Abstract
Toxin/antitoxin (TA) systems are prevalent in most bacterial and archaeal genomes, and one of the emerging physiological roles of TA systems is to help regulate pathogenicity. Although TA systems have been studied in several model organisms, few studies have investigated the role of TA systems in pseudomonads. Here, we demonstrate that the previously uncharacterized proteins HigB (unannotated) and HigA (PA4674) of Pseudomonas aeruginosa PA14 form a type II TA system in which antitoxin HigA masks the RNase activity of toxin HigB through direct binding. Furthermore, toxin HigB reduces production of the virulence factors pyochelin, pyocyanin, swarming, and biofilm formation; hence, this system affects the pathogencity of this strain in a manner that has not been demonstrated previously for TA systems.
Keywords: biofilm, HigA, HigB, pyochelin, pyocyanin, toxin/antitoxin system, virulence factors
Introduction
Pseudomonas aeruginosa is an opportunistic, Gram‐negative pathogen (Mace et al. 2008), and it is the primary cause of important chronic infections including those associated with cystic fibrosis (CF) (Moker et al. 2010), burn wound infections, bacterial keratitis, and urinary and peritoneal dialysis catheter infections (Mace et al. 2008). It is difficult to treat infections from P. aeruginosa because this species is highly tolerant to antibiotics (Lewis 2010). For example, late isolates of P. aeruginosa from CF patients can be high‐persister mutants with 100‐fold greater antibiotic tolerance (Mulcahy et al. 2010). The cause of antibiotic tolerance in many strains is thought to be due to activation of toxins of toxin/antitoxin (TA) systems (Jayaraman 2008; Lewis 2008) which causes dormancy by inactivating key metabolic functions like protein and ATP production (Kwan et al. 2013; Wood et al. 2013).
The first TA operons were discovered over 30 years ago (Ogura and Hiraga 1983) for stabilizing low copy number plasmids via postsegregational killing (Gerdes et al. 1986) and some additional roles of TA systems in cell physiology have become clear. For example, they are antiphage measures (Pecota and Wood 1996; Hazan and Engelberg‐Kulka 2004; Fineran et al. 2009), and TA systems slow metabolism to allow cells to withstand stress such as that from antibiotics (Cheng et al. 2014; Islam et al. 2015) and bile acid (Kwan et al. 2015). Furthermore, the expression of the toxin genes are induced under stress conditions (Aizenman et al. 1996; Sat et al. 2001; Hazan et al. 2004), and antitoxins like MqsA and DinJ directly control the stress response by regulating the stress response sigma factor RpoS (Wang et al. 2011; Hu et al. 2012). TA systems also have a role in biofilm formation (Ren et al. 2004; González Barrios et al. 2006; Kim et al. 2009).
Beyond phage inhibition, stress response, and biofilm formation, the role of TA systems in pathogenicity is also becoming more clear. Production of almost any active toxin to the point of making the cell dormant increases antibiotic tolerance (i.e., persistence) (Wang and Wood 2011), and deletion of some single TA systems decreases persistence (Dörr et al. 2010; Kim and Wood 2010). Although Mycobacterium tuberculosis has about 88 putative TA systems (Ramage et al. 2009), the nonpathogenic Mycobacterium smegmatis has only two putative TA systems, so pathogenicity might be related to the number of TA systems (Yamaguchi and Inouye 2011). As anticipated, inactivation of three of M. tuberculosis MazF/MazE TA systems reduced its persistence in vitro, its survival in macrophages, and its cell numbers in the spleen and lungs of guinea pigs (Tiwari et al. 2015). Similarly, TA systems play a role in the persistence of Salmonella typhimurium in macrophages in a mouse model for typhoid fever (Helaine et al. 2014). There are at least 11 type II TA systems in S. typhimurium, and several of them are conserved in other pathogenic bacteria but absent from other nonpathogenic strains (De la Cruz et al. 2013). Also, inactivation of three Vap‐type TA systems reduced virulence for nontypeable Haemophilus influenzae (NTHi) in a chinchilla model for otitis media (Ren et al. 2012), and inactivation of YoeB/YefM, Hha/TomB (García Contreras et al. 2008), and PasT/PasI TA systems are important for uropathogenic Escherichia coli infections in the bladder and spleen in murine models (Norton and Mulvey 2012). Hence, determining the function of TA systems and the target of these toxins is very important because of their roles in bacterial physiology and pathogenicity (Yamaguchi and Inouye 2011).
One prominent TA system found in many pathogens is HigB/HigA in which the antitoxin HigA masks the toxicity of the toxin HigB. Genes for the HigB/HigA TA system are found on the Rts1 plasmid originally isolated from Proteus vulgaris and are found in the chromosomes of the pathogens Vibrio cholera (Christensen‐Dalsgaard and Gerdes 2006; Budde et al. 2007), Streptococcus pneumonia (Chan et al. 2012), Acinetobactor baumanni (Jurenaite et al. 2013), S. typhimurium (De la Cruz et al. 2013), Yersinia pestis (Goulard et al. 2010), M. tuberculosis (Schuessler et al. 2013), E. coli CFT073 (Pandey and Gerdes 2005), and E. coli O157:H7 (Pandey and Gerdes 2005) and is also present in E. coli K12 (Christensen‐Dalsgaard et al. 2010). Additionally, higBA is widespread in P. aeruginosa clinical isolates (Williams et al. 2011). The HigB/HigA system has the uncommon gene arrangement with the toxin gene (higB) upstream of the antitoxin gene (higA), which is the reverse arrangement of most other TA systems (Tian et al. 1996).
HigB functions as an endoribonuclease in Proteus spp. (Hurley and Woychik 2009), V. cholera (Christensen‐Dalsgaard and Gerdes 2006), A. baumanni (Jurenaite et al. 2013), and E. coli K12 (Christensen‐Dalsgaard et al. 2010). Although the mechanism of how the SehA (S almonella e nterica Hig‐like) toxin works is unknown, it shares 40% identity with HigB from E. coli K12 (De la Cruz et al. 2013); hence, SehA is likely to be an endoribonuclease and have the same target as HigB (De la Cruz et al. 2013). SehAB was also found to play an important role in virulence in mice (De la Cruz et al. 2013).
Here, we identified and characterized the HigB/HigA system in the chromosome of P. aeruginosa PA14. The HigB/HigA system is shown to be a bona fide TA system. Moreover, the HigB/HigA system affects the virulence factors of the strain in a fashion that has not been demonstrated previously for TA systems, since activation of toxin HigB reduces pyocyanin, a toxin produced and secreted by P. aeruginosa, reduces the siderophore pyochelin, reduces swarming, and reduces biofilm formation. The closest HigB homolog to that we identify here in P. aeruginosa has only 34.3% identity (from V. cholera), so our findings are for a novel HigB/HigA TA system.
Experimental Procedures
Bacterial strains and culture conditions
The strains and plasmids used in this study are shown in Table 1. All strains were grown in lysogeny broth (LB) (Sambrook and Russell 2001) at 37°C except for the biofilm assay where M9 medium with 0.4% glucose and 0.4% casamino acids (Rodriguez and Tait 1983) was used. Chloramphenicol (30 μg/mL) was used to maintain pCA24N‐based plasmids (Kitagawa et al. 2005) in E. coli. To obtain the specific growth rates, the P. aeruginosa PA14 wild‐type and higA‐mutant strains were inoculated into LB medium at an initial turbidity of 0.05 at 600 nm, and the turbidity at 600 nm was measured every an hour. At least two replicates were performed.
Table 1.
