and Gibson Q. H.. 1967. The role of oxygen in the photoexcited luminescence of bacterial luciferase. J. Biol. Chem. 242:720–726. [PubMed] [Google Scholar]
- Heim, R. , Prashert D. C., and Tsien R. Y.. 1994. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl Acad. Sci. USA 91:12501–12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Z. , and Lutkenhaus J.. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34:82–90. [DOI] [PubMed] [Google Scholar]
- Hu, Z. , and Lutkenhaus J.. 2001. Topological regulation of cell division in E. coli spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7:1337–1343. [DOI] [PubMed] [Google Scholar]
- Hu, Z. , Gogol E. P., and Lutkenhaus J.. 2002. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc. Natl Acad. Sci. USA 99:6761–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamroškovič, J. , Pavlendová N., Muchová K., Wilkinson A. J., and Barák I.. 2012. An oscillating Min system in Bacillus subtilis influences asymmetric septation during sporulation. Microbiology 158:1972–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova, G. , Pidoux J., Ullmann A., and Ladant D.. 1998. A bacterial two‐hybrid system based on a reconstituted signal transduction pathway. Proc. Natl Acad. Sci. USA 95:5752–5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korachi, M. , Rupnik M., Blinkhorn A. S., Boote V., and Drucker D. B.. 2002. Comparison of polar lipid profiles of Clostridium difficile isolates from different geographical locations. Anaerobe 8:35–39. [Google Scholar]
- Kusters, R. , Dowhan W., and De Kruijff B.. 1991. Negatively charged phospholipids restore prePhoE translocation across phosphatidylglycerol depleted Escherichia coli inner membranes. J. Biol. Chem. 266:8659–8662. [PubMed] [Google Scholar]
- Lenarcic, R. , Halbedel S., Visser L., Shaw M., Wu L. J., Errington J., et al. 2009. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 28:2272–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, P. A. , Margolis P. S., Setlow P., Losick R., and Sun D.. 1992. Identification of Bacillus subtilis genes for septum placement and shape determination. J. Bacteriol. 174:6717–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, P. A. , Shim J. J., and Grossman A. D.. 1998. Effect of minCD on FtsZ ring position and polar septation in Bacillus subtilis . J. Bacteriol. 180:6048–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose, M. , Fischer‐Friedrich E., Ries J., Kruse K., and Schwille P.. 2008. Spatial regulators for bacterial cell division self‐organize into surface waves in vitro. Science 320:789–792. [DOI] [PubMed] [Google Scholar]
- López, C. S. , Heras H., Ruzal S. M., Sánchez‐Rivas C., and Rivas E. A.. 1998. Variations of the envelope composition of Bacillus subtilis during growth in hyperosmotic medium. Curr. Microbiol. 36:55–61. [DOI] [PubMed] [Google Scholar]
- Marston, A. L. , and Errington J.. 1999. Selection of the midcell division site in Bacillus subtilis through MinD–dependent polar localization and activation of MinC. Mol. Microbiol. 33:84–96. [DOI] [PubMed] [Google Scholar]
- Meselson, M. , and Yuan R.. 1968. DNA restriction enzyme from E. coli . Nature 217:1110–1114. [DOI] [PubMed] [Google Scholar]
- Miller, J. H. 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Monahan, L. G. , Liew A. T. F., Bottomley A. L., and Harry E. J.. 2014. Division site positioning in bacteria:one size does not fit all. Front. Microbiol. 5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick, J. E. , and Kearns D. B.. 2008. MinJ YvjD is a topological determinant of cell division in Bacillus subtilis . Mol. Microbiol. 70:1166–1179. [DOI] [PubMed] [Google Scholar]
- Pavlendová, N. , Muchová K., and Barák I.. 2010. Expression of Escherichia coli Min system in Bacillus subtilis and its effect on cell division. FEMS Microbiol. Lett. 302:58–68. [DOI] [PubMed] [Google Scholar]
- Ramirez‐Arcos, S. , Szeto J., Dillon J. A. R., and Margolin W.. 2002. Conservation of dynamic localization among minD and minE orthologues: oscillation of Neisseria gonorrhoeae proteins in Escherichia coli . Mol. Microbiol. 46:493–504. [DOI] [PubMed] [Google Scholar]
- Ransom, E. M. , Ellermeier C. D., and Weiss D. S.. 2015. Use of mCherry Red Fluorescent Protein for Studies of Protein Localization and Gene Expression in Clostridium difficile . Appl. Environ. Microbiol. 81:1652–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin, D. M. , and de Boer P. A. J.. 1999a. Rapid pole‐to‐pole oscillation of a protein required for directing division to the middle of Escherichia coli . Proc. Natl Acad. Sci. USA 96:4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin, D. M. , and de Boer P. A. J.. 1999b. MinDE‐dependent pole‐to‐pole oscillation of division inhibitor MinC in Escherichia coli . J. Bacteriol. 181:6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve, J. N. , Mendelson N. H., Coyne S. I., Hallock L. L., and Cole R. M.. 1973. Minicells of Bacillus subtilis . J. Bacteriol. 114:860–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlett, V. W. , and Margolin W.. 2015. The Min system and other nucleoid‐independent regulators of Z ring positioning. Front. Microbiol. 6:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Fritsch E. F., and Maniatis T.. 1989. Molecular Cloning: a Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Sapay, N. , Guermeur Y., and Deléage G.. 2006. Prediction of amphipathic in‐plane membrane anchors in monotopic proteins using a SVM classifier. BMC Bioinformatics 7:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saujet, L. , Pereira F. C., Serrano M., Soutourina O., Monot M., Shelyakin P. V., et al. 2013. Genome–wide analysis of cell type‐specific gene transcription during spore formation in Clostridium difficile . PLoS Genet. 9:e1003756. doi:10.1371/journal.pgen.1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer, P. , Millet J., and Aubert J. P.. 1965. Catabolic repression of bacterial sporulation. Proc. Natl Acad. Sci. USA 54:704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser, F. , Brannigan J. A., Lewis R. J., Wilkinson A. J., Youngman P., and Barák I.. 2000. A new mutation in spo0A with intragenic suppressors in the effector domain. FEMS Microbiol. Lett. 185:123–128. [DOI] [PubMed] [Google Scholar]
- Stragier, P. 2002. A gene odyssey: exploring the genomes of endospore‐forming bacteria Pp. 519–525 in Sonenshein L., Losick R. and Hoch J. A., eds. Bacillus subtilis and its relatives: from genes to cells. American Society for Microbiology, Washington, D. C.. [Google Scholar]
- Szeto, J. , Ramirez‐Arcos S., Raymond C., Hicks L. D., Kay C. M., and Dillon J. A.. 2001. Gonococcal MinD affects cell division in Neisseria gonorrhoeae and Escherichia coli and exhibits a novel self‐interaction. J. Bacteriol. 183:6253–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto, T. , Rowland S., Rothfield L., and King G.. 2002. Membrane localization of MinD is mediated by a C‐terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc. Natl Acad. Sci. USA 99:15693–15698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavva, V. S. , Collins G. B., and Dinkins R. D.. 2006. Targeted overexpression of the Escherichia coli MinC protein in higher plants results in abnormal chloroplasts. Plant Cell Rep. 25:341–348. [DOI] [PubMed] [Google Scholar]
- Thomaides, H. B. , Freeman M., El Karoui M., and Errington J.. 2001. Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation. Genes Dev. 15:1662–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocheva, E. I. , Matson E. G., Morris D. M., Moussavi F., Leadbetter J. R., and Jensen G. J.. 2011. Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell 146:799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touhami, A. , Jericho M., and Rutenberg A. D.. 2006. Temperature dependence of MinD oscillation in Escherichia coli:running hot and fast. J. Bacteriol. 188:7661–7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuner‐Lange, A. , Aguiluz K., van der Does C., Gómez‐Santos N., Harms A., Schumacher D., et al. 2013. PomZ, a ParA‐like protein, regulates Z‐ring formation and cell division in Myxococcus xanthus . Mol. Microbiol. 87:235–253. [DOI] [PubMed] [Google Scholar]
- Vavrová, L'. , Muchová K., and Barák I.. 2010. Comparison of different Bacillus subtilis expression systems. Res. Microbiol. 161:791–797. [DOI] [PubMed] [Google Scholar]
- Vecchiarelli, A. G. , Li M., Mizuuchi M., and Mizuuchi K.. 2014. Differential affiities of MinD and MinE to anionic phospholipid influence Min patterning dynamics in vitro. Mol. Microbiol. 93:453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemse, J. , and van Wezel G. P.. 2009. Imaging of Streptomyces coelicolor A32 with reduced autofluorescence reveals a novel stage of FtsZ localization. PLoS ONE 4:e4242. doi:10.1371/journal.pone.0004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L. J. , and Errington J.. 2011. Nucleoid occlusion and bacterial cell division. Nat. Rev. Microbiol. 10:8–12. [DOI] [PubMed] [Google Scholar]
- Youngman, P. , Perkins J. B., and Losick R.. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon‐borne erm gene. Plasmid 12:1–9. [DOI] [PubMed] [Google Scholar]
- Yutin, N. , and Galperin M. Y.. 2013. A genomic update on clostridial phylogeny:gram‐negative spore formers and other misplaced clostridia. Environ. Microbiol. 15:2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieske, K. , and Schwille P.. 2014. Reconstitution of self‐organizing protein gradients as spatial cues in cell‐free systems. eLife 3:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Multiple sequence alignment of Min proteins.
Video S1: Oscillation of YFP‐tagged MinDCd in the presence of MinECd, recorded in B.subtilis ΔminD Bs minD Cd minE Cd (IB1418). Scale bar represents 1 μm.
Video S2: Oscillation of YFP‐tagged MinDCd in the presence of MinECd, recorded in B.subtilis ΔminD Bs ΔminJ Bs minD Cd minE Cd (IB1546). Scale bar represents 5 μm.
Table S1: Plasmids used in this study and their construction.
Table S2: Primers used in this study.
Table S3: Cell length measurements.
Table S4: The sequence similarity/identity of Min proteins of selected members of Clostridia and Negativicutes compared with their counterparts in E.coli and B.subtilis. Similarity and identity values are derived from a BLAST query (Altschul et al. 1997). For MinC and MinD, the sequence of the B.subtilis proteins was used as reference, since these queries gave lower E‐values and higher query cover and identities than the E.coli sequences (not shown). The E.coli sequence was used as a reference for MinE, and B.subtilis sequences for a search of MinJ and DivIVA homologs. All listed organisms are endospore‐formers except E.coli.