Bacterial strains and plasmids used in this study
| Strains or plasmids | Description | Source |
|---|---|---|
| Strains | ||
| Escherichia coli TG1 | supE thi‐1 Δ(lac‐proAB) Δ(mcrB‐hsdSM)5, (r K − m K − ) F′ [traD36 proAB + lacI q lacZΔM15] | Sambrook et al. (1989) |
| Pseudomonas aeruginosa PA14 | Wild type | Liberati et al. (2006) |
| P. aeruginosa higA | GmR, MAR2xT7 transposon insertion | Liberati et al. (2006) |
| Plasmids | ||
| pCA24N | CmR; lacI q | Kitagawa et al. (2005) |
| pCA24N‐lacZ | CmR; lacI q PT5‐lac::His 6 ‐lacZ + | Kitagawa et al. (2005) |
| pCA24N‐His‐higB | CmR; lacI q PT5‐lac::His 6 ‐higB + | This study |
| pCA24N‐ higA‐FLAG | CmR; lacI q PT5‐lac::His 6 ‐higA‐flag + | This study |
| pCA24N‐ His‐higB‐higA‐flag | CmR; lacI q PT5‐lac:: His 6 ‐higB +, higA‐flag + | This study |
| pCA24N‐ His‐higB‐higAX‐FLAG | CmR; lacI q PT5‐lac:: His 6 ‐higB +, higA‐flag + with ATG (start codon) mutated to ACC and with ATG (16–18 nt relative to start codon) mutated to ACCG | This study |
GmR and CmR denote gentamycin and chloramphenicol resistance, respectively.
Bioinformatics search
The web‐based search tool RASTA (Sevin and Barloy‐Hubler 2007) was used to search for type II TA systems. The search was performed against the complete genome sequence of P. aeruginosa PA14 (Lee et al. 2006) with standard parameters.
Plasmid construction
Plasmid pCA24N(His‐higB) was constructed by amplifying the higB gene from the chromosome of P. aeruginosa PA14 (Lee et al. 2006) and cloning into pCA24N at the NotI and PstI restriction sites placed higB under the control of T5‐lac promoter. The higB gene was fused with His tag in the pCA24N plasmid at the N terminus. Plasmids pCA24N(higA‐FLAG) and pCA24N(His‐higB‐higA‐FLAG), Fig. 3B, were constructed by amplifying the higA gene from the chromosome of P. aeruginosa PA14 with the addition of a FLAG tag at the C terminus and cloning into pCA24N and pCA24N(His‐higB), respectively, at the PstI and HindIII restriction sites. All plasmids were verified by DNA sequencing. All oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA), and the primer sequences are listed in Table 2.
Figure 3.
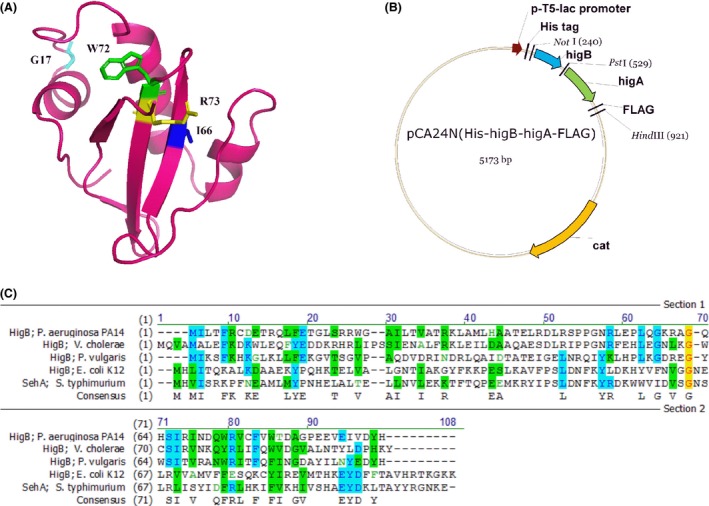
(A) Predictive modeled structure of HigB of Pseudomonas aeruginosa PA14 based on HigB structure of Proteus vulgaris using Phyre (Moker et al. 2010). The RNase conserved amino acids G17, I66, W72, and R73 that were identified in the mutagenesis experiment are in light blue, blue, green, and yellow, respectively. PyMOL was used to edit the modeled structure. (B) Plasmid map of pCA24N(higBA). The plasmid map showing the pCA24N backbone plasmid containing higB and higA genes and cloning sites (NotI, PstI, and Hind III). (C) Alignment of the toxin Hig B protein from P. aeruginosa PA14, Vibrio cholera, Proteus vulgaris, and Escherichia coli K12, and SehA from Salmonella typhimurium.
Table 2.
Oligonucleotides used for cloning and sequencing
| Primer name | Sequence (5′→3′) |
|---|---|
| Cloning and sequencing | |
| HigB‐NotI‐f | TTTTTTGCGGCCGCTAATTCTGACCTTTCG |
| HigB‐PstI‐r | TTTTTTCTGCAGTCAGTGGTAATCAACTATTTCGAC |
| HigA‐PstI‐f | TTTTTTTTCTGCAGATTAAAGAGGAGAAATTAACTATGAGGAGGTGGACCATGGC |
| HigA‐FLAG‐HindIII‐r | TTTTTGTCGACAAGCTTCTACTTGTCATCGTCGTCCTTGTAGTCCTTGTCATCGTCGTCCTTGTAGTCTCCGTGAGCAAGCAGCGGCTCA |
| HigA‐PstI‐start‐QC‐f | ACCACTGACTGCAGATTAAAGAGGAGAAATTAACTACCAGGAGGTGGACCACCGCTACCA |
| pCA24N‐f | GCCCTTTCGTCTTCACCTCG |
| pCA24N‐r | GAACAAATCCAGATGGAGTTCTGAGGTCATT |
| Northern blot | |
| ompA‐f | CACTGGCTGGTTTCGCTACCG |
| ompA‐r | ACCCATTTCAAAGCCAACATC |
| ompF‐f | AAGCGCAATATTCTGGCAGT |
| ompF‐r | TGCCACCGTAACTGTTTTCA |
All restriction enzyme sites are underlined. f indicates forward primer and r indicates reverse primer.
Site‐directed mutagenesis of higA
Plasmid pCA24N(His‐higB‐higA‐FLAG) was used as a PCR template to replace the start codon of HigA with Thr; since methionine residues exist at amino acid positions 1 and 6, both methionines were converted to threonine (i.e., M1T and M6T) in case either one is the start codon. The primers higA‐PstI‐start‐QC‐f and higA‐FLAG‐HindIII‐r (Table 2) were used in the PCR reaction to generate the mutations. The PCR product was cloned into pCA24N(His‐higB) using the PstI and HindIII sites. The resulting plasmid pCA24N(His‐higB‐higAX‐FLAG) was sequenced to confirm the mutations.
Random mutagenesis of higB toxin
The plasmid pCA24N(His‐higB) was used for the error‐prone PCR template. Using the pCA24N‐f and pCA24N‐r primers (Table 2), higB was randomly mutated under error‐prone conditions (0.5 mmol/L Mn2+ and 5 mmol/L Mg2+) (Cadwell and Joyce 1992). Mutated higB inserts were digested and cloned into pCA24N at the NotI and PstI restriction sites. Ligated product was electroporated into E. coli TG1. The electroporated population was plated on LB–chloramphenicol agars. To select for growth, recovered colonies were restreaked on LB–chloramphenicol agar with 1 mmol/L IPTG. Variants were chosen for DNA sequencing analysis and retransformation tests.
Toxicity assay
Overnight cultures of strains E. coli TG1/pCA24N, E. coli TG1/pCA24N(lacZ), E. coli TG1/pCA24N(His‐higB), E. coli TG1/pCA24N(higA‐FLAG), E. coli TG1/pCA24N(His‐higB‐higA‐FLAG), and E. coli TG1/pCA24N(His‐higB‐higAX‐FLAG) were inoculated into 25 mL of LB medium at an initial of turbidity of 0.05 at 600 nm. IPTG (0.01 mmol/L) was added after 1 h, and the turbidity was recorded to determine growth.
Western blot analysis and pull‐down assay
Escherichia coli TG1/pCA24N(His‐higB) and E. coli TG1/pCA24N(His‐higB‐higA‐FLAG) were inoculated into LB medium from the overnight culture at an initial turbidity of 0.05 at 600 nm, then 0.1 mmol/L of IPTG was added to produce HigB and HigA for 5 h. The cell pellets were resuspended in buffer (50 mmol/L NaCl and 20 mmol/L Tris, pH 7.4) with protease inhibitor, and the cells were lysed using a French Press (Thermo Electron, Waltham, MA); centrifugation (15,000g for 1 h at 4°C) was used to remove cell debris. The lysate was filtered through a 0.22‐μm membrane and loaded on a HisTrap HP column (GE Healthcare, Pittsburgh, PA, USA). HigB and associated proteins were eluted with a 5–500 mmol/L imidazole gradient. The fractions containing HigB and HigA were identified using western blot analysis using HRP‐conjugated anti‐His antibody and HRP‐conjugated anti‐FLAG antibody (Thermo Scientific, Waltham, MA, USA). Blotted proteins were detected using the chemiluminescence reagents from the SuperSignal West‐Pico Chemiluminescence kit (Thermo Scientific).
Northern blot analysis
Escherichia coli TG1/pCA24N(His‐higB) was inoculated from the overnight culture at an initial turbidity of 0.05 at 600 nm, then 0.5 mmol/L of IPTG was added to produce HigB for 3 h. The samples were collected at 0, 1 15, 30, and 60 min after induction. Total RNA was isolated using RNeasy Mini kit (Qiagen, Hilden, Germany). The ompA (outer membrane porin protein A) and ompF (outer membrane porin protein F) (Hurley and Woychik 2009) DNA probes were amplified from the chromosomal DNA of E. coli TG1/pCA24N(His‐higB) using primers ompA‐f and ompA‐r and ompF‐f and ompF‐r, respectively. The probes were labeled using Biotin 3′ End DNA Labeling Kit (Thermo Scientific). Northern blot analysis method was described previously (Sambrook et al. 1989). RNA levels were detected using Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific).
DNA microarrays
To isolate total RNA, the overnight culture of P. aeruginosa PA14 wild‐type and the higA‐mutant strains were inoculated in 25 mL of LB medium at an initial turbidity of 0.05 at 600 nm. The culture was collected in 2 mL tubes (four tubes) at a turbidity of 2.0 at 600 nm. RNAlater buffer (Applied Biosystems, Foster City, CA) (100 μL) was added immediately into the sample tubes to stabilize RNA, and the sample tubes were rapidly frozen in ethanol/dry ice. Cells were lysed using 0.1 mm zirconia/silica beads and a bead beater (Biospec, Bartlesville, OK) and total RNA was isolated using an RNeasy Mini kit (Qiagen) (Ren et al. 2004). cDNA synthesis, fragmentation, and hybridization to Affymetrix P. aeruginosa Genome array (Affymetrix, Santa Clara, CA, USA) were performed as previously described (González Barrios et al. 2006). The gene expression data are accessible through GEO accession number GSE74730.
Pyocyanin assay
The P. aeruginosa PA14 wild‐type, higA‐mutant, and negative control (phzM and phzS mutants) strains were inoculated at a 1/1000 dilution from the overnight culture in LB medium and incubated for 24 h. As described previously (Essar et al. 1990), a 1‐mL sample was centrifuged. The 800 μL of supernatant was extracted with 480 μL of chloroform. The sample was vortexed and centrifuged, then re‐extracted with 0.2 N HCl. The absorbance of this sample was measured at 520 nm. The OD values at 520 nm were normalized with bacteria growth to eliminate any possible growth affects. At least three replicates were performed.
Pyochelin assays
The P. aeruginosa PA14 wild‐type, higA‐mutant, and negative control (pchB and pchF mutants) strains were grown overnight in LB medium. As described previously (Takase et al. 2000), acetic acid (0.3 mL) and dichloromethane (1.5 mL) were added into 3 mL of each culture. The samples were vortexed briefly and centrifuged for 5 min. One milliliter of dichloromethane fractions (bottom layer) were collected and evaporated. The samples were resuspened in 10 μL of dichloromethane and applied onto silica thin‐layer plates for chromatography in chloroform–acetic acid–ethanol (90:5:2.5). The pyochelin spots were scraped from the plates, eluted with 1 mL of methanol, and measured using a spectrophotometer at 313 nm and using a spectrofluorimeter (excitation at 355 nm and emission at 430 nm). At least three replicates were performed.
Pyoverdine assays
The P. aeruginosa PA14 wild‐type, higA‐mutant, and negative control (pvdF mutant) strains were grown overnight in LB medium. For the chrome azurol S (CAS) agar plate assay (Owen and Ackerley 2011), 1 μL of the overnight culture was placed in the middle of CAS agar plate. After incubation at 37°C for 1 and 2 days, the plate was observed under UV light by the formation of a fluorescent zone around the cells (Yu et al. 2014). For CAS liquid assay (Yu et al. 2014), an overnight culture in LB was diluted to an initial turbidity of 0.1 at 600 nm in King's B medium (Yu et al. 2014). After 1 or 2 days incubation at 37°C, the samples were centrifuged, and the supernatant was used to measure the pyoverdine production using a fluorescence spectrometer (excitation at 405 nm and emission at 460 nm).
Biofilm assay using crystal violet
Biofilm formation was assayed in 96‐well polystyrene plates using 0.1% crystal violet staining as described previously (Fletcher 1977) with some modifications. Diluted overnight cultures at an initial turbidity of 0.05 at 600 nm were inoculated into 96‐well plates with M9 medium with 0.4% glucose and 0.4% casamino acids (Rodriguez and Tait 1983) and the bacteria were cultured for 48 h without shaking. After the crystal violet was added to each well, the wells were rinsed and dried, and ethanol was added to dissolve the crystal violet. The total biofilm formation samples were measured at 540 nm, whereas cell growth was measured at 620 nm. Biofilm formation was normalized by the bacterial growth to reduce any growth effect. At least two independent cultures were used for each strain.
Swarming assay
The P. aeruginosa PA14 wild‐type, higA‐mutant, and negative control (rhlR mutant) strains were grown overnight in LB medium. The culture (1 μL) was inoculated in the middle of fresh BM2 plates (Overhage et al. 2008) that were dry for 3 h before inoculation, and the plates were incubated for 18 h. The agar plate coverage was measured using ImageJ software http://http%5c%5c:(www.imagej.nih.gov/ij/). At least three replicates were performed.
Results
Identification of the putative HigB/HigA TA system
We identified a putative HigB/HigA system in the genome of P. aeruginosa PA14 by using the RASTA Bacteria program (Sevin and Barloy‐Hubler 2007). This program searches for type II TA systems, and the HigB/HigA system got the highest score (90%) which indicates it is likely to be a TA system (scores above 70% are indicated to likely be TA systems). HigB was not annotated previously. HigB is a small protein (92 amino acids), and HigA consists of 106 amino acids (Fig. 1A). Their genes overlap by one nucleotide, which is the same as the HigB/HigA system in P. vulgaris (Schureck et al. 2014), and gene overlap is a common characteristic of TA systems (Yamaguchi et al. 2011).
Figure 1.
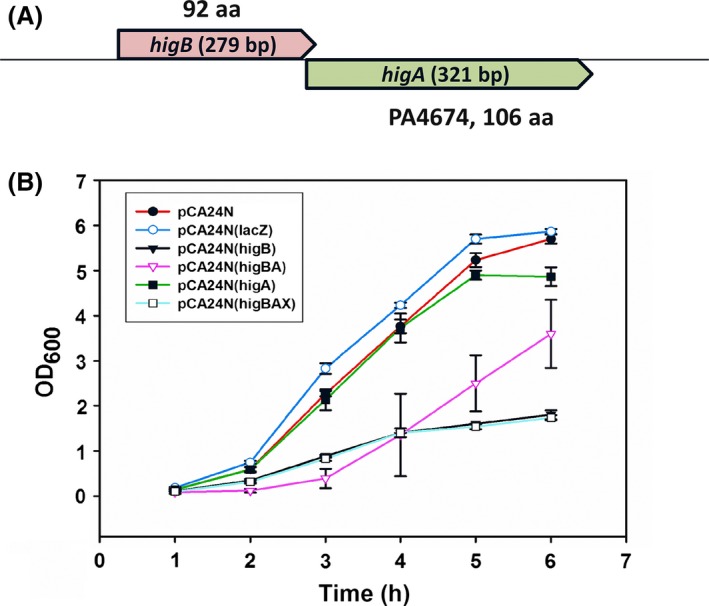
higBA loci and results showing HigB toxicity is masked by HigA. (A) The higBA operon of Pseudomonas aeruginosa PA14. The toxin gene higB (not annotated, 5514513‐5514791) and the antitoxin gene higA (PA4674, PA14_61840, 5514196‐5514516) were unveiled using RASTA software. (B) Overnight cultures of strains of Escherichia coli TG1/pCA24N (pCA24N, control), E. coli TG1/pCA24N (lacZ) (pCA24N(lacZ), control), E. coli TG1/pCA24N(His‐higB) (pCA24N(higB)), E. coli TG1/pCA24N(higA‐FLAG) (pCA24N(higA)), E. coli TG1/pCA24N(His‐higB‐higA‐FLAG) (pCA24N(higBA)), and E. coli TG1/pCA24N(His‐higB‐higAX‐FLAG), where “X” indicates the translation start signal was changed to a threonine codon so antitoxin HigA is not produced (pCA24N(higBAX)), were inoculated into 25 mL of LB medium at an initial of turbidity of 0.05 at 600 nm at 37°C. IPTG (0.01 mmol/L) was added after 1 h. The error bars shown are standard deviation from three independent cultures.
HigB and HigA form a TA system
To assess whether HigB functions as a toxin and whether HigA masks HigB toxicity, the activity of the two proteins were evaluated in E. coli. HigB toxin produced from plasmid pCA24N inhibited the growth in E. coli TG1 (Fig. 1B); hence, HigB showed strong toxin activity in this non‐native host. When both HigB and HigA were produced simultaneously, the toxicity of HigB was counteracted (Fig. 1B). Therefore, HigB/HigA is a bona fide TA system.
Antitoxin HigA binds to toxin HigB and functions as a protein
Antitoxin HigA could mask HigB toxicity as either RNA or as a protein; hence, the start codon of higA was converted to Thr and its effect on HigB toxicity was evaluated. Growth in E. coli showed that the translation mutation in higA abolished its antitoxin activity. Therefore, HigA functions as a protein antitoxin (Fig. 1B).
We then hypothesized that the HigA antitoxin directly binds toxin HigB to inactivate it. To verify the direct interaction between antitoxin HigA and toxin HigB, a pull‐down experiment was performed. The protein HigB from pCA24N(His‐higB) and from pCA24N(His‐higB‐higA‐FLAG) was tagged with six histidines at the N terminus, and the protein HigA from pCA24N(His‐higB‐higA‐FLAG) was tagged using the FLAG octapeptide at the C terminus.
Using the His‐tagged HigB protein to pull down the Flag‐tagged HigA antitoxin, a western blot was performed. Both proteins HigB and HigA were detected using His and FLAG tag antibodies, respectively; hence, antitoxin HigA interacts directly with toxin HigB (Fig. 2A). Therefore, HigB and HigA interact at the protein level and are a type II TA system (i.e., protein–protein TA system).
Figure 2.
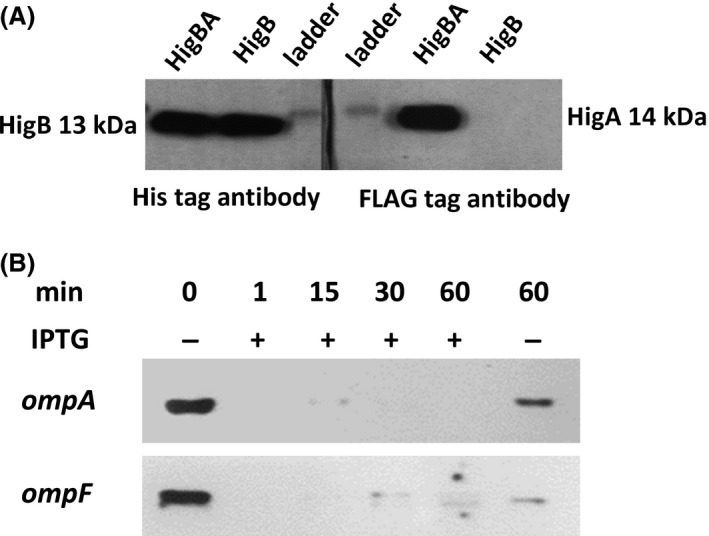
Western blot of HigB/HigA showing HigA binds to HigB and HigB degrades ompA and ompF mRNA in vivo. Escherichia coli TG1/pCA24N(His‐higB) and E. coli TG1/pCA24N(His‐higB‐higA‐FLAG) were grown in LB medium at 37°C at an initial turbidity of 0.05 at 600 nm, then 0.1 mmol/L of IPTG was added to produce HigB and HigA for 5 h. (A) The toxin HigB was tagged with six histidines, and the antitoxin HigA was tagged with the FLAG octapeptide. After producing both proteins via the E. coli TG1/pCA24N(His‐higB‐higA‐FLAG) strain, HigA binding to HigB was checked based on binding of HigB to the nickel column via its His tag. After purification of HigB, both HigB (left hand panel, using a His antibody) and HigA (right hand panel, using a FLAG antibody) from the HigB/HigA complex (HigBA) were detected independently in a denaturing gel. HigB production from the E. coli TG1/pCA24N(His‐higB) strain served as a positive control for HigB detection via the His antibody (left hand panel) as well as a negative control for the absence of HigA detection with the FLAG antibody (right hand panel). (B) RNA was isolated after higB was induced with 0.5 mmol/L of IPTG using the E. coli TG1/pCA24N(His‐higB) strain for 0, 1, 15, 30, and 60 min. Northern blot analysis was performed for ompA and ompF RNA detection.
Toxin HigB functions as an RNase
To determine the enzyme activity of HigB in E. coli, a Northern blot analysis for ompA and ompF was performed (Fig. 2B); these two loci encode large genes so they are used frequently in TA studies (Hurley and Woychik 2009). The degradation of the mRNAs was detectable immediately after induction (1 min) of higB. The level of the control mRNAs at the 60‐min time point without higB induction was also decreased due to low‐level activity of the plasmid promoter, but it was not as strong as in the samples with higB induction.
Toxin HigB catalytic sites
Error‐prone PCR of higB was performed to generate a library of HigB variants in E. coli to determine the residues required for its RNase activity. The HigB variants were selected on LB plates with 0.1 mmol/L of IPTG to produce the toxin; under these conditions, native HigB prevented the formation of colonies. Variants were selected based on their enhanced growth, and the substitutions were identified as G17D/A61E, V28D, R31H, Q57P, Q63H/W72C, H64P, I66N, R67W, W72R, and R73H. The Q63H, H64P, R67W, and W72C substitutions were found multiple times. These results indicate that the amino acids at these positions are crucial for the toxicity of HigB. We also aligned HigB sequences of P. aeruginosa PA14, P. vulgaris, and V. cholerae and found that most of the crucial amino acids we identified in the variants (G17, I66, W72, and R73, Fig. 3A) are conserved or nearby the conserved amino acids which are likely to be the active site of HigB (Schureck et al. 2014).
Toxin HigB reduces pyochelin production
To determine the physiological role of HigB/HigA in P. aeruginosa, a whole‐transcriptome experiment was performed for the higA antitoxin deletion mutant compared to the P. aeruginosa PA14 wild‐type strain. The rationale was that for the strain that lacks the antitoxin, the effect of the toxin could be discerned due to enhanced activity of the toxin. Notably, the specific growth rates of P. aeruginosa PA14 and the higA antitoxin deletion mutant strain were 1.2 and 0.8 h−1, respectively, so there is a modest decrease in growth upon activation of toxin HigB (Fig. 4A).
Figure 4.
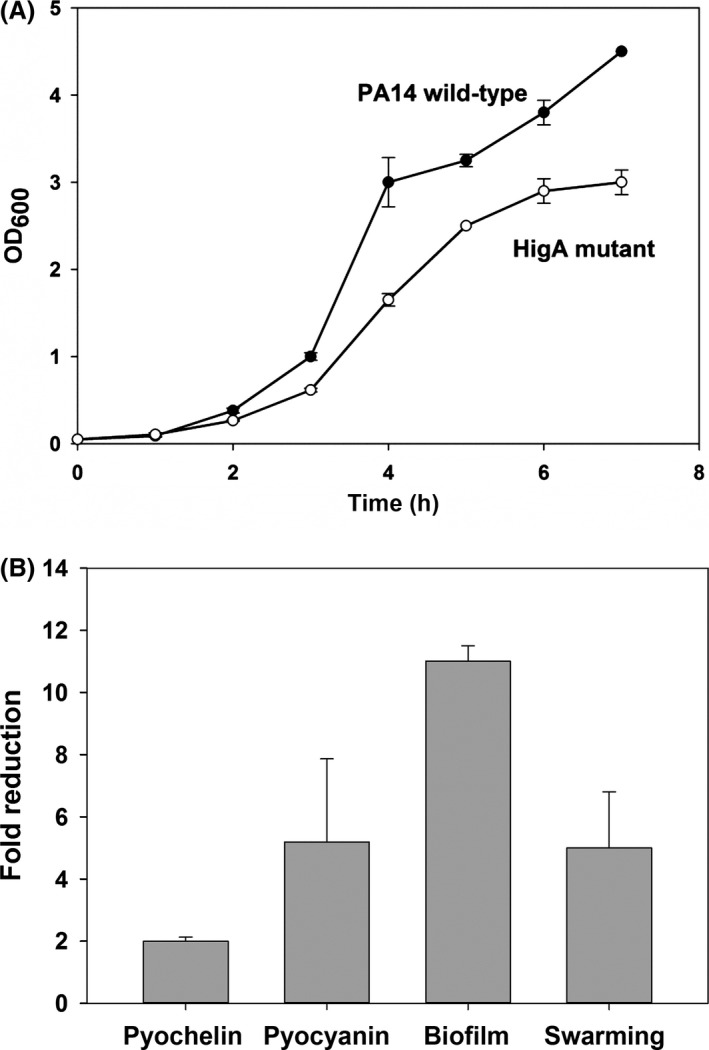
(A) Growth of Pseudomonas aeruginosa PA14 and the higA mutant. The strains were grown in LB medium at 37°C. At least two replicates were performed. (B) The higA mutation reduces pyochelin, pyocyanin, biofilm formation, and swarming. Pseudomonas aeruginosa PA14 and the higA mutant were grown in LB medium at 37°C. Results from two replicates are shown.
The microarray results (Table 3) indicate that deletion of the antitoxin induces toxin transcription by 28‐fold, as expected since antitoxins normally repress the TA operon (Brown et al. 2013). Furthermore, the PA2405‐2410 operon, which is part of the pyoverdine locus (Ravel and Cornelis 2003), has the most induced genes upon activating toxin HigB (i.e., deletion of higA); these genes probably encode proteins related to membrane transport. However, there was not much difference in pyoverdine production between the wild‐type strain and the higA mutant (data not shown) which collaborated previous work which reported that the mutations in this PvdS‐regulated PA2403‐PA2410 cluster did not affect pyoverdine production (Ravel and Cornelis 2003).
Table 3.
Summary of the largest fold changes in gene expression for the higA mutant versus the isogenic Pseudomonas aeruginosa PA14 wild type strain. Both strains were grown to a turbidity of 2.0 at 600 nm in LB medium
| Genes | Fold change | Description |
|---|---|---|
| higB 28.0 toxin gene | ||
| Part of the pyoverdine locus (Ravel and Cornelis 2003) | ||
| PA2405 | 9.2 | Hypothetical protein |
| PA2404 | 8.0 | Hypothetical protein; membrane proteins |
| PA2408 | 7.0 | Probable ATP‐binding component of ABC transporter, membrane proteins |
| PA2403 | 5.7 | Hypothetical protein; membrane proteins |
| PA2406 | 5.3 | Hypothetical protein |
| PA2409 | 4.0 | Probable permease of ABC transporter, membrane proteins/transport of small molecules |
| PA2407 | 3.5 | Probable adhesion protein, motility and attachment |
| PA2410 | 3.5 | Hypothetical protein |
| Related to pyoverdine and iron transportation | ||
| PA2398_fpvA | 7.0 | Ferripyoverdine receptor/transport of small molecules |
| PA4675 | 5.3 | Probable TonB‐dependent receptor/transport of small molecules |
| PA0805 | 4.0 | Hypothetical protein |
| Pyochelin genes | ||
| PA4230_pchB | −9.2 | Salicylate biosynthesis protein PchB/transport of small molecules; secreted factors (toxins, enzymes, alginate) |
| PA4231_pchA | −7.5 | Salicylate biosynthesis isochorismate synthase/secreted factors (toxins, enzymes, alginate); transport of small molecules |
| PA4229_pchC | −6.1 | Pyochelin biosynthetic protein PchC/transport of small molecules; secreted factors (toxins, enzymes, alginate) |
| PA4226_pchE | −4.9 | Dihydroaeruginoic acid synthetase/transport of small molecules; secreted factors (toxins, enzymes, alginate) |
| PA4224_pchG | −4.9 | Hypothetical protein; membrane proteins |
| PA4228_pchD | −4.6 | Pyochelin biosynthesis protein PchD/transport of small molecules; secreted factors (toxins, enzymes, alginate) |
| PA4225_pchF | −4.0 | Pyochelin synthetase/transport of small molecules; secreted factors (toxins, enzymes, alginate) |
| PA4227_pchR | −2.6 | Transcriptional regulator PchR/transcriptional regulators |
| Pyochelin‐related genes and nearby genes to the pch operon | ||
| PA4221_fptA | −6.1 | Fe(III)‐pyochelin receptor precursor/transport of small molecules |
| PA4223 | −4.0 | Probable ATP‐binding component of ABC transporter/membrane proteins; transport of small molecules |
| PA4218 | −4.0 | Probable transporter/membrane proteins; transport of small molecules |
| PA4220 | −3.7 | Hypothetical protein |
| PA4219 | −3.5 | Hypothetical protein/membrane proteins |
| PA4222 | −3.2 | Probable ATP‐binding component of ABC transporter/transport of small molecules |
| PA0985_pyoS5 | −3.2 | Probable colicin‐like toxin/membrane proteins; secreted factors (toxins, enzymes, alginate) |
Critically, the microarray data show that pyochelin‐related genes (pchA, pchB, pchC, pchD, pchE, pchF, pchG, pchR, and fptA), which encode proteins related to iron transfer, are repressed upon activating the toxin HigB. Pyochelin is a siderophore produced by P. aeruginosa which increases the growth and lethality of pathogenic bacteria (Cox 1982). FptA is the receptor for ferric pyochelin, and its expression is pyochelin dependent (Heinrichs and Poole 1996). A number of studies have shown that there is a correlation between pyochelin and virulence (Cox 1982; Sokol 1987; Wang et al. 1996). Pyochelin levels were assayed in the higA mutant and found to be reduced by 2.0 ± 0.1‐fold (Fig. 4B); hence, the microarray data were corroborated by the reduced pyochelin levels. The OD313 values for the pyochelin assay for P. aeruginosa PA14 and the higA mutant were 0.104 and 0.05, respectively. Therefore, the HigB toxin reduces virulence via a reduction in pyochelin, and HigA antitoxin represses its own transcription. These results were not affected by growth since the samples for pyochelin assay were collected in the stationary phase at the same turbidity.
Toxin HigB reduces pyocyanin production
Pyocyanin is a blue redox‐active secondary metabolite that interferes with multiple cellular functions and has crucial roles in P. aeruginosa infections (Lau et al. 2004). The higA antitoxin deletion mutant was less blue compared to the wild type; hence, chloroform extraction was performed to more accurately measure the amount of the pyocyanin in an overnight culture. The higA‐mutant strain produced 5 ± 3‐fold less pyocyanin than the P. aeruginosa PA14 wild‐type strain (Fig. 4B), which showed that the toxin HigB affected the pyocyanin level in P. aeruginosa. The OD520/OD600 values for the pyocyanin assay for P. aeruginosa PA14 and the higA mutant were 0.0086 ± 0.0012 and 0.0018 ± 0.00007, respectively. These results were not affected by growth because the samples were collected after 24 h at the same turbidity.
Toxin HigB reduces biofilm formation and swarming motility
Since TA systems are related to biofilm formation (Ren et al. 2004; González Barrios et al. 2006; Kim et al. 2009) as well as quorum sensing and motility (González Barrios et al. 2006), biofilm formation and swarming were investigated for the HigB/HigA TA system. After 48 h in M9 medium with 0.4% glucose and 0.4% casamino acids (Rodriguez and Tait 1983), the higA mutant had 11.0 ± 0.5‐fold less normalized biofilm than the P. aeruginosa PA14 wild‐type strain (Fig. 4B). The OD540/OD600 values for the biofilm assay for P. aeruginosa PA14 and the higA mutant were 1.3 ± 0.4 and 0.12 ± 0.04, respectively. These results were not affected by growth because the samples were collected after 48 h, and the planktonic cell densities were nearly identical. Furthermore, the higA mutant had 5 ± 2‐fold lower swarming motility than the P. aeruginosa PA14 wild‐type strain (Figs. 4B and 5). Hence, toxin HigB reduces both biofilm formation and swarming.
Figure 5.
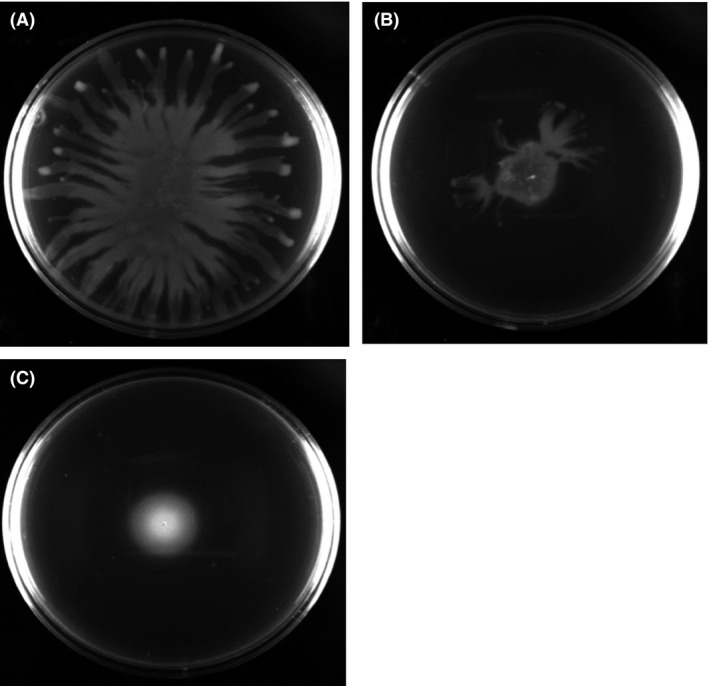
The higA mutant has less swarming compared to the wild‐type strain. Pseudomonas aeruginosa PA14, the higA mutant, and the negative control (rhlR mutant) were grown overnight in LB medium. The culture (1 μL) was inoculated into the middle of a BM2 plate (Overhage et al. 2008) that was dried for 3 h before inoculation, and the plates were incubated for 18 h. Swarming plates of (A) P. aeruginosa PA14 with 52 ± 8% area coverage, (B) the higA mutant with 11 ± 2% area coverage, and (C) the rlhR mutant (negative control) with 5 ± 0.4% area coverage. Results shown are from one of three representative independent cultures.
Discussion
We present additional evidence in this report that TA systems are involved in pathogenicity. Furthermore, we find that the HigB/HigA TA system of P. aeruginosa affects its virulence in a manner that is distinct from the way any other TA system has been linked to virulence, since here we found HigB/HigA affects virulence through pyochelin, pyocyanin, swarming (as well as through biofilm formation). We also demonstrate clearly that the P. aeruginosa HigB/HigA system is a type II bona fide TA system.
Pseudomonas aeruginosa produces two siderophores, pyoverdine and pyochelin (Ankenbauer and Quan 1994), and gene expression related to both siderophores was affected by the HigB/HigA TA system. Based on our microarray results, fpvA, which encodes the receptor of the siderophore ferripyoverdine, was induced in the higA mutant compared to the P. aeruginosa PA14 wild‐type strain; this would make cells with the HigB toxin activated more susceptible to pyocins S2 and S3 since these pyocins use the ferripyoverdine receptor (Denayer et al. 2007). In contrast, fptA, which encodes the receptor of the siderophore pyochelin, was reduced in the higA mutant. pyoS5, which encodes a toxin that uses the FptA ferripyochelin receptor to enter the cell (Elfarash et al. 2014), was also repressed.
The HigB toxin is prevalent in pseudomonads; for example, P. aeruginosa DK2, P. aeruginosa B136‐33, Pseudomonas stutzeri DSM 10701, Pseudomonas putida F1, Pseudomonas putida KT2440, Pseudomonas fluorescens F113, and Pseudomonas denitrificans all contain genes for the toxin. Although the genes for putative HigB/HigA TA systems have been found in many organisms, most of them have not been characterized. An exception is the HigB toxin from Proteus vulgaris which has a highly conserved residue N71 for controlling mRNA specificity by interacting with the 16S rRNA residue C1054 (Schureck et al. 2015). However, the HigB from P. aeruginosa has glutamine at this position. The HigB toxin protein we found in P. aeruginosa PA14 was aligned with the HigB from V. cholera, Proteus vulgaris, E. coli K12, and SehA from S. typhimurium to reveal that there is only 34.3%, 28%, 10.6%, and 8.7% identity, respectively (Fig. 3C). Hence, the P. aeruginosa system that we describe here is distinct since proteins that share about 20% identity are not related, in that only those with identities of 50% or greater are usually considered related in databases (Seffernick et al. 2001). Therefore, we have characterized a novel HigB/HigA TA family member and shown how the TA system affects virulence factors of an important opportunistic pathogen. This study also represents the first TA system that has been characterized in P. aeruginosa.
Conflict of Interest
None declared.
Acknowledgments
T. K. W. is the Biotechnology Endowed Professor at the Pennsylvania State University. We appreciate the help of Olga Vinogradova and Kenneth Hubbell in generating, screening, and sequencing some HigB variants.
MicrobiologyOpen 2016; 5(3): 499–511
References
- Aizenman, E. , Engelberg‐Kulka H., and Glaser G.. 1996. An Escherichia coli chromosomal ‘‘addiction module’’ regulated by 3′,5′‐bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankenbauer, R. G. , and Quan H. N.. 1994. Fpta, the Fe(Iii)‐pyochelin receptor of Pseudomonas aeruginosa ‐ a phenolate siderophore receptor homologous to hydroxamate siderophore receptors. J. Bacteriol. 176:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, B. L. , Lord D. M., Grigoriu S., Peti W., and Page R.. 2013. The Escherichia coli toxin MqsR destabilizes the transcriptional repression complex formed between the antitoxin MqsA and the mqsRA operon promoter. J. Biol. Chem. 288:1286–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde, P. P. , Davis B. M., Yuan J., and Waldor M. K.. 2007. Characterization of a higBA toxin‐antitoxin locus in Vibrio cholerae . J. Bacteriol. 189:491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell, R. C. , and Joyce G. F.. 1992. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2:28–33. [DOI] [PubMed] [Google Scholar]
- Chan, W. T. , Moreno‐Cordoba I., Yeo C. C., and Espinosa M.. 2012. Toxin‐antitoxin genes of the gram‐positive pathogen Streptococcus pneumoniae: so few and yet so many. Microbiol. Mol. Biol. Rev. 76:773–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H. Y. , Soo V. W. C., Islam S., McAnulty M. J., Benedik M. J., and Wood T. K.. 2014. Toxin GhoT of the GhoT/GhoS toxin/antitoxin system damages the cell membrane to reduce adenosine triphosphate and to reduce growth under stress. Environ. Microbiol. 16:1741–1754. [DOI] [PubMed] [Google Scholar]
- Christensen‐Dalsgaard, M. , and Gerdes K.. 2006. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol. Microbiol. 62:397–411. [DOI] [PubMed] [Google Scholar]
- Christensen‐Dalsgaard, M. , Jorgensen M. G., and Gerdes K.. 2010. Three new RelE‐homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 75:333–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, C. D. 1982. Effect of pyochelin on the virulence of Pseudomonas aeruginosa . Infect. Immun. 36:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Cruz, M. A. , Zhao W. D., Farenc C., Gimenez G., Raoult D., Cambillau C., et al. 2013. A toxin‐antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog. 9:e1003827. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Denayer, S. , Matthijs S., and Cornelis P.. 2007. Pyocin S2 (Sa) kills Pseudomonas aeruginosa strains via the FpvA type I ferripyoverdine receptor. J. Bacteriol. 189:7663–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr, T. , Vulic´ M., and Lewis K.. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli . PLoS Biol. 8:e1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfarash, A. , Dingemans J., Ye L. M., Hassan A. A., Craggs M., Reimmann C., et al. 2014. Pore‐forming pyocin S5 utilizes the FptA ferripyochelin receptor to kill Pseudomonas aeruginosa . Microbiology 160:261–269. [DOI] [PubMed] [Google Scholar]
- Essar, D. W. , Eberly L., Hadero A., and Crawford I. P.. 1990. Identification and characterization of genes for a 2nd anthranilate synthase in Pseudomonas aeruginosa ‐ interchangeability of the 2 anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran, P. C. , Blower T. R., Foulds I. J., Humphreys D. P., Lilley K. S., and Salmond G. P. C.. 2009. The phage abortive infection system, ToxIN, functions as a protein–RNA toxin–antitoxin pair. Proc. Natl. Acad. Sci. USA 106:894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, M. 1977. The effects of culture concentration and age, time, and temperature on bacterial attachment to polystyrene. Can. J. Microbiol. 23:1–6. [Google Scholar]
- García Contreras, R. , Zhang X.‐S., Kim Y., and Wood T. K.. 2008. Protein translation and cell death: the role of rare tRNAS in biofilm formation and in activating dormant phage killer genes. PLoS ONE 3:e2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes, K. , Bech F. W., Jorgensen S. T., Lobner‐Olesen A., Rasmussen P. B., Atlung T., et al. 1986. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 5:2023–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Barrios, A. F. , Zuo R., Hashimoto Y., Yang L., Bentley W. E., and Wood T. K.. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum‐sensing regulator (MqsR, B3022). J. Bacteriol. 188:305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulard, C. , Langrand S., Carniel E., and Chauvaux S.. 2010. The Yersinia pestis chromosome encodes active addiction toxins. J. Bacteriol. 192:3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan, R. , and Engelberg‐Kulka H.. 2004. Escherichia coli mazEF‐mediated cell death as a defense mechanism that inhibits the spread of phage P1. Mol. Genet. Genomics 272:227–234. [DOI] [PubMed] [Google Scholar]
- Hazan, R. , Sat B., and Engelberg‐Kulka H.. 2004. Escherichia coli mazEF‐mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186:3663–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs, D. E. , and Poole K.. 1996. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J. Bacteriol. 178:2586–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaine, S. , Cheverton A. M., Watson K. G., Faure L. M., Matthews S. A., and Holden D. W.. 2014. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 343:204–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Benedik M. J., and Wood T. K.. 2012. Antitoxin DinJ influences the general stress response through transcript stabilizer CspE. Environ. Microbiol. 14:669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley, J. M. , and Woychik N. A.. 2009. Bacterial toxin HigB associates with ribosomes and mediates translation‐dependent mRNA cleavage at A‐rich sites. J. Biol. Chem. 284:18605–18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, S. , Benedik M. J., and Wood T. K.. 2015. Orphan toxin OrtT (YdcX) of Escherichia coli reduces growth during the stringent response. Toxins 7:299–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman, R. 2008. Bacterial persistence: some new insights into an old phenomenon. J. Biosci. 33:795–805. [DOI] [PubMed] [Google Scholar]
- Jurenaite, M. , Markuckas A., and Suziedeliene E.. 2013. Identification and characterization of type II toxin‐antitoxin systems in the opportunistic pathogen Acinetobacter baumannii . J. Bacteriol. 195:3165–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y. , and Wood T. K.. 2010. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli . Biochem. Biophys. Res. Commun. 391:209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y. , Wang X., Ma Q., Zhang X.‐S., and Wood T. K.. 2009. Toxin‐antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J. Bacteriol. 191:1258–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa, M. , Ara T., Arifuzzaman M., Ioka‐Nakamichi T., Inamoto E., Toyonaga H., et al. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K‐12 ORF archive): unique resources for biological research. DNA Res. 12:291–299. [DOI] [PubMed] [Google Scholar]
- Kwan, B. W. , Valenta J. A., Benedik M. J., and Wood T. K.. 2013. Arrested protein synthesis increases persister‐like cell formation. Antimicrob. Agents Chemother. 57:1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan, B. W. , Lord D. M., Peti W., Page R., Benedik M. J., and Wood T. K.. 2015. The MqsR/MqsA toxin/antitoxin system protects Escherichia coli during bile acid stress. Environ. Microbiol. 17:3168–3181. [DOI] [PubMed] [Google Scholar]
- Lau, G. W. , Hassett D. J., Ran H. M., and Kong F. S.. 2004. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 10:599–606. [DOI] [PubMed] [Google Scholar]
- Lee, D. G. , Urbach J. M., Wu G., Liberati N. T., Feinbaum R. L., Miyata S., et al. 2006. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 7:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, K. 2008. Multidrug tolerance of biofilms and persister cells in Romeo T., ed. Bacterial Biofilms. Springer, Berlin. [DOI] [PubMed] [Google Scholar]
- Lewis, K. 2010. Persister cells. Annu. Rev. Microbiol. 64:357–372. [DOI] [PubMed] [Google Scholar]
- Liberati, N. T. , Urbach J. M., Miyata S., Lee D. G., Drenkard E., Wu G., et al. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 103:2833–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace, C. , Seyer D., Chemani C., Cosette P., Di‐Martino P., Guery B., et al. 2008. Identification of biofilm‐associated cluster (bac) in Pseudomonas aeruginosa involved in biofilm formation and virulence. PLoS ONE 3:e3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moker, N. , Dean C. R., and Tao J.. 2010. Pseudomonas aeruginosa increases formation of multidrug‐tolerant persister cells in response to quorum‐sensing signaling molecules. J. Bacteriol. 192:1946–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy, L. R. , Burns J. L., Lory S., and Lewis K.. 2010. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J. Bacteriol. 192:6191–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton, J. P. , and Mulvey M. A.. 2012. Toxin‐antitoxin systems are important for niche‐specific colonization and stress resistance of uropathogenic Escherichia coli . PLoS Pathog. 8:e1002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura, T. , and Hiraga S.. 1983. Mini‐F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 80:4784–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhage, J. , Bains M., Brazas M. D., and Hancock R. E. W.. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 190:2671–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, J. G. , and Ackerley D. F.. 2011. Characterization of pyoverdine and achromobactin in Pseudomonas syringae pv. phaseolicola 1448a. BMC Microbiol. 11:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, D. P. , and Gerdes K.. 2005. Toxin‐antitoxin loci are highly abundant in free‐living but lost from host‐associated prokaryotes. Nucleic Acids Res. 33:966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecota, D. C. , and Wood T. K.. 1996. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J. Bacteriol. 178:2044–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage, H. R. , Connolly L. E., and Cox J. S.. 2009. Comprehensive functional analysis of Mycobacterium tuberculosis toxin‐antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 5:e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravel, J. , and Cornelis P.. 2003. Genomics of pyoverdine‐mediated iron uptake in pseudomonads. Trends Microbiol. 11:195–200. [DOI] [PubMed] [Google Scholar]
- Ren, D. , Bedzyk L. A., Thomas S. M., Ye R. W., and Wood T. K.. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515–524. [DOI] [PubMed] [Google Scholar]
- Ren, D. , Walker A. N., and Daines D. A.. 2012. Toxin‐antitoxin loci vapBC‐1 and vapXD contribute to survival and virulence in nontypeable Haemophilus influenzae . BMC Microbiol. 12:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, R. L. , and Tait R. C.. 1983. Recombinant DNA techniques: an introduction. Benjamin/Cummings Publishing, Menlo Park, CA. [Google Scholar]
- Sambrook, J. F. , and Russell D. W.. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- Sambrook, J. , Fritsch E. F., and Maniatis T.. 1989. Molecular cloning, a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sat, B. , Hazan R., Fisher T., Khaner H., Glaser G., and Engelberg‐Kulka H.. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 183:2041–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuessler, D. L. , Cortes T., Fivian‐Hughes A. S., Lougheed K. E. A., Harvey E., Buxton R. S., et al. 2013. Induced ectopic expression of HigB toxin in Mycobacterium tuberculosis results in growth inhibition, reduced abundance of a subset of mRNAs and cleavage of tmRNA. Mol. Microbiol. 90:195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schureck, M. A. , Maehigashi T., Miles S. J., Marquez J., Cho S. E., Erdman R., et al. 2014. Structure of the Proteus vulgaris HigB‐(HigA)(2)‐HigB toxin‐antitoxin complex. J. Biol. Chem. 289:1060–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schureck, M. A. , Dunkle J. A., Maehigashi T., Miles S. J., and Dunham C. M.. 2015. Defining the mRNA recognition signature of a bacterial toxin protein. Proc. Natl. Acad. Sci. USA 112:13862–13867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seffernick, J. L. , de Souza M. L., Sadowsky M. J., and Wackett L. P.. 2001. Melamine deaminase and atrazine chlorohydrolase: 98 percent identical but functionally different. J. Bacteriol. 183:2405–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevin, E. W. , and Barloy‐Hubler F.. 2007. RASTA‐Bacteria: a web‐based tool for identifying toxin‐antitoxin loci in prokaryotes. Genome Biol. 8:R155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol, P. A. 1987. Surface expression of ferripyochelin‐binding protein is required for virulence of Pseudomonas aeruginosa . Infect. Immun. 55:2021–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase, H. , Nitanai H., Hoshino K., and Otani T.. 2000. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect. Immun. 68:1834–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Q. B. , Hayashi T., Murata T., and Terawaki Y.. 1996. Gene product identification and promoter analysis of hig locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 225:679–684. [DOI] [PubMed] [Google Scholar]
- Tiwari, P. , Arora G., Singh M., Kidwai S., Narayan O. P., and Singh R.. 2015. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 6:6059. [DOI] [PubMed] [Google Scholar]
- Wang, X. , and Wood T. K.. 2011. Toxin/Antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 77:5577–5583. 10.1128/AEM.05068-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. Y. , Mushegian A., Lory S., and Jin S. G.. 1996. Large‐scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc. Natl. Acad. Sci. USA 93:10434–10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. X. , Kim Y., Hong S. H., Ma Q., Brown B. L., Pu M. M., et al. 2011. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 7:359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. J. , Halvorsen E. M., Dwyer E. M., DiFazio R. M., and Hergenrother P. J.. 2011. Toxin‐antitoxin (TA) systems are prevalent and transcribed in clinical isolates of Pseudomonas aeruginosa and methicillin‐resistant Staphylococcus aureus . FEMS Microbiol. Lett. 322:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, T. K. , Knabel S. J., and Kwan B. W.. 2013. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 79:7116–7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, Y. , and Inouye M.. 2011. Regulation of growth and death in Escherichia coli by toxin‐antitoxin systems. Nat. Rev. Microbiol. 9:779–790. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, Y. , Park J. H., and Inouye M.. 2011. Toxin‐antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45:61–79. [DOI] [PubMed] [Google Scholar]
- Yu, X. , Chen M., Jiang Z., Hu Y., and Xie Z.. 2014. The two‐component regulators GacS and GacA positively regulate a nonfluorescent siderophore through the Gac/Rsm signaling cascade in high‐siderophore‐yielding Pseudomonas sp. strain HYS. J. Bacteriol. 196:3259–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]


